There are only few reports about travel-associated, imported tropical hepatitis E virus (HEV) genotype 1 infections within Western travellers. We describe the clinical course of a single outbreak of hepatitis E in a German travellers group returning from India and compare the results of two commercial HEV-seroassays.
Material and MethodsAfter identifying hepatitis E in an index patient returning from a journey to India all 24 members of this journey were tested for anti-HEV-IgG and IgM using two commercial seroassays (Wantai and Mikrogen), for HEV-RNA by PCR and HEV-Ag by an antigen-assay (Wantai).
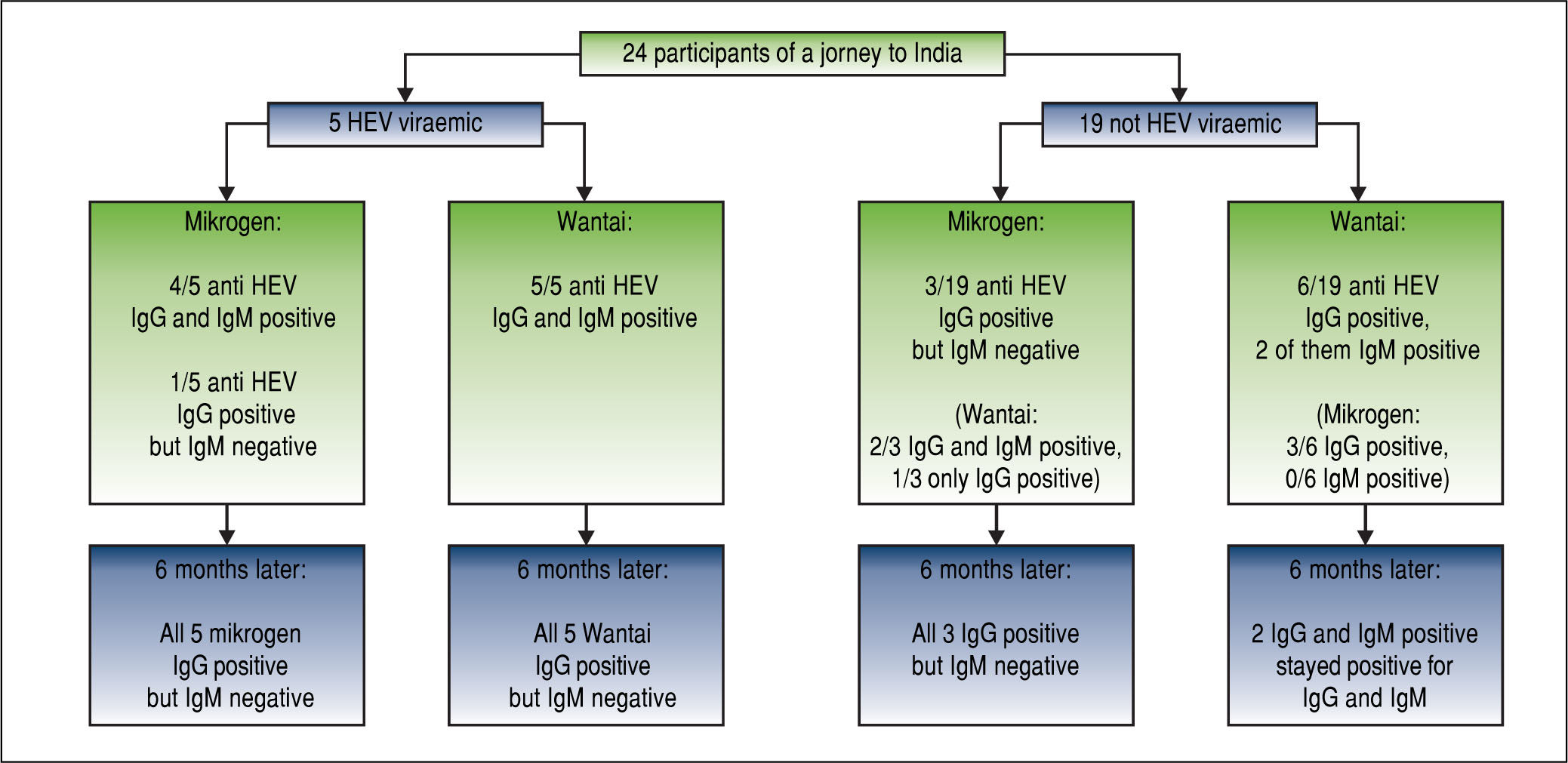
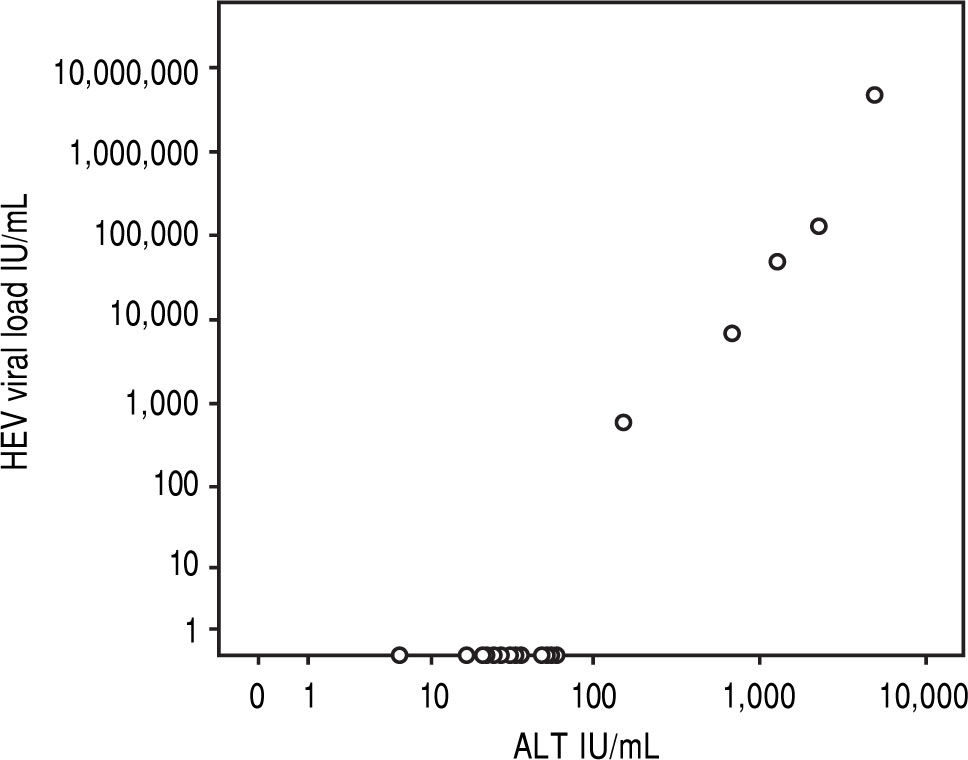
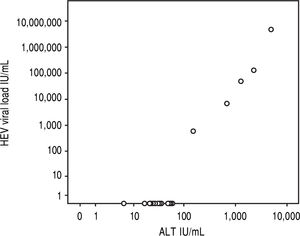
Results5/24 (21%) individuals were viraemic with viral loads between 580-4,800,000 IU/mL. Bilirubin and ALT levels in these patients ranged from 1.3-14.9 mg/dL (mean 7.3 mg/dL, SD 5.6 mg/dL) and 151-4,820 U/L (mean 1,832U/L, SD 1842U/L), respectively and showed significant correlations with viral loads (r = 0.863, p < 0.001; r = 0.890, p < 0.001). No risk factor for food-borne HEV-transmission was identified. All viraemic patients (5/5) tested positive for anti-HEV-IgG and IgM in the Wantai-assay but only 4/5 in the Mikrogen-as-say. Wantai-HEV-antigen-assay was negative in all patients. Six months later all previously viraemic patients tested positive for anti-HEV-IgG and negative for IgM in both assays. However, two non-viremic individuals who initially tested Wantai-IgM-positive stayed positive indicating false positive results.
ConclusionsDespite the exact number of exposed individuals could not be determined HEV genotype 1 infections have a high manifestation rate of more than 20%.The Wantai-antigen-test failed, the Wantai-IgM-rapid-test and the Mikrogen-IgM-recomblot showed a better performance but still they cannot replace real-time PCR for diagnosing ongoing HEV-infections.
Hepatitis E virus (HEV) infections in the tropics are caused by genotypes 1 and 2 while genotypes 3 and 4 mostly occur in industrialized countries.1,2
In tropical regions HEV causes 20 million infections, 3 million symptomatic infections and 70,000 deaths (especially during pregnancy) each year.3 The disease is transmitted by contaminated drinking water. No chronic HEV-genotype 1 or 2 infections have been reported so far.
In contrast to the tropical HEV-genotypes, genotype 3 and 4 infections cause sporadic cases in the Western World and are presumably transmitted by pork or blood products.4-6 Fatal courses frequently oc-cured in elderly men and patients with underlying liver diseases.4,5 Chronic HEV-genotype 3 and 4 infections have only been described in immunosup-pressed individuals.4
Typically, hepatitis E in tropical regions manifest in habitants of living under reduced hygieneic conditions or in travellers exposed to the risk of HEV acquisition by violation of general travel safety advice, e.g. swimming in open water or drinking unbottled water.1,2,4
Here we describe an outbreak of hepatitis E in a group of travellers returning from India and we evaluated different serological assays in this setting.
Material and MethodsAfter diagnosing acute hepatitis E in a patient who returned from a journey to India (6th to 16th December 2013) six weeks beforehand (patient #1) all 24 passengers who attended this journey were also investigated in January 2014. Names and contact addresses were provided by the index patient after gaining informed consent of all travellers. Participants completed standardized questionnaires assessing the type of food consumed in India and possible symptoms of acute hepatitis E.
All participants sent blood samples taken by their own physician to our laboratory for testing for HEV-RNA by real-time PCR (LLoD 12 IU/mL)7 and for HEV-antibodies (IgG and IgM) by using the Wantai Diagnostics ELISAs (Wantai Bejing, China) and the Mikrogen-recomblot (Mikrogen, Hilden, Germany) according to manufacturers’ instructions. For the Wantai-IgM-assay we used the rapid test version. Furthermore blood samples were tested by the Wantai-HEV-antigen-assay for the presence of HEV-antigen as described previously.11 Recently an advanced version of this antigen-assay has been developed (“Wantai-Antigen-As-say-Plus”) and kindly provided to us by the manufacturer (Wantai through Sanbio, Uden, Netherlands).
RNA was extracted of HEV-PCR positive samples by Qiasymphony and sequenced using the nested primer set MJ-C as previously described.8 For phylogenetic analysis of HEV 10 orthohepevirus A sequences9 covering ORF1, positions 4263 to 4557, were retrieved from GenBank. The sequences were aligned in Geneious 6.1.8 and trimmed manually as required. The unrooted maximum likelihood tree was created in Geneious using the Tamura Nei substitution model with 4 gamma rate categories and invariant sites. Bootstrap analysis was done using 1000 replicates as previously published.10
Patients testing positive for HEV-RNA or anti-HEV-IgM were re-tested six months later for anti-HEV-IgG and IgM by the Wantai-assays and the Microgen-recomblot.
Clinical courses and resultsThis outbreak occurred in a travel group (n = 24) travelling to Delhi for a short term journey. The participants visited Delhi and consumed food in their hotel. None of the participants swam in open water or visited rural areas of India.
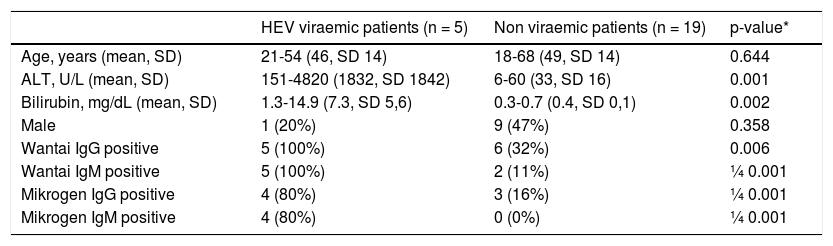
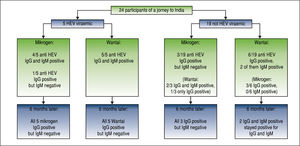
Five patients (patient #1-5) were found to have an acute viraemic HEV-infection (genotype 1) six weeks after returning from India (Figure 1). All patients cleared the infection spontaneously, no patient developed acute liver failure or was treated with ribavirin. The German health authorities had been informed about the outbreak in this group tour and investigations were initiated. However, to the best of our knowledge, no further case of hepatitis E has been documented in another group visiting India during the same period. Comparison of patients characteristics with non-viraemic travellers is depicted in table 1.
Characteristics of HEV viraemic and non-viraemic patients.
| HEV viraemic patients (n = 5) | Non viraemic patients (n = 19) | p-value* | |
|---|---|---|---|
| Age, years (mean, SD) | 21-54 (46, SD 14) | 18-68 (49, SD 14) | 0.644 |
| ALT, U/L (mean, SD) | 151-4820 (1832, SD 1842) | 6-60 (33, SD 16) | 0.001 |
| Bilirubin, mg/dL (mean, SD) | 1.3-14.9 (7.3, SD 5,6) | 0.3-0.7 (0.4, SD 0,1) | 0.002 |
| Male | 1 (20%) | 9 (47%) | 0.358 |
| Wantai IgG positive | 5 (100%) | 6 (32%) | 0.006 |
| Wantai IgM positive | 5 (100%) | 2 (11%) | ¼ 0.001 |
| Mikrogen IgG positive | 4 (80%) | 3 (16%) | ¼ 0.001 |
| Mikrogen IgM positive | 4 (80%) | 0 (0%) | ¼ 0.001 |
- •
Patient #1 was a 53 years old female presenting with repeated episodes of abdominal pain during the journey and after her return.She had no jaundice or further symptoms. Her bilirubin level was almost normal (1.3 mg/dL), while her ALT was strongly increased (2246 IU/mL). She had a high HEV viral load (130,000 IU/ mL). Patient #1 was the index patient. Further investigation of the travel group let us identify patients #2-5.
- •
Patient #2 was a 54 years old woman without any symptoms. Her infection was only detected as she was tested as a part of this investigation. She had a strongly elevated level of ALT (685 IU/mL) and a low HEV viral load of 6800 IU/mL.
- •
Patients #3-5 belong to the same family. Patient #3 was a 51 years old woman, who showed no other symptom besides jaundice (bilirubin 6.5mg/dL; ALT 1257 IU/mL). She had a moderate viral load (49,000 IU/mL). Patient #4 was her 52 years old husband who suffered from jaundice, myalgia and nausea. His bi-lirubin-level and his ALT-level were strongly raised (14.9 mg/dL and 4820 IU/mL). He had the highest HEV viral load of all HEV-infected patients (4,800,000 IU/mL). Patient #5 was the 21 years old daughter of patient #3 and #4. This patient had the lowest viral load of all viraemic patients (580 IU/mL). She suffered from nausea and jaundice (bilirubin 6.3 mg/dL, ALT 151 IU/mL).
In the overall cohort, viral load showed significant correlations with levels of bilirubin or ALT (r = 0.863, p < 0.001; r = 0.890, p < 0.001, Figure 2). After exclusion of HEV-RNA negative patients, there was also a good correlation between HEV viral load and ALT in viraemic patients (r = 0.917, p = 0.029) while there was no significant correlation with bilirubin (r = 0.895, p = 0.105). All patients tested negative by the Wantai-antigen-assay, while 2/5 (#1 and #4, with the highest viral loads) tested positive for HEV by the novel Wantai-Antigen-Assay-Plus.
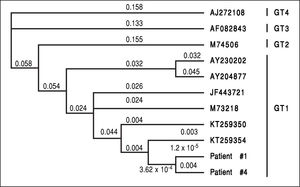
In two patients (patient #1 and patient #4, the patients with the highest viral load) it was possible to sequence the HEV genome (200 nucleotids). Both sequences were identical indicating exposure to the same HEV-strain as origin of infection (Figure 3). Unfortunately it was not possible to determine the sequence in the three remaining patients due to low virus loads.
All five viraemic patients tested positive for anti-HEV-IgG and IgM using the Wantai-sero-tests. Only 4/5 patients tested positive for anti-HEV-IgG and IgM with the Mikrogen-assays (Figure 1), while patient #2 tested positive in the Wantai-assays, but negative in the Mikrogen-as-says. Six months later all five patients with previous hepatitis E tested positive for anti-HEV-IgG in the Wan-tai-assay and the Mikrogen-recomblot and negative in both IgM-assays, repectively (Figure 1).
3/19 individuals who were initially negative for HEV-RNA tested positive by the Mikrogen-IgG-test but negative by the Mikrogen-IgM-test (Figure 1). These three and three further non-viraemic individuals tested also positive in the Wantai-IgG-assay, two of them were additionally positive in the Wantai-IgM-test. These two individuals remained Wantai-IgM and IgG positive and Mikrogen-anti-HEV-IgG positive and IgM negative on follow-up.
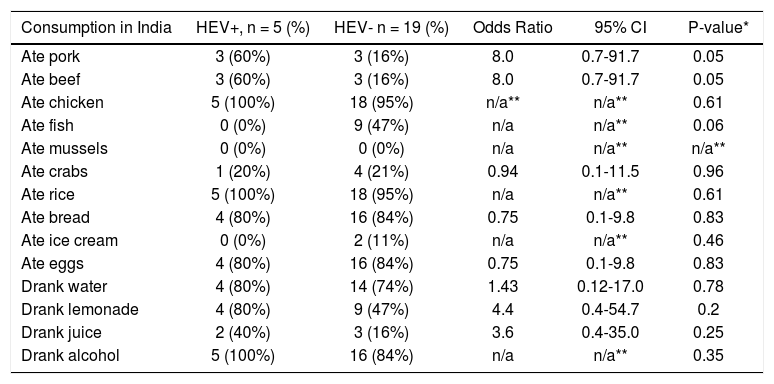
Unfortunately, no clear risk factor for HEV exposure could be identified by analysing the questionnaires filled out by the participants (Table 2). However, consumption of beef or pork showed a weak association with HEV infection (Table 2).
Analysis of risk factors for hepatitis E.
| Consumption in India | HEV+, n = 5 (%) | HEV- n = 19 (%) | Odds Ratio | 95% CI | P-value* |
|---|---|---|---|---|---|
| Ate pork | 3 (60%) | 3 (16%) | 8.0 | 0.7-91.7 | 0.05 |
| Ate beef | 3 (60%) | 3 (16%) | 8.0 | 0.7-91.7 | 0.05 |
| Ate chicken | 5 (100%) | 18 (95%) | n/a** | n/a** | 0.61 |
| Ate fish | 0 (0%) | 9 (47%) | n/a | n/a** | 0.06 |
| Ate mussels | 0 (0%) | 0 (0%) | n/a | n/a** | n/a** |
| Ate crabs | 1 (20%) | 4 (21%) | 0.94 | 0.1-11.5 | 0.96 |
| Ate rice | 5 (100%) | 18 (95%) | n/a | n/a** | 0.61 |
| Ate bread | 4 (80%) | 16 (84%) | 0.75 | 0.1-9.8 | 0.83 |
| Ate ice cream | 0 (0%) | 2 (11%) | n/a | n/a** | 0.46 |
| Ate eggs | 4 (80%) | 16 (84%) | 0.75 | 0.1-9.8 | 0.83 |
| Drank water | 4 (80%) | 14 (74%) | 1.43 | 0.12-17.0 | 0.78 |
| Drank lemonade | 4 (80%) | 9 (47%) | 4.4 | 0.4-54.7 | 0.2 |
| Drank juice | 2 (40%) | 3 (16%) | 3.6 | 0.4-35.0 | 0.25 |
| Drank alcohol | 5 (100%) | 16 (84%) | n/a | n/a** | 0.35 |
None of the infected patients were swimming in rivers or lakes and none of them consumed unbottled water.
DiscussionThis hepatitis E outbreak within a travel group became overt four weeks after the group returned and all members of this group tour have been tested four to six weeks after their return. The fact that more than 20% of the returning travelers developed a clinically relevant vi-raemic hepatitis E, demonstrates the high clinical manifestation index of tropical HEV-infections. It cannot be determined exactly how many individuals had been in contact with HEV. However, if more than 20% developed acute hepatitis E the manifestation rate is above 20%. It cannot be ruled out that more individuals of this particular group of travellers had contact with HEV, cleared the infection, yet did not seroconvert. This high manifestation rate is in line with previous reports, demonstrating that HEV-genotype 1 and 2 infections lead to symptomatic cases in 16% of infections.3 In contrast in HEV-genotype 3 or 4 infections far less than 2% of persons develop symptomatic hepatitis.4,12,13
However, all viraemic patients had high ALT-levels of more than four times the upper limit of normal (Figure 2). This indicates the high virulence of this particular virus strain. We were able to sequence the virus characterizing it as HEV genotype 1 (Figure 3, the HEV genotype 1 subtype was not able to determine due to overlapping sequences with different subtypes). Of note, it has previously been speculated that some HEV-strains in industrialized countries are less virulent than tropical strains.14
There was no association between HEV-infection and age or gender in contrast to a previous outbreak on a cruise ship.15 Perhaps our cohort is too small to detect such an association. Unfortunately, the ultimate source of this particular outbreak could not be identified. However, none of the participants swam in open water or visited rural areas of India. Of note, a number of the travellers visited a small village in Rajhastan during this trip. However, three of the patients with acute hepatitis E infection did not participate at this, excluding this event as source of the infection. Furthermore, the group visited the Taj Mahal, without any food consumption. The majority of the trip the group stayed in Delhi and food consumption was restricted to the five-star hotel, so the infected patients most likely acquired the infection in the hotel. Investigation of the questionnaires did not identify a clear foodborne risk factor for HEV-transmission. There is some evidence that Hepatitis E was associated with consumption of pork or beef (OR 8.0, 95% CI 0.7-91.7, p = 0.05). Perhaps these dishes have been contaminated during preparation in the kitchen. However, the sample size was not large enough to make obtain robust estimates or perform multivariate analyses. On the other hand, since none of the cases consumed fish, mussels or ice cream, these foods can be ruled out as the source. It is interesting to speculate whether different chefs were responsible for the different dishes. However, this is hypothetical and the exact chain of infection will never be clarified. In this particular outbreak, classical ways of HEV-transmission in developing countries, e.g. swimming or consumption of unbottled water, are unlikely. The group of travellers has not been apparently exposed to reduced hygienic conditions, as the group lived in a high-grade hotel.
Additionally to clinical aspects our study demonstrates the value of two commercial seroassays. The Wantai-IgM-rapid-test has a high sensitivity to predict ongoing HEV-infections since all viraemic patients tested positive in this assay. In contrast, the Mikrogen-recomblot overlooked one viraemic patient. However, two non-viraemic individuals tested positive for anti-HEV-IgM in the Wantai-IgM-rapid-test and stayed positive six months after the outbreak indicating a lower specifity of this assay.
The Wantai-antigen-test failed to detect any viraemic patient whereas the novel version of this test, the Wantai-Antigen-Assay-Plus, had a slightly better validity and 2/5 viraemic patients tested positive in this assay. This result is plausible as the two patients positive in this antigen-test were the patients with the highest viral loads. However, this detection rate is far away from being satisfactory.
Both IgG-assays (Mikrogen and Wantai) proved to be useful and reliable for the diagnosis of HEV: all previously viraemic patients tested anti-HEV-IgG positive (and IgM negative) six months after infection. However, anti-HEV IgG testing does not help to solve the question of current, ongoing infection, thus it is important to highlight the value of the quantitative real time PCR for the diagnosis of an acute HEV infection.
The observation that 6/19 (32%) non-viraemic individuals in the present study tested positive for anti-HEV-IgG using the Wantai-Assay is well in line with previous studies that demonstrated a seroprevalence-rate of 30% in healthy people in Germany using this assay.16,17
The present study contains several relevant novel findings:
- •
21% of the group travelling to India acquired hepatitis E indicating a high manifestation rate of this HEV-strain. Unfortunately, it cannot be determined how many of the travellers had contact with HEV despite an absence of detectable viraemia or seroconversion.
- •
Since two additional patients had unspecific reactions in the Wantai-IgM-rapid-test and the Mikrogen-IgM-recomblot tested negative in one viraemic patient both assays cannot replace real-time PCR.
- •
The use of the Wantai-antigen-test in genotype 1 infected patients is very limited.
- •
While the exact chain of infection could not been clarified in the present case series, the risk of acquiring HEV infection even in luxury hotels could be suggested.
- •
ALT: alanine aminotransferase.
- •
HEV: hepatitis E virus.
- •
IgG: immunoglobulin G.
- •
IgM: immunoglobulin M.
- •
LLOD: lower limit of detection.
- •
PCR: polymerase chain reaction.
- •
SD: standard deviation.
- •
ULN: upper limit of normal.
The authors thank all studied patients for participating in this study. AWL, ML, JSzW have funding by the German Research agency (DFG) and the German Center for Infection (DZIF).
Conflict of Interest- •
Funding: No funding.
- •
Competing interests: None.
- •
Ethical approval: Not necessary (according to our ethical court).