Sporothrix globosa es un hongo patógeno recientemente descrito. Esta especie, morfológicamente similar a Sporothrix schenckii, se ha descrito en EE.UU., Europa y Asia. En este trabajo se investigaron las relaciones filogenéticas de 32 aislamientos clínicos y ambientales, identificados morfológicamente como S. schenckii, procedentes de México, Guatemala y Colombia, mediante análisis cladístico de secuencias parciales del gen de la calmodulina usando los métodos de máxima parsimonia y neighbor-joining. El estudio reveló que uno de los 25 aislamientos de México (4%), uno de los tres aislamientos de Guatemala (33%) y dos de los cuatro aislamientos de Colombia (50%) correspondían a S. globosa, mientras que los demás aislamientos pertenecían a S. schenckii sensu stricto. La presencia de S. globosa en México, América Central y del Sur se describe por primera vez.
Sporothrix globosa, reported from the USA, Europe, and Asia, is a recently described pathogenic species morphologically similar to Sporothrix schenckii. In this study, the phylogenetic affinities of 32 clinical and environmental isolates morphologically identified as S. schenckii, from Mexico, Guatemala, and Colombia, were assessed by cladistic analysis of partial sequences of the calmodulin gene using the maximum parsimony and neighbor-joining methods. The study revealed that one out of 25 isolates from Mexico (4%), one out of three isolates from Guatemala (33.3%), and two out of four isolates from Colombia (50%) belonged to S. globosa, while the other isolates belonged to S. schenckii sensu stricto. This is the first record of S. globosa from Mexico, and Central and South America.
Sporotrichosis is a chronic infectious disease that typically involves the skin and subcutaneous tissue.7,12 Cases of arthritis, meningitis, and other deep-seated forms have been reported less frequently.1,3,4,5,10,25 Infection is acquired via traumatic implantation or less frequently by inhalation of propagules of the etiological agent living in soil, plant material, and other substrata.14,19 Sporotrichosis is distributed worldwide, but most cases occur in temperate, warm, and tropical countries. The largest number of reports comes from North America, but it is also common in areas of Central and South America, Asia, and South Africa.8,12
For several decades, sporotrichosis has been attributed to a single pathogen, Sporothrix schenckii Hektoen & Perkins, an anamorphic fungus related to the ascomycetous genus Ophiostoma H. & P. Syd.2 However, isolates identified as S. schenckii showed a great deal of phenotypic6,12 and genetic9,13,20 variability, suggesting that this taxon was a species complex. In a recent phylogenetic study based on the analysis of sequences of the chitin-synthase, ß-tubulin, and calmodulin (CAL) genes, numerous isolates phenotypically identified as S. schenckii were tested.17 The strains were distributed into at least six distinct groups, which were considered as putative phylogenetic species. Later, the same authors found diagnostic features to separate phenotypically and genetically three of these clades, which were formally proposed as new species. They were Sporothrix brasiliensis Marimon, Gené, Cano & Guarro, Sporothrix globosa Marimon, Gené, Cano & Guarro, and Sporothrix mexicana Marimon, Gené, Cano & Guarro.16
Since variations in antifungal susceptibility have been demonstrated among the different species of the S. schenckii complex, their identification is clinically relevant.18 Furthermore, considering that the taxonomy of the fungi causing sporotrichosis has been reevaluated, it becomes necessary to identify clinical isolates at the species level in order to study their epidemiology and geographical distribution, and to determine if different clinical patterns are associated with each of these taxa. Recently we have had the opportunity of studying numerous isolates from Mexico, Colombia, and Guatemala and our interest was to assess if a given species is predominantly implicated in cases of human infection in these countries, or, by contrast, a range of species can be present. To do this, we analyzed partial sequences of the CAL locus, which had previously proven to be the most informative gene.16,17
Materials and methodsFungal isolatesThirty-two clinical and environmental isolates morphologically identified as S. schenckii, from Colombia, Guatemala, and Mexico were included in this study (tabla 1). These isolates were obtained from culture collections located at the Servicio de Dermatología y Departamento de Micología, in Hospital General de México, Mexico, and at the Departamento de Microbiología y Parasitología, Facultad de Medicina, Universidad Autónoma de México, Mexico. The isolates were subcultured on potato dextrose agar (PDA; Difco Laboratories, USA) plates and incubated at 25°C for 14 days in the dark. Isolates were stored at 4–7°C and in slant cultures submerged in mineral oil at room temperature.
Table 1. Collection number, fungal species, source, origin, and EMBL accession numbers of the isolates studied.
| Isolate no. | Species | Source | Origin | EMBL accession no. |
| FMR 9549 | S. schenckii | Human, lymphocutaneous sporotrichosis | Mexico | – |
| FMR 9550 | S. schenckii | Human, lymphocutaneous sporotrichosis | Mexico | – |
| FMR 9551 | S. schenckii | Human, lymphocutaneous sporotrichosis | Mexico | – |
| FMR 9553 | S. schenckii | Human, lymphocutaneous sporotrichosis | Mexico | – |
| FMR 9554 | S. schenckii | Human, lymphocutaneous sporotrichosis | Mexico | – |
| FMR 9555 | S. schenckii | Human, lymphocutaneous sporotrichosis | Mexico | – |
| FMR 9556 | S. globosa | Human, lymphocutaneous sporotrichosis | Mexico | FM179331 |
| FMR 9557 | S. schenckii | Human, fixed cutaneous sporotrichosis | Mexico | – |
| FMR 9559 | S. schenckii | Human, fixed cutaneous sporotrichosis | Mexico | – |
| FMR 9560 | S. schenckii | Human, lymphocutaneous sporotrichosis | Mexico | – |
| FMR 9561 | S. schenckii | Human, fungaemia | Mexico | – |
| FMR 9562 | S. schenckii | Human, lymphocutaneous sporotrichosis | Mexico | FM179332 |
| FMR 9563 | S. schenckii | Human, lymphocutaneous sporotrichosis | Mexico | – |
| FMR 9564 | S. schenckii | Human, lymphocutaneous sporotrichosis | Mexico | – |
| FMR 9565 | S. schenckii | Human, fixed cutaneous sporotrichosis | Mexico | – |
| FMR 9566 | S. schenckii | Human, lymphocutaneous sporotrichosis | Mexico | – |
| FMR 9567 | S. schenckii | Human, lymphocutaneous sporotrichosis | Mexico | – |
| FMR 9568 | S. schenckii | Human, fixed cutaneous sporotrichosis | Mexico | – |
| FMR 9570 | S. schenckii | Human, lymphocutaneous sporotrichosis | Mexico | – |
| FMR 9572 | S. schenckii | Human, lymphocutaneous sporotrichosis | Mexico | – |
| FMR 9616 | S. schenckii | Human, fixed cutaneous sporotrichosis, hand | Colombia | – |
| FMR 9617 | S. globosa | Human, fixed cutaneous sporotrichosis, wrist | Colombia | FM179329 |
| FMR 9619 | S. globosa | Human, fixed cutaneous sporotrichosis, cheek | Colombia | FM179330 |
| FMR 9620 | S. schenckii | Human, lymphocutaneous sporotrichosis, arm | Colombia | – |
| FMR 9621 | S. schenckii | Human, lymphocutaneous sporotrichosis | Mexico | – |
| FMR 9622 | S. schenckii | Soil under Coffea sp. | Mexico | – |
| FMR 9624 | S. globosa | Human, lymphocutaneous sporotrichosis, finger | Guatemala | FM207489 |
| FMR 9625 | S. schenckii | Human, fixed cutaneous sporotrichosis, forearm | Guatemala | – |
| FMR 9626 | S. schenckii | Human, lymphocutaneous sporotrichosis foot | Guatemala | – |
| FMR 9629 | S. schenckii | Soil | Mexico | – |
| FMR 9631 | S. schenckii | Soil | Mexico | FM179333 |
| FMR 9632 | S. schenckii | Soil | Mexico | – |
FMR, Facultat de Medicina i Ciències de la Salut, Reus, Spain.
DNA extraction, amplification, and sequencing of the CAL locus of the 32 isolates were performed as described previously,16,17 using primers CL1 and CL2A.21
DNA sequence alignmentsThe program Autoassembler vers. 1.40 (Applied Biosystems) was used to obtain consensus sequences from the two complementary sequences of each isolate. The consensus sequences of the 32 isolates sequenced here were aligned with CAL sequences of 39 other isolates of S. schenckii sensu stricto and the related species S. brasiliensis, S. globosa, S. mexicana, and Sporothrix albicans S.B. Saksena determined in a previous study,16 using ClustalX vers. 1.81,24 followed by manual adjustments with a text editor.
Phylogenetic analysisA phylogenetic analysis was performed by the maximum parsimony method using the PAUP* version 4.0b10 software.23 Briefly, the most parsimonious trees were obtained after 100 heuristic searches with random sequence addition and tree bisection–reconnection branch-swapping algorithms, collapsing zero-length branches and saving all minimal-length trees (MulTrees). Also, a neighbor-joining phylogeny22 was constructed using the Kimura-2-parameter substitution model with pairwise deletion of gaps, as implemented in the MEGA3 computer program.11 The robustness of branches was assessed by bootstrap analysis of 1000 replicates.
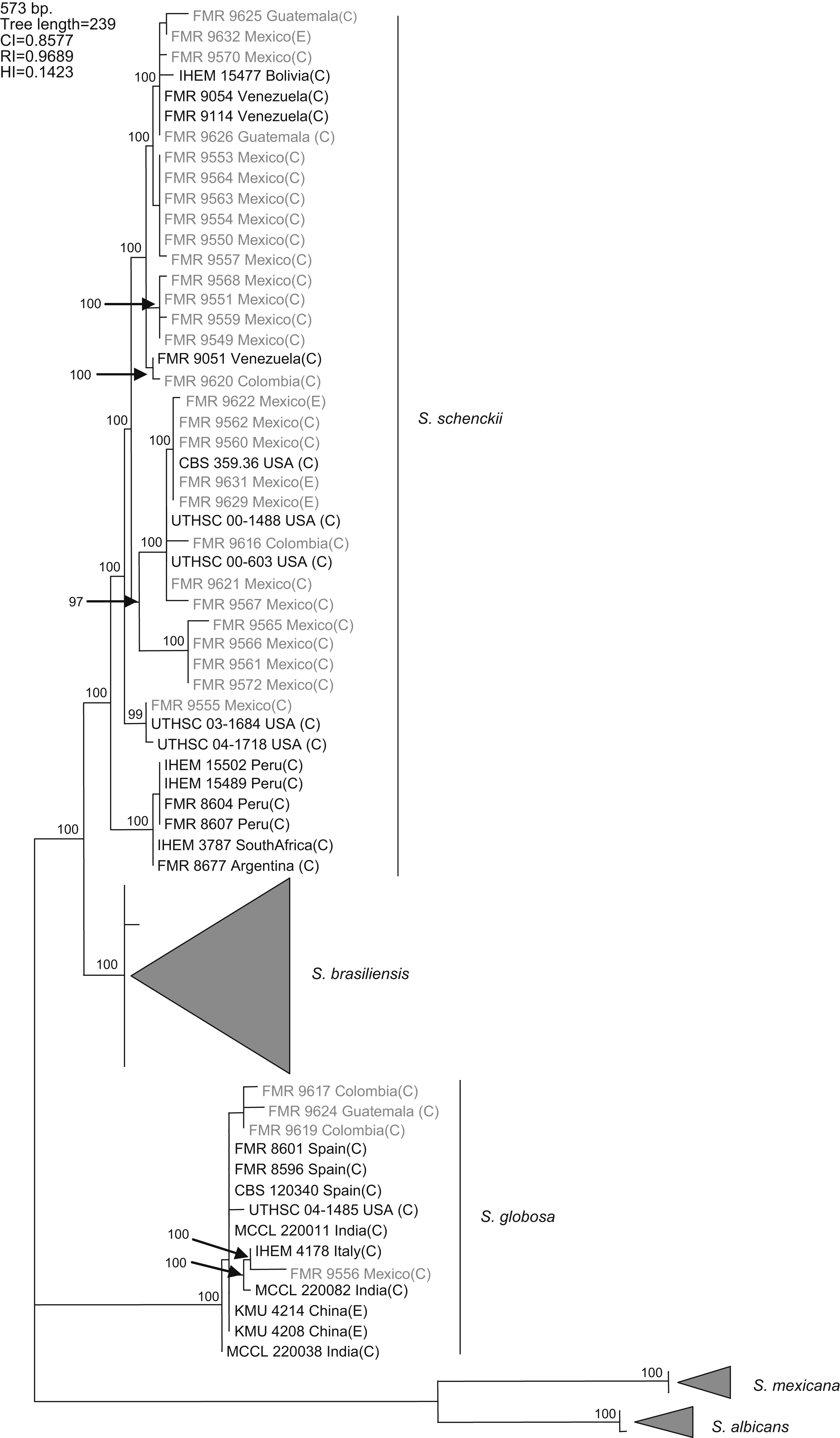
Results and discussionWith the primers used, a fragment of 573bp of the CAL locus was amplified and sequenced. The complete alignment included 71 sequences, 32 generated in this study and 39 retrieved from a previous study,16 the latter belonging to S. schenckii s. str. (15), S. brasiliensis (10), S. globosa (10), S. mexicana (2), and S. albicans (2), resulting in a data set of 573 characters, including 403 constant, 160 variable parsimony informative (39.7%), and 10 variable parsimony noninformative sites. Cladistic analysis by the neighbor-joining and maximum parsimony methods generated trees with identical topology. Maximum parsimony analysis of the CAL data set yielded 5000 trees, 239 steps in length, in which 20 nodes received 100% bootstrap support. One of the most parsimonious trees is shown in figura 1. The 71 sequences were distributed into the five main groups detected in previous studies,16,17 representing S. brasiliensis, S. schenckii sensu stricto, S. globosa, S. mexicana, and S. albicans. One out of 25 isolates from Mexico (4%), one out of three isolates from Guatemala (33.3%), and two out of four (50%) isolates from Colombia grouped into the S. globosa clade, which also included another 10 sequences belonging to isolates from India, China, Italy, USA, and Spain. The other isolates from Colombia, Mexico, and Guatemala grouped into the S. schenckii s. str. clade, which also included sequences of another 15 isolates from Bolivia, Venezuela, USA, Peru, South Africa, and Argentina. Since none of the sequences generated in this study grouped into the S. mexicana, S. brasiliensis or S. albicans clades, the isolates belonging to these clades are not detailed in figura 1. The 24 isolates from Mexico in the S. schenckii clade were distributed among 14 different haplotypes. Two clinical (FMR 9560 and FMR 9562) and two environmental isolates from Mexico (FMR 9629 and FMR 9631) exhibited the same haplotype as the type strain of S. schenckii, CBS 359.36. The S. schenckii isolates from Guatemala and Colombia were distributed among four different haplotypes. The S. globosa isolates from Mexico, Guatemala, and Colombia were distributed among four haplotypes different from that of the type strain.
Figure 1. One of the 5000 most parsimonious trees obtained from heuristic searches based on an analysis of the CAL locus. Bootstrap values above 80% are indicated at the nodes. Type strains are indicated in bold type. Isolates for which new CAL sequences were generated during this study are highlighted in blue. CI, consistency index; RI, retention index; HI, homoplasy index; (E), environmental isolate; (C), clinical isolate; FMR, Facultat de Medicina i Ciències de la Salut, Reus, Spain; IHEM, The BCCM/IHEM Biomedical Fungi and Yeasts Collection, Brussels, Belgium; CBS, Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands; UTHSC, Fungus Testing Laboratory, University of Texas Health Science Center; MCCL, Mycology Culture Collection Laboratory, Postgraduate Institute of Medical Education and Research, Chandigarh, India; KMU, Kanazawa Medical University, Ishikawa, Japan.
A previous study revealed the existence of differences in the geographical distribution among the members of the S. schenckii complex, including widespread as well as geographically restricted species.16S. brasiliensis and S. mexicana occurred only in Brazil and Mexico, respectively, and these taxa grouped all the isolates from Brazil (N=29) and Mexico (N=2) tested in that study. Based on these observations, we first thought the 25 isolates from Mexico studied here could follow the same pattern of close phylogenetic affinity observed in isolates from Brazil. However, none of these isolates grouped into the S. mexicana clade, a species originally reported from soil and from carnation leaves. On the other hand, S. schenckii and S. globosa are widespread fungi showing transoceanic distributions.16 Until now, 36 isolates of S. globosa have been reported from United Kingdom, Spain, Italy, China, Japan, USA, and India. This is the first record of this species from Mexico, and Central and South America.
An experimental model of disseminated infection by different Sporothrix species in immunocompetent mice showed that, while S. schenckii s. str. and S. brasiliensis were able to kill immunocompetent animals inoculated intravenously, S. globosa did not cause any apparent lesion, suggesting that it might be less virulent than the former species.15 Interestingly, in contrast with S. schenckii s. str. and S. brasiliensis, which have been associated with both localized and invasive disease,16,17 no cases of invasive infections have been attributed to S. globosa. This apparently lower virulence might be related to the inability of the fungus to grow at 37°C.16
Marimon et al.16 proposed as key features for the differentiation of the clinically relevant Sporothrix species the presence or absence of pigmented sessile conidia, growth rates at different temperatures, and carbohydrate assimilation test results. Although morphologically similar to other taxa within the S. schenckii complex, S. globosa is the only member of the complex unable to grow at 37°C on PDA. Moreover, among the four Sporothrix species with pigmented sessile conidia treated, only S. globosa showed the combination of positive sucrose and negative raffinose assimilations. These easily diagnosed features allow the identification of S. globosa and related taxa by simple inexpensive laboratory procedures.
Corresponding author.