This study aimed to determine the roles of microRNA (miR)-122 in the activation of hepatic stellate cells (HSCs) and liver cirrhosis.
MethodsRat primary HSCs were incubated with transforming growth factor-beta (TGF-β), during which miR-122 and EphB2 expression was measured. miR-122 mimic and/or pcDNA3.1 EphB2 was transfected into TGF-β-induced HSCs. A mouse model of liver cirrhosis was established via an intraperitoneal injection of carbon tetrachloride (CCl4), followed by the injection of miR-122 agomir. Levels of serum alanine transaminase (ALT) and aspartate aminotransferase (AST) were measured. Fibronectin (FN), alpha smooth muscle actin (α-SMA), Collagen I, miR-122, and EphB2 expression was evaluated in liver tissues and HSCs. Cell proliferation was measured using CCK-8 assay. Interactions between miR-122 and EphB2 were assessed using dual luciferase reporter assay.
ResultsmiR-122 (0.15-fold) was downregulated and EphB2 (mRNA: 5.06-fold; protein: 2.35-fold) was upregulated after TGF-β induction of HSCs. Overexpressed miR-122 decreased proliferation and EphB2 (mRNA: 0.46-fold; protein: 0.62-fold), FN (mRNA: 0.45-fold; protein: 0.64-fold), α-SMA (mRNA: 0.48-fold; protein: 0.51-fold), and Collagen I (mRNA: 0.44-fold; protein: 0.51-fold) expression in HSCs, which was abrogated by EphB2 upregulation. miR-122 expression was reduced by 0.21-fold and serum ALT and AST levels were enhanced in mice following 8-week CCl4 induction along with increased expression of FN, α-SMA, and Collagen I in liver tissues, which was blocked by miR-122 overexpression. Moreover, EphB2 was a target gene of miR-122.
ConclusionmiR-122 curtails HSC proliferation and activation by targeting EphB2 and suppresses liver cirrhosis in mice.
Chronic liver disease is prevalent worldwide, with a global incidence of 1.5 billion cases in 2017 [1]. Liver cirrhosis represents the final stage of chronic liver disease, and liver fibrosis is considered a converging point for the deterioration of liver architecture [2]. Hepatic stellate cell (HSC) activation assumes a vital role in the pathological process of liver fibrosis [3]. However, the exact mechanisms underlying liver fibrosis remain uncharacterized [4]. microRNAs (miRs) are small noncoding RNAs that can post-transcriptionally mediate gene expression by targeting specific mRNAs and may participate in the progression of liver fibrosis ([5,6]). In fact, miR-122 exerts anti-fibrotic effects by enhancing the action of adipose tissue-derived mesenchymal stem cells [7]. Leptin aggravates liver fibrosis by reducing miR-122 expression in HSCs [8]. The involvement of miR-122 in liver fibrosis is probably associated with the upregulation of BCL2 [9]. However, the mechanisms of action of miR-122 in liver fibrosis are complex and largely unknown.
The membrane-bound Ephrin type-B receptor 2 (EphB2) belongs to the largest subfamily of tyrosine kinase receptors and is involved in tumors [10]. This gene is implicated in the fibrosis of multiple organs. For example, EphB2 knockdown restrains the generation of astroglial-fibrotic scars after spinal cord injury in rats [11]. In addition, EphB2 is remarkably upregulated in idiopathic pulmonary fibrosis, which is characterized by fibroblast proliferation and extracellular matrix accumulation [12]. Furthermore, EphB2 aggravates liver fibrosis and inflammation by activating HSCs [13]. Moreover, miR-451 and miR-185 synergistically exhibit anti-fibrotic effects in the liver and impair HSC activation by co-targeting EphB2 [14]. Starbase database [15] utilized in our research predicted a binding site of miR-122 on EphB2. Based on the aforesaid observations, we hypothesized that miR-122 might control HSC activation and liver cirrhosis by regulating EphB2 expression. Thus, the research aimed to identify interactions between miR-122 and EphB2 and their effects on HSC activation and proliferation in vitro and liver cirrhosis in vivo.
2Materials and methods2.1Laboratory animalsMale rats with better physical health indicators relative to female rats were used for animal experiments. Male C57BL/6J mice (n = 54; weighing 20-22 g; aged 6-8 weeks) were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China) and male Sprague Dawley rats (n = 5; weighing 200-300 g; aged 6-8 weeks) from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). These animals were fed in the animal center of our hospital as per clean grade mouse or rat feeding requirements with dietary standards referring to the nutritional standards of the American Institute for Nutrition (AIN93). In addition, these animals were housed at 22-24°C and with 50% ± 5% humidity, a 12/12 h light-dark cycle, and ad libitum access to food and water. All animal experiments complied with the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines and was conducted in accordance with the UK Animals (Scientific Procedures) Act, 1986, and associated guidelines, EU Directive 2010/63/EU for Animal Experiments. The experimental designs were ratified by the animal ethics committee of our hospital (Approval No. 2021930).
2.2A mouse model of liver cirrhosisAfter one week of acclimatization, the mice were randomly assigned to control (n = 12) or model (n = 42) groups. The mouse model of liver cirrhosis was induced via intraperitoneal injection of carbon tetrachloride (CCl4) diluted with olive oil (1 mL/kg; twice weekly for 8 weeks) at 1:3 (v/v). Mice in the control group were intraperitoneally injected with olive oil (1 mL/kg). Six mice in the CCl4 group were euthanized at 2, 4, 6, and 8 weeks after CCl4 injection. The remaining 18 mice in the CCl4 group were injected with CCl4 for 8 weeks, among which 12 mice were also injected via the caudal vein with 62.5 nmol scramble-miR (agomir NC group, n = 6) or miR-122 agomir (miR-122 agomir group, n = 6) twice weekly from 10 days after CCl4 injection, and the remaining 6 mice were only intraperitoneally injected with a mixture of CCl4 and olive oil. All mice were euthanized eight weeks later.
2.3Animal treatmentMice fasted for one day, after which, 1 mL of blood was obtained from each mouse through eyeball removal and the livers were isolated via rapid dissection. The liver tissues were completely excised and washed with sterile saline. A piece of liver tissue from the same site was fixed in 4% paraformaldehyde (room temperature), embedded in paraffin, and sectioned. In addition, three to five small pieces of liver tissues were placed in a labeled cryogenic tube, placed in a portable liquid nitrogen container, and then immediately stored at -80°C.
2.4Serological detectionBlood extracted from mice was allowed to stand for 12 h before centrifugation at 3,600 rpm for 15 min. The upper layer of serum was isolated, and the levels of alanine transaminase (ALT) and aspartate aminotransferase (AST) were determined using an AU1000 automated biochemical detector (Olympus Corporation, Tokyo, Japan) according to the operation procedure.
2.5Hematoxylin and eosin (HE) stainingParaffin sections were dewaxed and then washed successively with xylene (I) (5 min), xylene (II) (5 min), gradient ethanol of 100% (2 min), 95% (1 min), 80% (1 min), and 75% ethanol (1 min), and distilled water (2 min). The sections were then stained with hematoxylin for 5 min and rinsed with tap water. After differentiation with hydrochloric acid/ethanol for 30 s and immersion in tap water for 15 min, the sections were placed in eosin solution for 2 min. Afterward, the sections were dehydrated and cleared using 95% ethanol (I) for 1 min, 95% ethanol (II) for 1 min, 100% ethanol (I) for 1 min, 100% ethanol (II) for 1 min, xylene-carbonic acid (3:1) for 1 min, xylene (I) for 1 min, and xylene (II) for 1 min. The sections were sealed in neutral balsam and then examined using a microscope (Olympus Corporation).
2.6Masson stainingSections were stained with a mixture of potassium dichromate (0.1 g/mL) and trichloroacetic acid (0.1 g/mL) for 10 min and then washed three times with distilled water (3 min for each). After immersion in 0.05 g/mL celestine blue for 5 min, the sections were washed once with 0.01 g/mL glacial acetic acid, rinsed once with acid fuchsin and Ponceau red (2:1), stained with brilliant green dye for 1 min, and rinsed three times with glacial acetic acid. The sections were then dehydrated and sealed.
2.7ImmunohistochemistryParaffin sections were placed in an oven at 67°C for 2 h. After dewaxing, 3% hydrogen peroxide (H2O2) was added to the sections to block endogenous peroxidase, and antigens were retrieved in 0.01 mol/L citrate buffer. After three washes with phosphate buffered saline, nonspecific antibody binding was blocked with 5% bovine serum albumin at room temperature for 2 h. Afterward, the sections were incubated with primary antibodies (Abcam, Cambridge, UK) against fibronectin (FN; 1:100, ab2413), alpha smooth muscle actin (α-SMA; 1:100, ab5694), and Collagen I (1:100, ab270993) overnight at 4°C. Biotin-labeled secondary antibody was re-probed with the sections for 1 h at room temperature. Then, diaminobenzidine was added and cells were photographed using a biological microscope. Positive cells were statistically analyzed using the Image J software.
2.8Isolation and culture of HSCsThe purity of HSCs isolated from healthy male Sprague Dawley rats as described [16] was > 95%. Rat primary HSCs were cultured in low-glucose Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific Inc., Waltham, MA, USA) with 20% fetal bovine serum (FBS; Biological Industries, Kibbutz Beit-Haemek, Israel) at 37°C under a 5% CO2 atmosphere. The medium was changed every 48 h.
2.9Cell transfectionRat primary HSCs were activated by incubation with transforming growth factor-beta (TGF-β, 5 ng/mL) for 24 h and then cultured in FBS-free DMEM for 6 h. miR-122 mimic, pcDNA3.1 EphB2, and their negative controls (NCs, GenePharma, Shanghai, China) were transfected into the 3rd to 5th passage of the activated HSCs at a final concentration of 100 nM for 48 h using Lipofectamine 2000 (Thermo Fisher Scientific Inc.). Total RNA and protein were harvested for quantitative real-time polymerase chain reaction (qRT-PCR) and western blotting, respectively.
2.10Cell counting kit (CCK)-8 assayHSCs were seeded into 96-well plates with three replicate wells. One plate was taken daily for five consecutive days from the first day after seeding for CCK-8 assay of cell proliferation. The CCK-8 reagent (GlpBio, Montclair, CA, USA) was placed at room temperature until it completely melted. Thereafter, 10 µL CCK-8 reagents were added to each well and the cells were incubated at 37°C for 2 h. The optical density (OD) at 450 nm was determined using a microplate reader (Thermo Fisher Scientific Inc.).
2.11Quantitative real-time polymerase chain reaction (qRT-PCR)TRIzol reagents (15596-026, Invitrogen, Carlsbad, CA, USA) were applied to isolate total RNA from liver tissues and HSCs (A260/280 absorbance ratio between 1.8-2.1, and RNA integrity value 18S/28S of approximately 1:2). The OD value was measured using an ultraviolet spectrophotometer 3 times, and the RNA concentration was calculated according to the formula A260nm OD value × 40 × 50/1000 = µg/µL RNA (the RNA concentration of 0.2-1 µg/µL). RNA was reversely transcribed into cDNA using a reverse transcription kit (RR047A, TaKaRa, Tokyo, Japan). Primers synthesized by Sangon (Shanghai, China) were detailed in Table 1. Then, 10 μL cDNA underwent real-time fluorescence quantitative PCR using SYBR® Green Real-time PCR Master Mix (QPK-201, TOYOBO, Shanghai, China) on a Roche 480 real-time fluorescence quantitative PCR instrument. The thermal cycling parameters were as follows: 95°C for 10 s, 45 cycles of 95°C for 5 s, 60°C for 10 s, and 72°C for 10 s, and finally 72°C for 5-min extension. With U6 and GAPDH as the internal references for miR and mRNA, respectively, the relative gene expression was calculated using the 2−ΔΔCt method, in which ΔΔCt = ΔCt experimental group (Ct target gene - Ct reference gene) - ΔCt control group (Ct target gene - Ct reference gene). Quantitative PCR was set up with 3 replicates per reaction.
Primer sequences
Note: F, forward; R, reverse.
Total protein was obtained from HSCs using Radioimmunoprecipitation Assay (RIPA) lysis buffer, and the protein concentration (4-30 µg/µL) was measured using bicinchoninic acid kits (Beyotime, Shanghai, China). Total protein was mixed with sample loading buffer (5 ×) and denatured in the boiled water for 5 min. The protein was loaded (15 μL and 40 µg per well) and then resolved using 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis. The protein was electroblotted onto polyvinylidene fluoride membranes at 300 mA, and then the membranes were blocked with 5% skim milk at room temperature for 1 h. The membranes were incubated with the primary antibodies against EphB2 (ab216629, 1:1000), Collagen I (ab270993, 1:1,000), FN (ab2413, 1:2,000), α-SMA (ab5694, 1:1,000; all from Abcam), and GAPDH (#5174, 1:1,000; Cell Signaling Technology, Danvers, MA, USA) at 4°C overnight and washed with Tris-buffered saline Tween (TBST). The membranes were then incubated with secondary antibodies (ab6728, ab6721, 1:20000, Abcam) for 1 h. The membranes were washed with TBST, and then a chemiluminescent solution was evenly dropped onto the membranes to develop color. The results were analyzed using Image J with GAPDH as the internal reference.
2.13Dual luciferase reporter assayThe binding site of miR-122 to the 3’untranslated region (3’UTR) of EphB2 mRNA was predicted using the online bioinformatics tool StarBase (http://starbase.sysu.edu.cn/). The wild-type (wt) and mutant (mut) sequences of the binding sites (wt-EphB2-3’UTR and mut-EphB2-3’UTR) were inserted into pmirGLO vectors (pmirGLO-EphB2-wt and pmirGLO-EphB2-mut). These two reporter vectors were co-transfected into HSCs with miR-122 mimic, and the cells were collected 48 h later. Firefly and Renilla luciferase activities were measured to calculate the relative luciferase activity according to the instructions of the dual luciferase reporter assay kit (RG027; Beyotime).
2.14Statistical analysisAll experiments were repeated three times unless otherwise stated. Data were statistically analyzed using SPSS (version 18.0; IBM Corp., Armonk, NY, USA) and GraphPad Prism 6.0 (GraphPad Software Inc., San Diego, CA, USA). Data were expressed as means ± standard deviation. Two groups were compared using t-tests. Multiple groups were compared using one-way analysis of variance tests, followed by Dunnett multiple comparison post hoc tests. Values were considered statistically significant at P < 0.05.
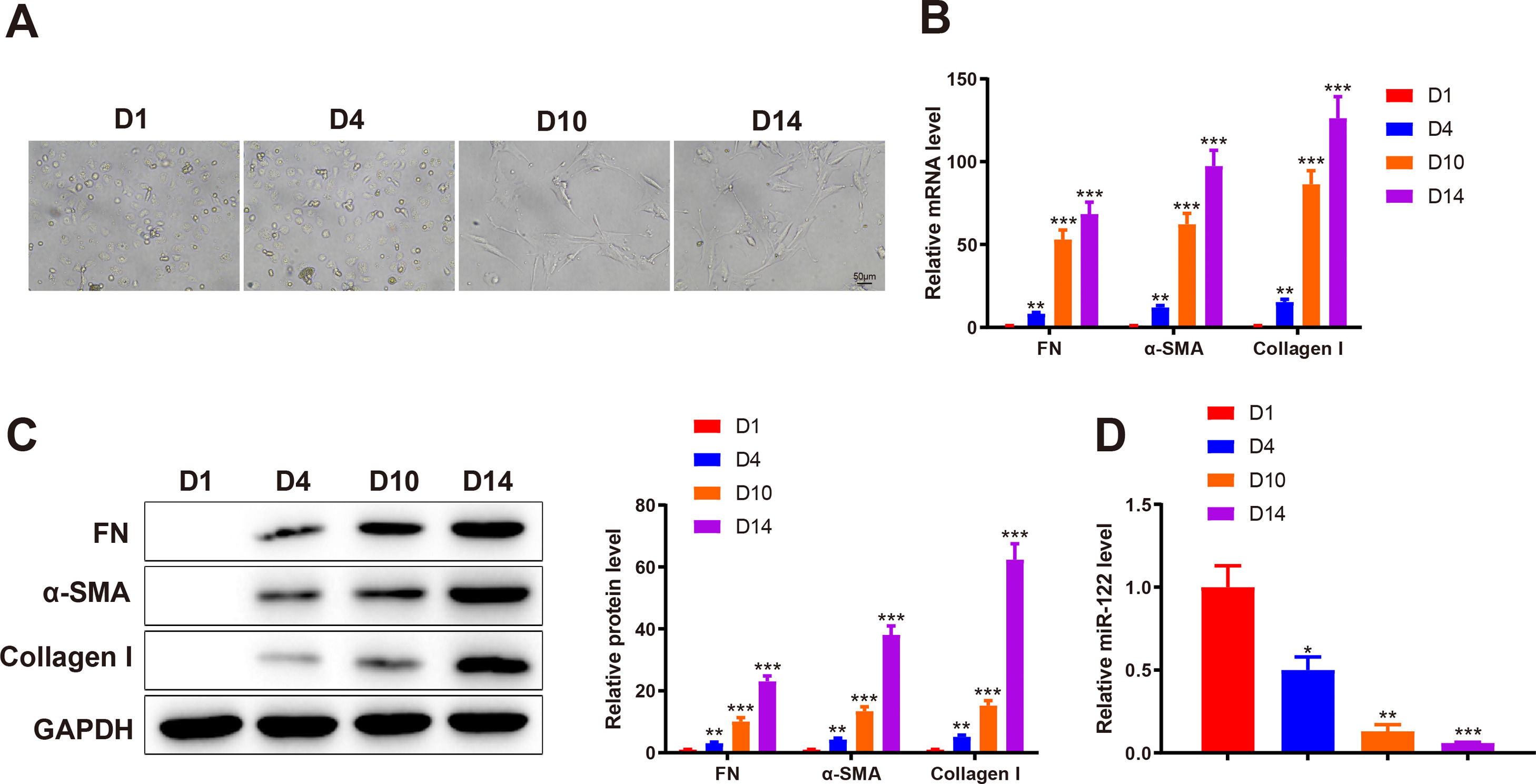
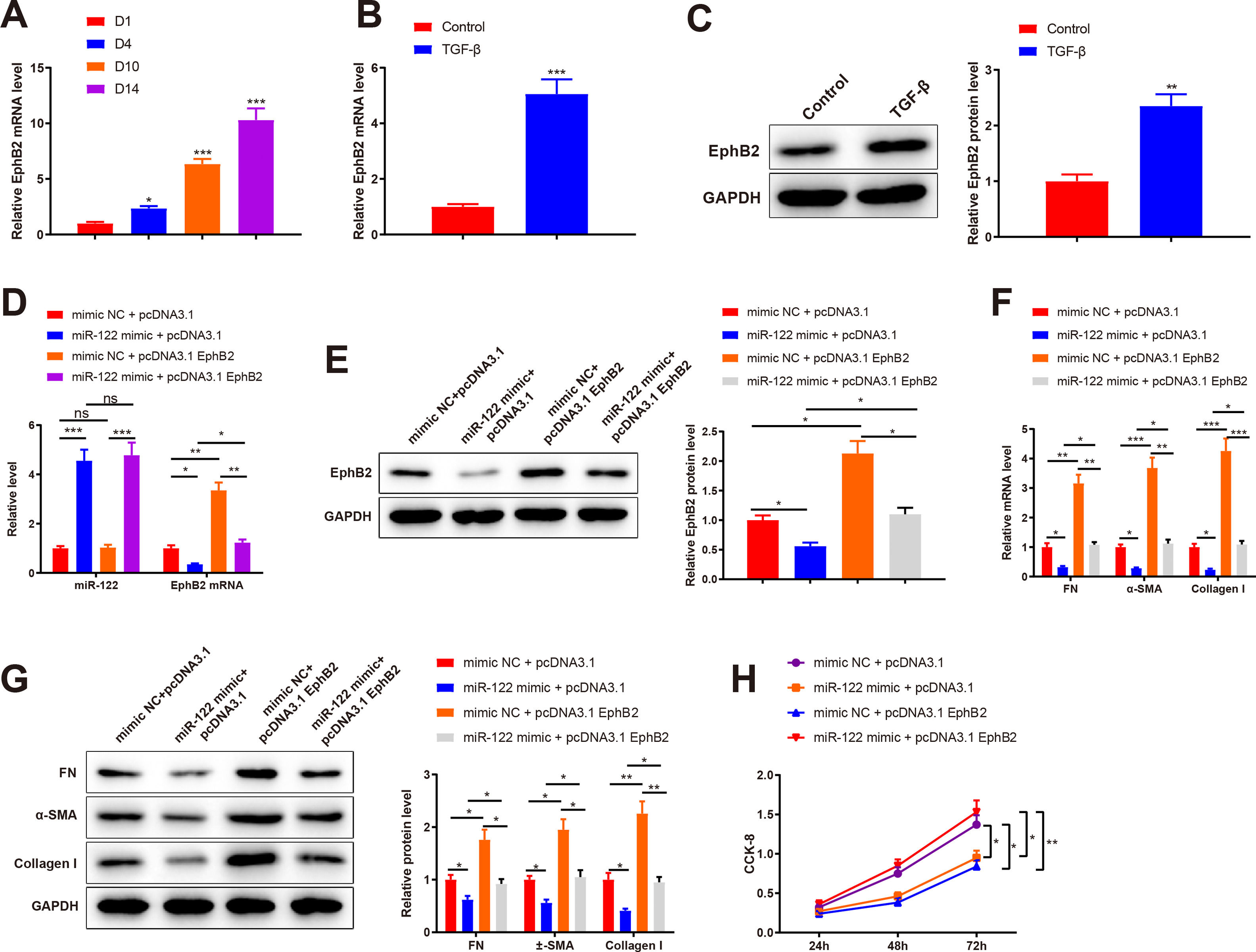
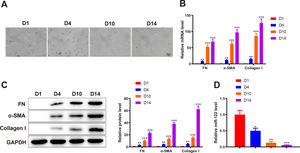
3Results3.1miR-122 was decreasingly expressed during HSC activationTo identify abnormally expressed miRs during HSC activation, primary HSCs were isolated from the livers of healthy rats. Initially, primary HSCs were round with lipid droplets and then changed to a star shape during culture in vitro, suggesting that HSCs were activated (Fig. 1A). In addition, the markers of HSC activation, including FN (mRNA: 8.34-fold, 53.12-fold, and 68.45-fold; protein: 3.13-fold, 10.12-fold, and 23.12 -fold), α-SMA (mRNA: 12.02-fold, 62.34-fold, and 97.45-fold; protein: 4.31-fold, 13.45-fold, and 38.12-fold), and Collagen I (mRNA: 15.34-fold, 86.45-fold, and 126.34-fold; protein: 5.15-fold, 15.20-fold, and 62.34-fold) expression was gradually enhanced, which further confirmed HSC activation (P < 0.05 ; Fig. 1B and C). qRT-PCR results showed that miR-122 (0.5-fold, 0.13-fold, and 0.061-fold) was gradually downregulated during HSC activation (P < 0.05; Fig. 1D). These findings indicated that miR-122 played a role in HSC activation.
miR-122 expression is reduced during HSC activation
(A) HSCs observed using a light microscope (× 200). (B and C) Expression of FN, α-SMA, and Collagen I in HSCs detected using qRT-PCR (B) and western blotting (C). (D) Expression of miR-122 assessed using qRT-PCR. N = 3. *P < 0.05, **P < 0.01, ***P < 0.001, vs. D1. α-SMA, alpha smooth muscle actin; FN, fibronectin; HSCs, hepatic stellate cells.
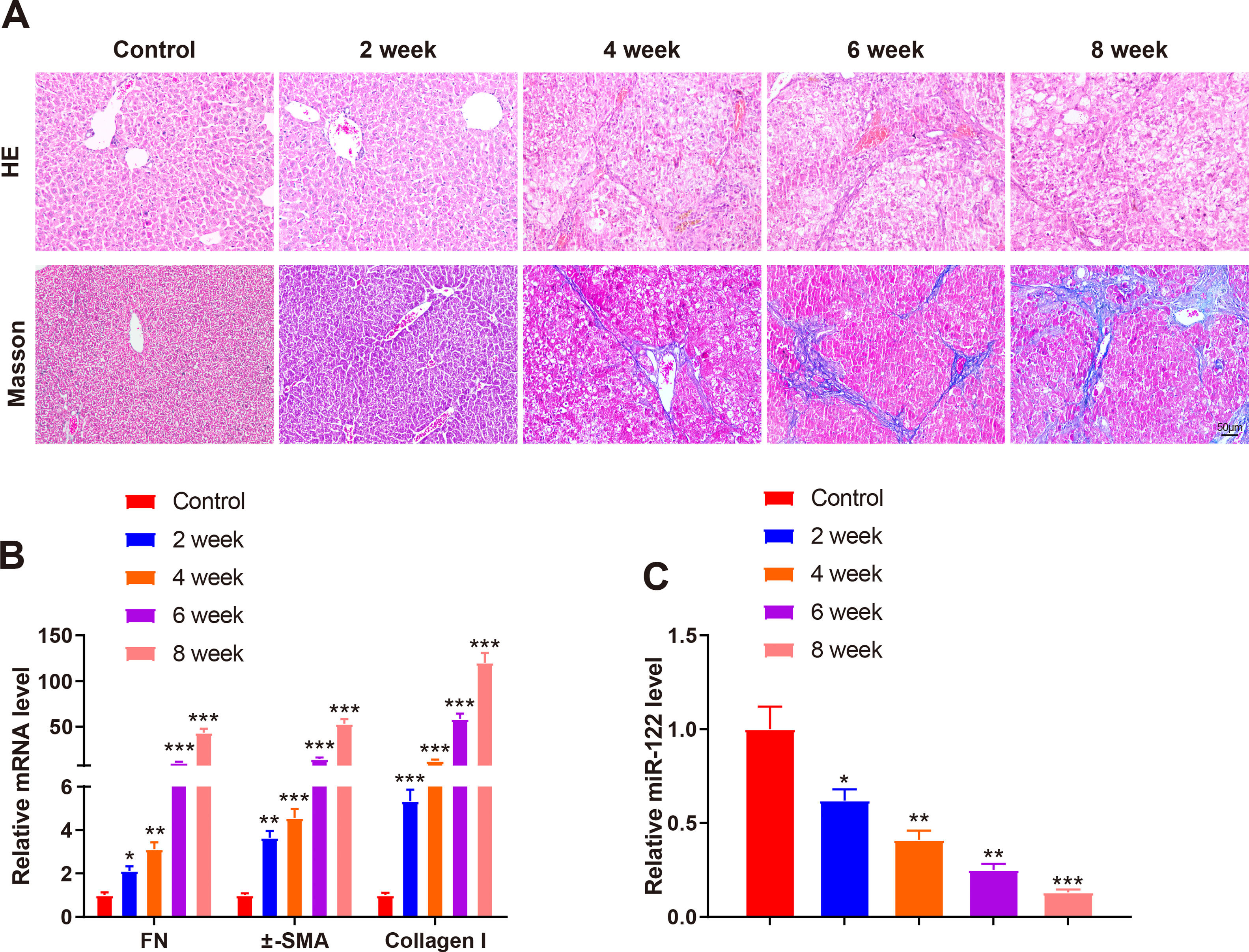
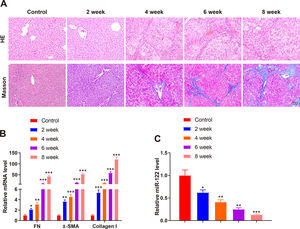
A mouse model of liver cirrhosis was induced by CCl4, and control mice were injected with olive oil to ascertain miR-122 expression in the livers of mice with liver cirrhosis. According to the results of HE and Masson staining, extensive fatty degeneration and necrosis occurred in the livers of mice treated with CCl4 for 2 weeks, accompanied by inflammatory infiltration and mild fibrosis. As the injection time prolonged, the liver gradually developed aggravated fibrosis and fibrotic nodules, with abundant inflammatory factor infiltration (Fig. 2A). qRT-PCR findings manifested that FN (2.12-fold, 3.12-fold, 10.23-fold, and 43.21-fold), α-SMA (3.65-fold, 4.56-fold, 14.35-fold, and 53.12-fold), and Collagen I (5.34-fold, 12.34-fold, 58.34-fold, and 120.34-fold) expression was gradually enhanced by prolonged exposure to CCl4 (Fig. 2B, P < 0.05). miR-122 (0.62-fold, 0.41-fold, 0.25-fold, and 0.13-fold) expression decreased as liver cirrhosis became aggravated (Fig. 2C, P < 0.05). These findings suggested that miR-122 expression decreased in the livers of mice with CCl4-induced liver cirrhosis.
miR-122 expression is decreased in mice with liver cirrhosis
(A) Staining with HE and Masson trichrome to determine pathological changes in liver tissues of mice after CCl4 induction (× 200). (B) Expression of FN, α-SMA, and Collagen I in HSCs assessed using qRT-PCR after CCl4 induction. (C) Expression of miR-122 after CCl4 induction determined using qRT-PCR. *P < 0.05, **P < 0.01, ***P < 0.001, vs. the control group. α-SMA, alpha smooth muscle actin; FN, fibronectin; HE, hematoxylin and eosin; HSCs, hepatic stellate cells.
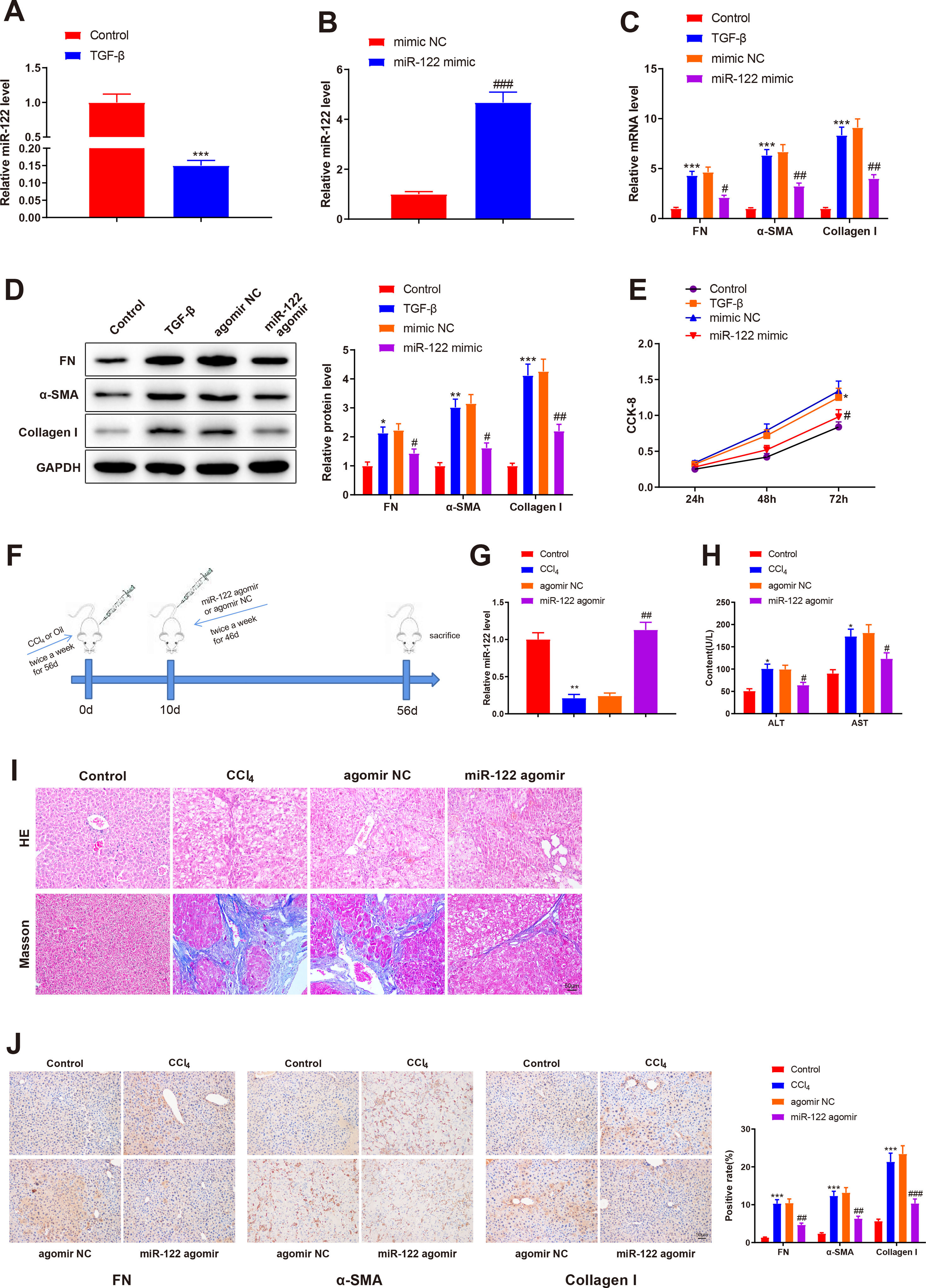
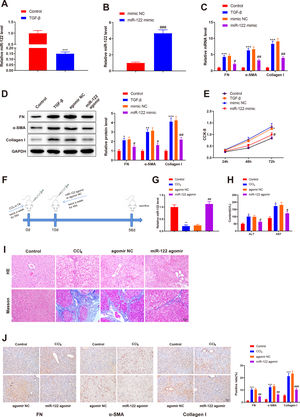
To investigate the role of miR-122 in liver cirrhosis, HSCs were induced using TGF-β for 24 h. miR-122 level reduced by 0.15-fold after TGF-β stimulation (Fig. 3A, P < 0.05). To further explore the role of miR-122 in HSCs, miR-122 mimic was transfected into TGF-β-induced HSCs, and qRT-PCR analysis was performed to confirm the transfection efficacy, the results of which manifested that miR-122 expression elevated by 4.68-fold (P < 0.05, Fig. 3B). The expression of FN (mRNA: 4.32-fold; protein: 2.13-fold), α-SMA (mRNA: 6.34-fold; protein: 3.02-fold), and Collagen I (mRNA: 8.34-fold; protein: 4.12-fold) in the TGF-β group was enhanced compared with the control group (Fig. 3C and D, P < 0.05), suggesting a pro-fibrotic effect of TGF-β. Moreover, the levels of FN (mRNA: 0.45-fold; protein: 0.64-fold), α-SMA (mRNA: 0.48-fold; protein: 0.51-fold), and Collagen I (mRNA: 0.44-fold; protein: 0.51-fold) were decreased in the miR-122 mimic group relative to the mimic NC group, indicating that miR-122 restrained HSC activation (Fig. 3C and D, P < 0.05). CCK-8 results revealed distinctly increased cell viability in the TGF-β group and decreased cell viability in the miR-122 mimic groups (Fig. 3E, P < 0.05). To evaluate whether miR-122 overexpression could restrict cirrhosis in vivo, a liver cirrhotic mouse model was induced by CCl4 and treated with miR-122 agomir (Fig. 3F). The expression of miR-122 was decreased by 0.21-fold in CCL4-treated mice, whereas injection with miR-122 agomir enhanced miR-122 expression by 4.70-fold in the liver of cirrhotic mice (Fig. 3G, P< 0.05). Serum ALT and AST levels were remarkably increased in the CCl4 group versus the control group but were significantly lower in the miR-122 agomir group than in the agomir NC group (Fig. 3H, P < 0.05). As suggested by the results of HE staining and Masson staining, the mice in the CCl4 group developed substantial fibrotic changes in the liver compared with the control group, and fibrotic changes were markedly attenuated in the miR-122 agomir group in contrast to the agomir NC group (Fig. 3I). The immunohistochemical results of FN, α-SMA, and Collagen I further confirmed that miR-122 agomir relieved liver fibrosis in mice (Fig. 3J). In conclusion, miR-122 upregulation attenuated CCl4-induced liver cirrhosis in mice and suppressed TGF-β-induced proliferation and activation of HSCs.
miR-122 inhibits HSC proliferation and activation and reduces cirrhosis in mice
(A) Expression of miR-122 in HSCs after TGF-β stimulation detected using qRT-PCR. (B) Expression of miR-122 in HSCs after miR-122 mimic treatment measured using qRT-PCR. (C-D) Expression of FN, α-SMA, and Collagen I in HSCs after TGF-β stimulation and miR-122 mimic treatment determined using qRT-PCR (C) and western blotting (D). (E) Proliferation of HSCs after TGF-β stimulation and miR-122 mimic treatment determined using CCK-8 method. (F) Carbon tetrachloride was injected into mice to establish a liver cirrhosis model and miR-122 agomir was injected into models through caudal vein. (G) Expression of miR-122 in liver tissues of mice after CCl4 induction and miR-122 agomir treatment measured using qRT-PCR. (H) Serum ALT and AST levels of mice after CCl4 induction and miR-122 agomir treatment quantified using assay kits. (I) Representative images of staining with HE and Masson trichrome of pathological changes in liver fibrosis in mice after CCl4 induction and miR-122 agomir treatment (× 200). (J) Levels of FN, α-SMA, and Collagen I in liver tissues of mice after CCl4 induction and miR-122 agomir detected using immunohistochemistry (× 200). N = 3 for cell experiments. N = 6 mice/group. *P < 0.05, **P < 0.01, ***P < 0.001, vs. control; #P < 0.05, ##P < 0.01, ###P < 0.001, vs. mimic NC (agomir NC). FN, fibronectin; α-SMA, alpha smooth muscle actin; ALT, alanine transaminase; AST, aspartate aminotransferase; HE, hematoxylin and eosin; HSCs, hepatic stellate cells; NC, negative control.
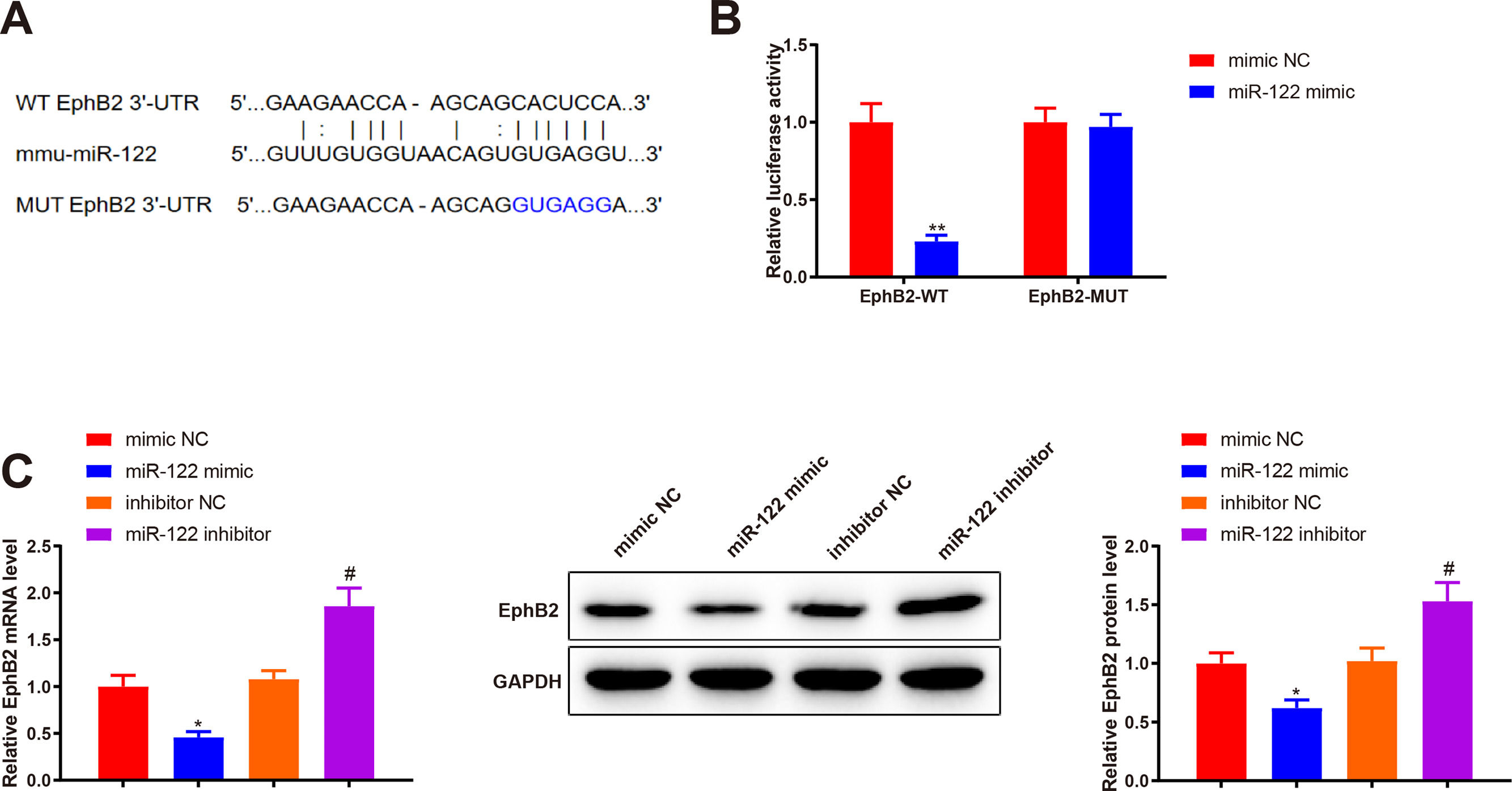
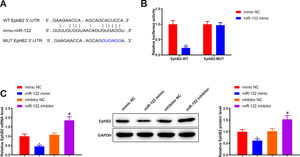
Bioinformatics analysis was implemented to predict the downstream target genes of miR-122 and clarify the molecular mechanism by which miR-122 mediated HSC activation and proliferation. Starbase predicted a binding site of miR-122 to the 3’UTR of EphB2 (Fig. 4A). Our study assumed that miR-122 was involved in HSC activation and proliferation by binding to EphB2. Dual luciferase reporter assay displayed that miR-122 mimic inhibited the luciferase activity of the wt reporter vectors by 0.23-fold (P < 0.05) but did not alter the luciferase activity of mut reporter vectors (Fig. 4B, P > 0.05). To further verify interactions between EphB2 and miR-122, EphB2 mRNA and protein levels were detected after miR-122 mimic or inhibitor treatment. The results exhibited that miR-122 mimic decreased EphB2 expression (mRNA: 0.46-fold; protein: 0.62-fold), whereas miR-122 inhibitor enhanced EphB2 expression (mRNA: 1.72-fold; protein: 1.5-fold) (Fig. 4C, P < 0.05), indicating that EphB2 was a target gene of miR-122.
EphB2 is directly targeted by miR-122
(A) StarBase predicting the binding site between miR-122 and 3’UTR of EphB2. (B) Interaction between EphB2 and miR-122 verified using dual luciferase reporter assay. (C) Expression of EphB2 determined using qRT-PCR and western blotting after miR-122 overexpression or knockdown. N = 3, *P < 0.05, **P < 0.01, vs. mimic NC; #P < 0.05, vs. inhibitor NC. EphB2, Ephrin type-B receptor 2; NC, negative control.
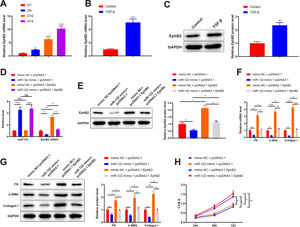
qRT-PCR results depicted elevated EphB2 expression during HSC activation (2.35-fold, 6.34-fold, and 10.31-fold) (Fig. 5A, P < 0.05). Thus, EphB2 expression was measured before and after TGF-β stimulation, which found increased EphB2 expression (mRNA: 5.06-fold; protein: 2.35-fold) in TGF-β-induced HSCs (Fig. 5B-C, P < 0.05). Therefore, we speculated that miR-122 modulated EphB2 to inhibit proliferation and activation in HSCs.
miR-122 inhibits HSC proliferation and activation by downregulating EphB2
(A) Level of EphB2 during HSC activation quantified using qRT-PCR. (B) Level of EphB2 in HSCs after TGF-β stimulation quantified using qRT-PCR. (C) Expression of EphB2 in HSCs after TGF-β stimulation determined by western blotting. (D) Expression of miR-122 and EphB2 in TGF-β-induced HSCs after treatment with miR-122 mimic and pcDNA3.1 EphB2 measured by qRT-PCR. (E) Expression of EphB2 in TGF-β-induced HSCs after treatment with miR-122 mimic and pcDNA3.1 EphB2 tested by western blotting. (F and G) FN, α-SMA, and Collagen I expression in TGF-β-induced HSCs after treatment with miR-122 mimic and pcDNA3.1 EphB2 examined using qRT-PCR (F) and western blotting (G). (H) Cell proliferation in TGF-β-induced HSCs after treatment with miR-122 mimic and pcDNA3.1 EphB2 determined using CCK-8. N = 3. *P < 0.05, **P < 0.01, ***P < 0.001, vs. D1, control, mimic NC + pcDNA3.1, and mimic NC + pcDNA3.1. α-SMA, alpha smooth muscle actin; EphB2. EphB2, Ephrin type-B receptor 2; FN, fibronectin.
Next, a rescue experiment was conducted to verify this speculation, in which miR-122 mimic, pcDNA3.1 EphB2, or miR-122 mimic + pcDNA3.1 EphB2 was transfected into HSCs induced by TGF-β. Compared with the NC + pcDNA3.1 group, miR-122 expression (4.56-fold) was increased and EphB2 expression (mRNA: 0.35-fold; protein: 0.56-fold) was decreased in the miR-122 mimic + pcDNA3.1 group, whilst miR-122 expression was unchanged and EphB2 expression (mRNA: 3.35-fold; protein: 2.13-fold) was elevated in the mimic NC + pcDNA3.1 EphB2 group (Fig. 5D-E, P < 0.05). In contrast to the miR-122 mimic + pcDNA3.1 group, miR-122 expression was unchanged and EphB2 (mRNA: 3.51-fold; protein: 1.96-fold) expression was elevated in the miR-122 mimic + pcDNA3.1 EphB2 group. Meanwhile, the expression of miR-122 (4.64-fold) and EphB2 (mRNA: 0.36-fold; protein: 0.51-fold) was respectively higher and lower in the miR-122 mimic + pcDNA3.1 EphB2 group than in the mimic NC + pcDNA3.1 EphB2 group (Fig. 5D and E, P < 0.05).
According to the results of qRT-PCR and western blotting, FN (mRNA: 0.32-fold; protein: 0.62-fold), α-SMA (mRNA: 0.28-fold; protein: 0.56-fold), and Collagen I (mRNA: 0.23-fold; protein: 0.41-fold) expression was reduced in the miR-122 mimic + pcDNA3.1 group (P < 0.05 vs. the mimic NC + pcDNA3.1 group). However, FN (mRNA: 3.16-fold; protein: 1.76-fold), α-SMA (mRNA: 3.68-fold; protein: 1.95-fold), and Collagen I (mRNA: 4.26-fold; protein: 2.26-fold) expression was enhanced in the mimic NC + pcDNA3.1 EphB2 group (P < 0.05 vs. the mimic NC + pcDNA3.1 group). Similarly, the miR-122 mimic + pcDNA3.1 EphB2 group had elevated FN (mRNA: 3.38-fold; protein: 1.48-fold), α-SMA (mRNA: 4-fold; protein: 1.88-fold), and Collagen I (mRNA: 4.74-fold; protein: 2.32-fold) expression (Fig. 5F and G, P < 0.05 vs. the miR-122 mimic + pcDNA3.1 group). The proliferation of activated HSCs was consistently decreased in the miR-122 mimic + pcDNA3.1 group versus the mimic NC + pcDNA3.1 group, but increased in the mimic NC + pcDNA3.1 EphB2 group and the miR-122 mimic + pcDNA3.1 EphB2 group in contrast to the mimic NC + pcDNA3.1 or miR-122 mimic + pcDNA3.1 group (Fig. 5H, P < 0.05). In comparison with the mimic NC + pcDNA3.1 EphB2 group, FN (mRNA: 0.34-fold; protein: 0.52-fold), α-SMA (mRNA: 0.30-fold; protein: 0.53-fold), and Collagen I (mRNA: 0.26-fold; protein: 0.42-fold) expression and cell proliferation were reduced in the miR-122 mimic + pcDNA3.1 EphB2 group (Fig. 5F-H, P < 0.05). Collectively, miR-122 diminished EphB2 expression, thus inhibiting HSC proliferation and activation.
4DiscussionThe activation of HSCs significantly impacts hepatic fibrosis, which is generally considered a convergent stage during the progression of liver cirrhosis [17]. miRs assume crucial roles in the regulation of HSC activation and the development of liver cirrhosis ([18,19]). Despite general agreement that miR-122 functions as an inhibitory orchestrator of HSC activation, the exact mechanisms remain poorly identified [20]. Therefore, based on bioinformatics prediction, the present research set out to ascertain the specific mechanism of miR-122 in HSC activation and liver cirrhosis.
The present in vitro and in vivo evidence suggested that miR-122 was downregulated during HSC activation and in CCl4-induced animal models. These results are consistent with the findings of a prior research that downregulated miR-122 could be regarded as a non-invasive biomarker for liver fibrosis in hepatitis C virus-related chronic liver disease [21]. Therefore, we overexpressed miR-122 in TGF-β-induced HSCs and liver cirrhotic mice. Our results revealed that miR-122 overexpression reduced FN, α-SMA, and Collagen I levels and the proliferation of HSCs. In addition, we also found that the injection with miR-122 agomir ameliorated liver cirrhosis in CCl4-induced mice. FN, α-SMA, and Collagen I are the markers of liver fibrosis [22]. As reported, liver cirrhosis is featured by liver fibrosis [23]. Moreover, miR-122 suppresses liver fibrosis by targeting pro-fibrotic B cell lymphoma 2 in mice and human HSCs [9]. Yu and colleagues observed that miR-122 is involved in nuclear paraspeckle assembly transcript 1 (NEAT1)-regulated liver cirrhosis [24]. miR-122 might regulate the epithelial-mesenchymal transition of HSCs mediated by the TGF-β1/Smad pathway [25]. miR-122 is inversely related to TGF-β1 expression in bile duct ligation-induced liver fibrosis rat models [26]. Also, mounting evidence elucidated that miR-122 downregulation led to HSC activation and proliferation ([8,27]). miRs have been documented to bind to the 3′UTR of target mRNAs to manipulate their expression [28]. Specifically, the targeting relationship between miR-122 and EphB2 was predicted and validated by Starbase database prediction and dual luciferase reporter assay. Moreover, our findings revealed upregulated EphB2 during HSC activation, and that EphB2 upregulation nullified repressed HSC proliferation and activation caused by miR-122 overexpression in vitro. Consistently, Mimche et al. observed that EphB2 was upregulated and activated in mouse HSCs treated with CCl, which accelerated HSC activation [13]. Furthermore, a prior research unraveled that EphB2 facilitated liver fibrosis and inflammation in CCl4-induced animal models [13]. The downregulation of EphB2 contributes to a significant reduction in malaria-related liver fibrosis in mice [29]. Also, Chen et al. showed that miR-451 and miR-185 synergistically inhibit liver fibrosis by regulating pro-fibrotic EphB2 [14]. As per the above findings, we reasoned that the anti-fibrotic impacts of miR-122 on liver cirrhosis in cells and mice might be achieved by targeting EphB2.
In summary, the findings in our research unveiled that miR-122 suppressed proliferation and activation of HSCs by targeting EphB2. Also, animal experiments in our research elaborated that miR-122 upregulation repressed liver cirrhosis in mice. Our results provide valuable novel insight into the molecular mechanisms through which miR-122 influences HSC activation and proliferation, which may serve as a potential strategy for the treatment of liver cirrhosis. However, based on the results of animal and cell experiments in this report, we would further dissect the expression profile of miR-122 in patient samples to evaluate the correlation of miR-122 with liver cirrhosis and reveal the potential therapeutic target of miR-122.
Author contributionsYL is the guarantor of integrity of the entire study and contributed to the experimental studies and manuscript preparation. YFJ contributed to the experimental studies. BH contributed to the manuscript preparation and statistical analysis. WHW contributed to the experimental studies. MXC contributed to the experimental studies and manuscript preparation. QQZ contributed to the analysis and interpretation of data. JM contributed to the literature research. WHW contributed to the data acquisition and literature research. YFJ contributed to the statistical analysis. YL drafted the article and revised it critically. All authors read and approved the final manuscript.
FundingThis research was funded by the grants from the National Key R&D Program of China (No.2019YFE0190800); Key R&D Program of Hunan province (No.2020SK2083); National Natural Science Foundation of China (No.81500455); National Natural Science Foundation of China (No.82070646); National Natural Science Foundation of China (No.81974079).