Polymorphic variant rs738409 within the PNPLA3 gene associates with alcoholic liver cirrhosis (ALC) in heavy drinkers of various ancestry but has not yet been established in the Russian population characterized by high incidence of ALC. PNPLA3 rs738409 involvement in the inflammatory process has been proposed as one of the mechanisms of liver dysfunction. Relationship between the PNPLA3 polymorphism and the biochemical markers of inflammation in patients with ALC remains unclear. The current study revealed the association between the rs738409 polymorphism, liver cirrhosis and serum cytokines in heavy drinkers in the Russian population.
Materials and methodsThe serum levels of IL6, IL10, IL8, and CCL2 along with PNPLA3 rs738409 polymorphism were determined in heavy drinkers (AA, n=71) and heavy drinkers with diagnosed liver cirrhosis (ALC, n=110). All of the recruited individuals were Caucasians and belonged to the Russian population.
ResultsHeavy drinkers carrying PNPLA3 rs738409 CG or CG+GG genotypes as compared with CC genotype carriers or G allele as compared with C allele carriers had significant risk of ALC. In ALC levels of interleukins and CCL2 increased as compared with AA. PNPLA3 rs738409 CC carriers had lower cirrhosis stage as compared with CG+GG carriers, however there were no differences of IL6, IL10, IL8 or CCL2 levels between G allele carriers and non-carriers in heavy drinkers.
ConclusionThus, in the Russian population heavy drinkers carrying PNPLA3 rs738409 G allele are at higher risk of ALC, however the presence of rs738409 allele does not influence the serum cytokine levels.
Excessive alcohol consumption may associate with development of alcoholic liver disease (ALD) gradually progressing from alcoholic fatty liver to alcoholic steatohepatitis which can eventually lead to cirrhosis in 8–20% of heavy drinkers [1]. Genetic background is a main risk factor underlying alcoholic liver cirrhosis (ALC) with heritability ranging from 21 to 67% [2]. According to World Health Organization report alcohol consumption in Russian population has a harmful pattern with a high rate of alcohol related death due to liver cirrhosis [3], however reliable genetic markers of ALC in this ethnic group have not yet been established.
Several studies of heavy drinkers of various ancestry validated robust associations between the polymorphic locus rs738409 within patatin-like phospholipase domain-containing 3 (PNPLA3) gene and the risk of the entire spectrum ALD including cirrhosis. Particularly, PNPLA3 rs738409 variant is strongly associated with ALC in Mestizo (mixed European and Native American ancestry) [4], Caucasians [5] and North Indians [6]. In addition, meta-analysis of data pooled from ten studies also demonstrated the increased risk of ALC associated with PNPLA3 rs738409 [7]. Moreover genome-wide association study confirms PNPLA3 rs738409 as an important risk locus for ALC at a genome-wide level of significance [8].
PNPLA3 is a lipid droplet-associated protein that has been shown to have predominant hydrolase activity toward triglycerides and retinyl esters and acyltransferase activity toward phospholipids [9,10]. Polymorphism rs738409 in PNPLA3 gene is a missense cytosine to guanine (C>G) change resulting in isoleucine to methionine substitution at position 148 (I148M) of the polypeptide. In general, I148M substitution abolishes enzymatic activity of PNPLA3 thereby increasing cellular content of substrates [11–13]. Additionally, I148M variant disrupts ubiquitylation and proteasomal degradation of PNPLA3, resulting in accumulation of PNPLA3 148M protein and impaired mobilization of triglyceride from lipid droplets [14].
In addition to ALD multiple genetic studies have validated the association of rs738409 locus with a broad spectrum of liver diseases of different etiologies such as non-alcoholic steatohepatitis, fibrosis, cirrhosis, and hepatocellular carcinoma [9,10]. Although association of PNPLA3 rs738409 with a development of ALC was reported for subjects of various ancestry [2] to date there are no published data addressing the relationship between PNPLA3 rs738409 ALC in the Russian population.
Liver inflammation accompanies alcohol abuse (AA) and associated ALD. Chronic excessive alcohol use initiates and aggravates inflammatory process by different mechanisms including gut leakiness of microbial products, sensitization of immune cells to stimulation, and by activating innate immune pathways [15]. Cytokines are the main mediators of liver inflammation under chronic AA conditions. As shown by several clinical studies cytokines, such as interleukins IL6, IL10, IL8, and C–C motif chemokine ligand 2 (CCL2), are elevated in the circulation and liver of patients with ALD, and animal studies demonstrated that IL6 and IL10 play a protective role while IL8 and CCL2 oppositely induce ALD [16].
In vitro studies show that PNPLA3 rs738409 variant define expression of interleukins such as IL6 [17] and IL8 [18,19]. At the same time, animal studies demonstrate that PNPLA3 rs738409 has been mechanistically associated with inflammation in the liver [20]. To date only a single work of Nischalke et al. [19] documents the association between PNPLA3 rs738409 polymorphism and the serum levels of IL8 in patients with ALC. In this paper we undertook an attempt to find the relationship between rs738409 polymorphism, liver cirrhosis and the serum levels of IL6, IL10, IL8 or CCL2 in the Russian population of heavy drinkers.
2Methods2.1SubjectsIndividuals of Caucasian ancestry with excessive alcohol consumption were recruited from Vinogradov Moscow City Clinical Hospital No. 64 and Clinic affiliated with Serbsky National Medical Research Center for Psychiatry and Drug Addiction, Moscow, Russia. During the period from 2016 to 2020, two hundred forty eligible patients were enrolled into the study. Subjects with verified cancer, history of myocardial infarction, congenital or acquired valvular abnormalities or positive for HIV, viral hepatitis B or C were excluded from the study. Patients were divided into alcohol abusers without clinically significant signs of liver or cardio-vascular diseases (AA, n=71) and alcohol-related liver cirrhosis (ALC, n=110) groups. Cardiovascular pathology is another consequence of excessive alcohol consumption. PNPLA3 rs738409 polymorphism scarcely associates with cardiovascular diseases. Additionally, inflammatory process does not obviously underlie cardiovascular disease in heavy drinkers. Thus, we found reasonable the inclusion of an independent comparison group of heavy drinkers with established alcohol-related cardiovascular disease (ACVD) with no liver involvement (n=59). All patients were considered as heavy alcohol drinkers as they had a history of alcohol consumption greater than 100g per day for at least 10 years. AA was demonstrated by self-reported interview using the CAGE and AUDIT questionnaires. The diagnosis of liver cirrhosis was based on the clinical presentation of hepatic encephalopathy and jaundice, the presence of portal hypertension, ascites and hepatomegaly as determined by ultrasonography, the presence of esophageal varices according to upper gastrointestinal endoscopy, and METAVR stage F4 according to ultrasound elastography. Liver cirrhosis was staged by calculation of Child-Pugh score. Dilated cardiomyopathy was defined by the established criteria of left ventricular dilation and reduced ejection fraction on echocardiography. Diagnosis of stable angina was based on the patient's history, physical examination, chest X-ray, electrocardiography and echocardiography. Stages of heart failure were classified according to NYHA (New York Heart Association). The study protocol followed the Declaration of Helsinki, and was approved by the Institutional Ethics Committee. All subjects provided written informed consent for research studies.
2.2BiochemistryThe serum levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), gamma-glutamyltranspeptidase (GGT) and alkaline phosphatase (ALP) were measured on SAPPHIRE 400 biochemical analyzer (Hirose Electronic System Co., Ltd., Japan) with compatible kits of reagents (Randox Laboratories Ltd., UK). Serum levels of cytokines were determined by commercially available ELISA EIA kits (Bender MedSystems GmbH, Austria): IL6 (#BMS213), IL10 (#BMS215HS), IL8 (#BMS204), and CCL2 (#BMS281) on Stat Fax 2100 Microplate Reader (Awareness Technology Inc., USA) according to the manufacturer's instructions. Content of the protein of interest was quantified against a standard curve with a detection limit <2.5pg/mL calibrated with known amounts of the protein. Concentrations were expressed as pg per mL of serum.
2.3GenotypingGenomic DNA was isolated from the whole blood using the spin column-based DNA purification kit “K-sorb” (#EX-514; Syntol, Russia) according to the manufacturer's instructions and stored at −20°C before use. The genotyping of PNPLA3 rs738409 variant was conducted by predesigned and validated TaqMan SNP Genotyping Assay (#4351379_C______7241_10; Thermo Fisher Scientific, USA) containing primers and probe, and master mix containing hot start Taq DNA-polymerase (#PK145; Evrogen, Russia). Real-time PCR was performed on ANK-48 thermocycler (Institute for Analytical Instrumentation RAS and Bauman Moscow State Technical University, Russia) using the following protocol: initial denaturation at 95°C for 120s; (1) 40s at 63°C; (2) 15s at 95°C for 45 cycles. No-template (negative) controls for each PCR run were included. Genotype discrimination was performed by using ANK_Expert Software ver. 1.0.5.100 (Syntol, Russia). After finishing the genotyping 20% of randomly chosen samples were re-tested to corroborate the correct allelic discrimination and 100% of concordance was observed.
2.4StatisticsStatistical analysis was performed using IBM SPSS Statistics version 23.0 (IBM Corp., USA) or Prism 6 for Windows (GraphPad Software Inc., USA). Chi-square test was used to compare categorical variables between the groups. After testing for normality by means of Lilliefors test, continuous variables were shown as the mean±standard deviation or median (lower quartile; upper quartile) for normal and abnormal distributed parameters, respectively. The comparison of continuous variables between groups was performed by one-way ANOVA followed by Tukey HSD post hoc multiple comparison test or one-way ANOVA on ranks (Kruskal–Wallis) followed by Dunn post hoc multiple comparison test depending on the distribution of continuous variables in the groups. The Hardy–Weinberg equilibrium of expected and observed genotype distribution was analyzed by χ2 test. The genotypes and allele frequencies were evaluated using the χ2 test. The association between PNPLA3 rs738409 variant and risk of somatic complications in heavy drinkers was estimated by computing odds ratios (ORs) and confidence intervals (CIs). Contingency tables were used for calculation of χ2 and crude ORs. Adjusted ORs were estimated using logistic regression analysis with gender, age, presence of hypertension and diabetes as co-variates. Statistical significance was assumed at two-sided p values at <0.05 level.
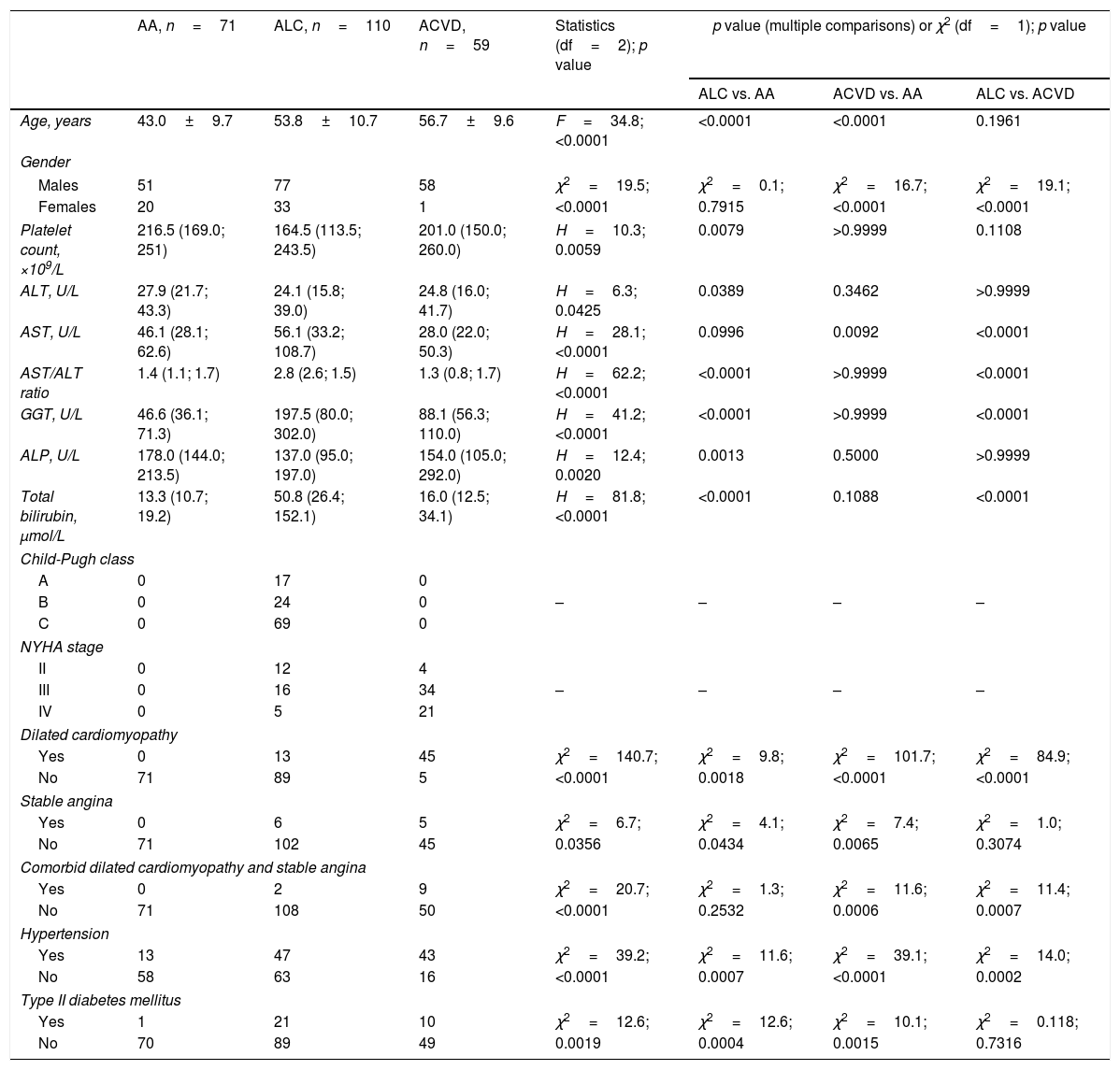
3ResultsThe demographics, laboratory and clinical characteristics of experimental groups are presented in Table 1. The patients in the ALC and ACVD groups were older as compared with heavy drinkers without comorbidities. Females and males were equally represented in the AA and ALC groups while the ACVD almost completely consisted of males. Platelet count, ALT and ALP activities were lower in patients with ALC as compared with AA group. AST to ALT ratio, GGT activity and total bilirubin level were significantly higher in ALC patients as compared with both AA and ACVD. AA and ACVD patients had no apparent signs of liver cirrhosis. However, a part of patients within ALC group had clinical signs of dilated cardiomyopathy or stable angina or both. ACVD patients had significantly higher incidence of dilated cardiomyopathy and/or stable angina as compared with ALC patients. There was higher frequency of arterial hypertension in patients with ALC and ACDV than in AA group, and in ACVD group as compared with ALC. Moreover, both ALC and ACVD patients had higher incidence of type II diabetes mellitus as compared with AA patients.
Demographics, clinical and laboratory characteristics of heavy drinkers.
| AA, n=71 | ALC, n=110 | ACVD, n=59 | Statistics (df=2); p value | p value (multiple comparisons) or χ2 (df=1); p value | |||
|---|---|---|---|---|---|---|---|
| ALC vs. AA | ACVD vs. AA | ALC vs. ACVD | |||||
| Age, years | 43.0±9.7 | 53.8±10.7 | 56.7±9.6 | F=34.8; <0.0001 | <0.0001 | <0.0001 | 0.1961 |
| Gender | |||||||
| Males | 51 | 77 | 58 | χ2=19.5; | χ2=0.1; | χ2=16.7; | χ2=19.1; |
| Females | 20 | 33 | 1 | <0.0001 | 0.7915 | <0.0001 | <0.0001 |
| Platelet count, ×109/L | 216.5 (169.0; 251) | 164.5 (113.5; 243.5) | 201.0 (150.0; 260.0) | H=10.3; 0.0059 | 0.0079 | >0.9999 | 0.1108 |
| ALT, U/L | 27.9 (21.7; 43.3) | 24.1 (15.8; 39.0) | 24.8 (16.0; 41.7) | H=6.3; 0.0425 | 0.0389 | 0.3462 | >0.9999 |
| AST, U/L | 46.1 (28.1; 62.6) | 56.1 (33.2; 108.7) | 28.0 (22.0; 50.3) | H=28.1; <0.0001 | 0.0996 | 0.0092 | <0.0001 |
| AST/ALT ratio | 1.4 (1.1; 1.7) | 2.8 (2.6; 1.5) | 1.3 (0.8; 1.7) | H=62.2; <0.0001 | <0.0001 | >0.9999 | <0.0001 |
| GGT, U/L | 46.6 (36.1; 71.3) | 197.5 (80.0; 302.0) | 88.1 (56.3; 110.0) | H=41.2; <0.0001 | <0.0001 | >0.9999 | <0.0001 |
| ALP, U/L | 178.0 (144.0; 213.5) | 137.0 (95.0; 197.0) | 154.0 (105.0; 292.0) | H=12.4; 0.0020 | 0.0013 | 0.5000 | >0.9999 |
| Total bilirubin, μmol/L | 13.3 (10.7; 19.2) | 50.8 (26.4; 152.1) | 16.0 (12.5; 34.1) | H=81.8; <0.0001 | <0.0001 | 0.1088 | <0.0001 |
| Child-Pugh class | |||||||
| A | 0 | 17 | 0 | ||||
| B | 0 | 24 | 0 | – | – | – | – |
| C | 0 | 69 | 0 | ||||
| NYHA stage | |||||||
| II | 0 | 12 | 4 | ||||
| III | 0 | 16 | 34 | – | – | – | – |
| IV | 0 | 5 | 21 | ||||
| Dilated cardiomyopathy | |||||||
| Yes | 0 | 13 | 45 | χ2=140.7; | χ2=9.8; | χ2=101.7; | χ2=84.9; |
| No | 71 | 89 | 5 | <0.0001 | 0.0018 | <0.0001 | <0.0001 |
| Stable angina | |||||||
| Yes | 0 | 6 | 5 | χ2=6.7; | χ2=4.1; | χ2=7.4; | χ2=1.0; |
| No | 71 | 102 | 45 | 0.0356 | 0.0434 | 0.0065 | 0.3074 |
| Comorbid dilated cardiomyopathy and stable angina | |||||||
| Yes | 0 | 2 | 9 | χ2=20.7; | χ2=1.3; | χ2=11.6; | χ2=11.4; |
| No | 71 | 108 | 50 | <0.0001 | 0.2532 | 0.0006 | 0.0007 |
| Hypertension | |||||||
| Yes | 13 | 47 | 43 | χ2=39.2; | χ2=11.6; | χ2=39.1; | χ2=14.0; |
| No | 58 | 63 | 16 | <0.0001 | 0.0007 | <0.0001 | 0.0002 |
| Type II diabetes mellitus | |||||||
| Yes | 1 | 21 | 10 | χ2=12.6; | χ2=12.6; | χ2=10.1; | χ2=0.118; |
| No | 70 | 89 | 49 | 0.0019 | 0.0004 | 0.0015 | 0.7316 |
Continuous variables were shown as the mean±standard deviation (age) or median (lower quartile; upper quartile). The comparison of continuous variables between groups was performed by one-way ANOVA followed by Tukey HSD post hoc multiple comparison test or one-way ANOVA on ranks (Kruskal–Wallis) followed by Dunn post hoc multiple comparison test. Chi-square test was used to compare categorical variables between the groups. Abbreviations: AA – alcohol abuse; ACVD – alcohol-related cardiovascular disease; ALC – alcoholic liver cirrhosis; ALT – alanine aminotransferase; AST – aspartate aminotransferase; ALP – alkaline phosphatase; GGT – gamma-glutamyltranspeptidase; NYHA – New York Heart Association.
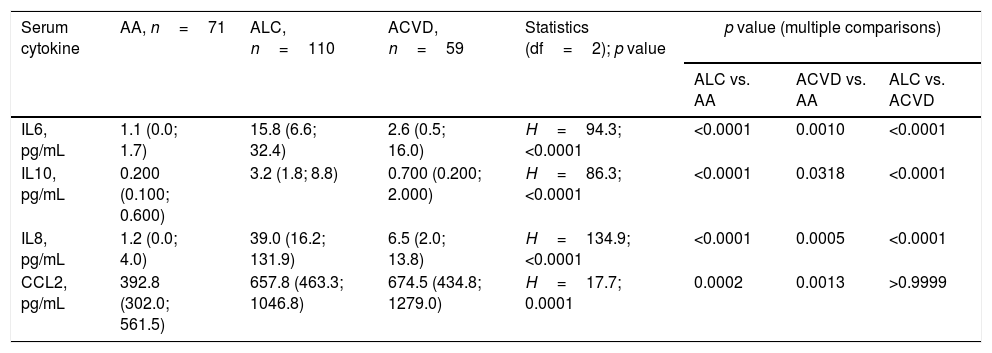
According to Kruskal–Wallis test serum levels of all the cytokines studied differed between groups (Table 2). Subsequent Dunn post hoc multiple comparison test revealed elevation of interleukins both in ALC and ACVD patients as compared with AA. In addition, in ACVD patients levels of interleukins were lesser as compared with ALC patients. As per Dunn post hoc multiple comparison test levels of CCL2 were elevated in both ALC and ACVD patients, as compared with AA, however there was no difference in CCL2 levels between ALC and ACVD groups.
Serum levels of cytokines elevates in heavy drinkers.
| Serum cytokine | AA, n=71 | ALC, n=110 | ACVD, n=59 | Statistics (df=2); p value | p value (multiple comparisons) | ||
|---|---|---|---|---|---|---|---|
| ALC vs. AA | ACVD vs. AA | ALC vs. ACVD | |||||
| IL6, pg/mL | 1.1 (0.0; 1.7) | 15.8 (6.6; 32.4) | 2.6 (0.5; 16.0) | H=94.3; <0.0001 | <0.0001 | 0.0010 | <0.0001 |
| IL10, pg/mL | 0.200 (0.100; 0.600) | 3.2 (1.8; 8.8) | 0.700 (0.200; 2.000) | H=86.3; <0.0001 | <0.0001 | 0.0318 | <0.0001 |
| IL8, pg/mL | 1.2 (0.0; 4.0) | 39.0 (16.2; 131.9) | 6.5 (2.0; 13.8) | H=134.9; <0.0001 | <0.0001 | 0.0005 | <0.0001 |
| CCL2, pg/mL | 392.8 (302.0; 561.5) | 657.8 (463.3; 1046.8) | 674.5 (434.8; 1279.0) | H=17.7; 0.0001 | 0.0002 | 0.0013 | >0.9999 |
Data presented as median (lower quartile; upper quartile). One-way ANOVA on ranks (Kruskal–Wallis H test) followed by Dunn post hoc multiple comparison test was used for comparison of continuous variables between groups. Abbreviations: AA – alcohol abuse; ACVD – alcohol-related cardiovascular disease; ALC – alcoholic liver cirrhosis; IL – interleukin; CCL – C–C motif chemokine ligand.
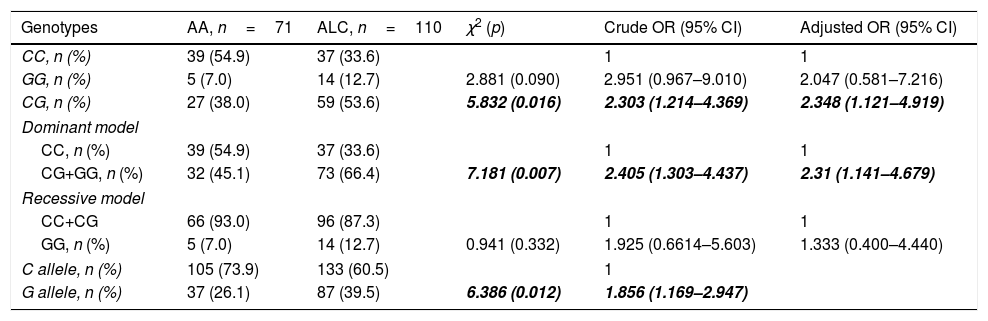
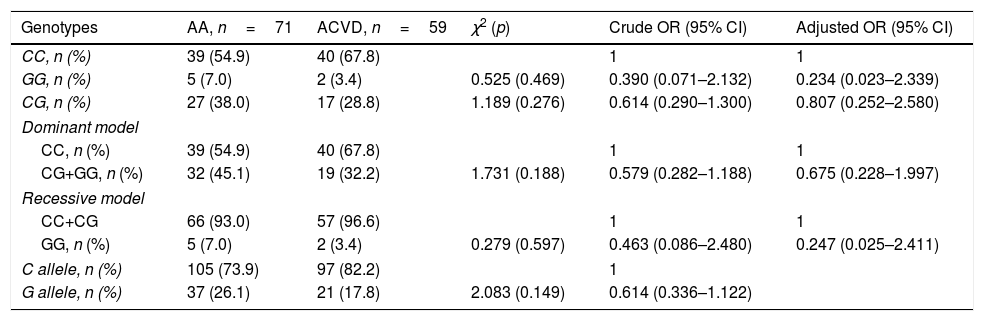
Distribution of PNPLA3 rs738409 genotypes in the experimental groups was in accordance with the Hardy–Weinberg equilibrium (AA: χ2=0.012, p=0.994; ALC: χ2=1.631, p=0.442; ACVD: χ2=0.014, p=0.993). According to the χ2 test, PNPLA3 rs738409 CG and CG+GG genotypes as well as allele G were more frequent in ALC patients as compared with heavy drinkers without comorbidity (Table 3). Heavy drinkers carried the PNPLA3 rs738409 CG (codominant genetic model) or CG+GG (dominant genetic model) genotypes had significantly higher risk of liver cirrhosis as compared with CC genotype carriers (Table 3). Carriers of G allele were also at risk of liver cirrhosis as compared with the carriers of C allele (Table 3). Adjustment for age, gender and the presence of hypertension or diabetes did not affect the increased risk of liver cirrhosis in heavy drinkers carried CG or CG+GG genotypes or G allele (Table 3). In contrast to ALC, PNPLA3 rs738409 genotypes and alleles distributed equally in ACVD and AA groups and did not influence the risk of cardiovascular disease (Table 4).
PNPLA3 rs738409 associates with increased risk of liver cirrhosis in heavy drinkers.
| Genotypes | AA, n=71 | ALC, n=110 | χ2 (p) | Crude OR (95% CI) | Adjusted OR (95% CI) |
|---|---|---|---|---|---|
| CC, n (%) | 39 (54.9) | 37 (33.6) | 1 | 1 | |
| GG, n (%) | 5 (7.0) | 14 (12.7) | 2.881 (0.090) | 2.951 (0.967–9.010) | 2.047 (0.581–7.216) |
| CG, n (%) | 27 (38.0) | 59 (53.6) | 5.832 (0.016) | 2.303 (1.214–4.369) | 2.348 (1.121–4.919) |
| Dominant model | |||||
| CC, n (%) | 39 (54.9) | 37 (33.6) | 1 | 1 | |
| CG+GG, n (%) | 32 (45.1) | 73 (66.4) | 7.181 (0.007) | 2.405 (1.303–4.437) | 2.31 (1.141–4.679) |
| Recessive model | |||||
| CC+CG | 66 (93.0) | 96 (87.3) | 1 | 1 | |
| GG, n (%) | 5 (7.0) | 14 (12.7) | 0.941 (0.332) | 1.925 (0.6614–5.603) | 1.333 (0.400–4.440) |
| C allele, n (%) | 105 (73.9) | 133 (60.5) | 1 | ||
| G allele, n (%) | 37 (26.1) | 87 (39.5) | 6.386 (0.012) | 1.856 (1.169–2.947) | |
χ2 and crude ORs values were established by means of contingency tables. Logistic regression analysis with gender, age, presence of hypertension and diabetes as co-variates was used to calculate adjusted ORs. Statistically significant values are in bold italic. Abbreviations: AA – alcohol abuse; ALC – alcoholic liver cirrhosis; OR – odds ratio; PNPLA – patatin-like phospholipase domain containing.
PNPLA3 rs738409 does not associate with risk of cardiovascular disease in heavy drinkers.
| Genotypes | AA, n=71 | ACVD, n=59 | χ2 (p) | Crude OR (95% CI) | Adjusted OR (95% CI) |
|---|---|---|---|---|---|
| CC, n (%) | 39 (54.9) | 40 (67.8) | 1 | 1 | |
| GG, n (%) | 5 (7.0) | 2 (3.4) | 0.525 (0.469) | 0.390 (0.071–2.132) | 0.234 (0.023–2.339) |
| CG, n (%) | 27 (38.0) | 17 (28.8) | 1.189 (0.276) | 0.614 (0.290–1.300) | 0.807 (0.252–2.580) |
| Dominant model | |||||
| CC, n (%) | 39 (54.9) | 40 (67.8) | 1 | 1 | |
| CG+GG, n (%) | 32 (45.1) | 19 (32.2) | 1.731 (0.188) | 0.579 (0.282–1.188) | 0.675 (0.228–1.997) |
| Recessive model | |||||
| CC+CG | 66 (93.0) | 57 (96.6) | 1 | 1 | |
| GG, n (%) | 5 (7.0) | 2 (3.4) | 0.279 (0.597) | 0.463 (0.086–2.480) | 0.247 (0.025–2.411) |
| C allele, n (%) | 105 (73.9) | 97 (82.2) | 1 | ||
| G allele, n (%) | 37 (26.1) | 21 (17.8) | 2.083 (0.149) | 0.614 (0.336–1.122) | |
χ2 and crude ORs values were established by means of contingency tables. Logistic regression analysis with gender, age, presence of hypertension and diabetes as co-variates was used to calculate adjusted ORs. Abbreviations: AA – alcohol abuse; ACVD – alcohol-related cardiovascular disease; OR – odds ratio; PNPLA – patatin-like phospholipase domain containing.
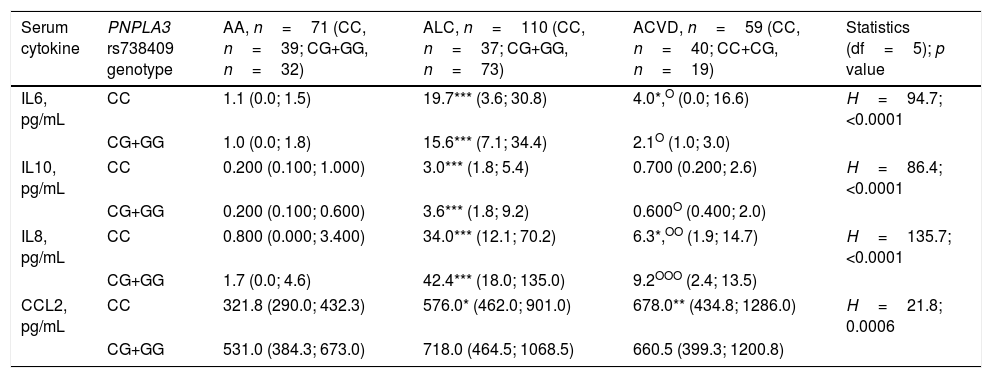
Next, we divided groups according to PNPLA3 rs738409 G allele carriage to test the hypothesis that the presence of G allele may affect the serum levels of IL6, IL10, IL8 or CCL2 in heavy drinkers. According to Kruskal–Wallis test serum levels of all the cytokines studied differed between groups (Table 5). However subsequent Dunn post hoc multiple comparison test failed to support the initial hypothesis since there were no differences in the levels of IL6, IL10, IL8 or CCL2 between G allele carriers and non-carriers in heavy drinkers irrespective of comorbidity. In general, division of patients according to the presence PNPLA3 rs738409 G allele did not affects trends obtained before genotyping. Post hoc multiple comparisons revealed elevation of interleukins in ALC patients as compared with alcohol heavy drinkers without comorbidity both in G allele carriers and non-carriers. IL6 and IL8 but not IL10 were elevated only in ACVD non-carriers of G allele as compared with alcohol heavy drinkers without comorbidity. Additionally, interleukins in ACVD patients were decreased as compared with ALC patients independently of genotype, excepting IL10 in non-carriers. Post hoc multiple comparison test demonstrated elevation of CCL2 levels only in ALC and ACVD G allele non-carriers as compared with heavy drinkers of the same genotype. Similarly to the results with the cytokines, no laboratory or clinical tests showed any association with the presence of PNPLA3 rs738409 in heavy drinkers irrespective of comorbidity (data not shown). Child-Pugh score in ALC patients was an exception: according to Mann–Whitney U-test (p=0.02) PNPLA3 rs738409 CC carriers had lower Child-Pugh score – 9.0 (8.0; 11.0) points as compared with CG+GG carriers – 11 (9.0; 12.0) points.
PNPLA3 rs738409 G allele does not associate with serum cytokines in heavy drinkers.
| Serum cytokine | PNPLA3 rs738409 genotype | AA, n=71 (CC, n=39; CG+GG, n=32) | ALC, n=110 (CC, n=37; CG+GG, n=73) | ACVD, n=59 (CC, n=40; CC+CG, n=19) | Statistics (df=5); p value |
|---|---|---|---|---|---|
| IL6, pg/mL | CC | 1.1 (0.0; 1.5) | 19.7*** (3.6; 30.8) | 4.0*,O (0.0; 16.6) | H=94.7; <0.0001 |
| CG+GG | 1.0 (0.0; 1.8) | 15.6*** (7.1; 34.4) | 2.1O (1.0; 3.0) | ||
| IL10, pg/mL | CC | 0.200 (0.100; 1.000) | 3.0*** (1.8; 5.4) | 0.700 (0.200; 2.6) | H=86.4; <0.0001 |
| CG+GG | 0.200 (0.100; 0.600) | 3.6*** (1.8; 9.2) | 0.600O (0.400; 2.0) | ||
| IL8, pg/mL | CC | 0.800 (0.000; 3.400) | 34.0*** (12.1; 70.2) | 6.3*,OO (1.9; 14.7) | H=135.7; <0.0001 |
| CG+GG | 1.7 (0.0; 4.6) | 42.4*** (18.0; 135.0) | 9.2OOO (2.4; 13.5) | ||
| CCL2, pg/mL | CC | 321.8 (290.0; 432.3) | 576.0* (462.0; 901.0) | 678.0** (434.8; 1286.0) | H=21.8; 0.0006 |
| CG+GG | 531.0 (384.3; 673.0) | 718.0 (464.5; 1068.5) | 660.5 (399.3; 1200.8) | ||
Data presented as median (lower quartile; upper quartile). One-way ANOVA on ranks (Kruskal–Wallis H test) followed by Dunn post hoc multiple comparison test was used for comparison of continuous variables between groups: * – <0.05, ** – <0.005, *** – <0.0005 vs. AA (same genotype); O – <0.05, OO – <0.005, OOO – <0.0005 vs. ALC (same genotype). Abbreviations: AA – alcohol abuse; ACVD – alcohol-related cardiovascular disease; ALC – alcoholic liver cirrhosis; IL – interleukin; CCL – C–C motif chemokine ligand; PNPLA – patatin-like phospholipase domain containing.
This is the first report of PNPLA3 rs738409 association with risk of liver cirrhosis in heavy drinkers in the Russian population. There are few reported genetic risk factors of ALC for heavy drinkers in Russia [21,22], so establishment and validation of additional genetic risk factors is of great demand. As demonstrated on a multicenter sample consisted from two independent cohorts of Caucasians from Germany relative to alcoholic patients without liver damage PNPLA3 rs738409 G allele carriers represent a genetically defined subpopulation of high-risk individuals susceptible to ALC as compared with C allele carriers with OR 1.86 (1.44–2.40) [5]. Similarly, PNPLA3 rs738409 GG+CG genotype carriers, as compared with CC carriers, and GG genotype carriers, as compared with CG+CC carriers, had increased risk of ALC with ORs 2.01 (1.44–2.81) and 2.79 (1.55–5.04), respectively [5]. PNPLA3 G allele carriage among at-risk alcohol drinkers from Italy is an independent risk factor for developing cirrhosis with hazard ratio 1.53 (1.07–2.19) [23]. According to the meta-analysis of ten studies that included mainly subjects of Caucasian ethnicity, OR of the rs738409 CG genotype compared with CC genotype was 2.09 (1.79–2.44) on comparing patients with ALC with healthy controls. In similar comparison, OR for the rs738409 GG genotype was 3.37 (2.49–4.575) [7]. Thus, the association of PNPLA3 rs738409 G allele with the risk of ALC in the Russian population is in agreement with the previous reports performed on Caucasians from European populations.
The main question is how PNPLA3 rs738409 affects liver functioning and accelerates the progression of ALD to cirrhosis. PNPLA3 I148M substitution resulting from rs738409 G allele abolishes hydrolase activity toward triglycerides [11]. Administration of viruses expressing PNPLA3 148M in the livers of mice associates with a dramatic increase in the number of lipid droplets and in the tissue levels of triglycerides and cholesterol esters [11]. Knock-in mice carrying PNPLA3 148M sequence variant display steatosis, liver inflammation and fibrosis upon steatogenic diet [20,24]. In knock-in mutant mice homozygous for the PNPLA3 148M sequence variant fed a high-sucrose diet PNPLA3 silencing reduced liver inflammation and fibrosis, these responses were accompanied by reduced liver levels of CCL2 [20].
We have demonstrated elevated levels of interleukins IL6, IL10, IL8 and CCL2 in ALC patients. These data are in strong agreement with the previous reports. Indeed, levels of IL6 [25], IL10 [26], IL8 [25,27] in the circulation and CCL2 in the liver tissue [28] have been found to be significantly increased in ALC patients as compared with healthy controls. Contrary to the expectations, PNPLA3 rs738409 in our study has not been associated with the serum levels of IL6, IL10, IL8 or CCL2. We could not replicate data obtained by Nischalke et al. who demonstrated that patients with liver cirrhosis had significantly increased IL8 serum levels in carriers of the PNPLA3 rs738409 G allele as compared with non-carriers [19]. We may propose some explanations for this inconsistency. First consideration is that individual cytokines arise from a number of seemingly unrelated cells [29] so levels of cytokines in circulation and in the liver tissue may not correlate. In fact, depending on context some authors report significant correlation between serum and tissue levels of cytokines [30] whereas some authors do not [31]. Second, PNPLA3 rs738409 may act through cytokines other than investigated in this study. In addition to elevated IL8 level, PNPLA3 148M variant in ALC patients and stimulated hepatocytes associates with increased levels of C-X-C motif chemokine ligand (CXCL1) [19]. Stable overexpression of PNPLA3 148M variant in LX-2 hepatic stellate cells induces expression and/or release not only cytokines studied here CCL2 and IL8, but also CCL5, granulocyte-macrophage colony-stimulating factor (GM-CSF) and CXCL1 [18]. Overexpression of PNPLA3 148M upregulated TNF expression and NF-kB-related inflammation in HepG2 hepatoma cells treated with palmitic acid [32].
In the present study we demonstrated that PNPLA3 rs738409 specifically associates with the risk of ALC in heavy drinkers but as expected it does not associate with cardiovascular diseases. It has been demonstrated that among the 94708 individuals in the Copenhagen Studies, PNPLA3 rs738409 does not associate with risk of ischaemic heart disease; the corresponding ORs were 1.00 (0.95–1.04) for CG and 0.95 (0.86–1.04) for GG vs. CC genotypes [33]. Interestingly, PNPLA3 rs738409 may even protect against the development of heart diseases. The presence of the PNPLA3 rs738409 G allele inversely associates with significant coronary heart disease [34]. Moreover, non-alcoholic fatty liver disease patients who carry the PNPLA3 rs738409 CG+GG genotype have the relative lower risk of coronary heart disease than CC genotype carriers with OR value of 0.6 (0.40–0.90) [35].
Meanwhile the present study has some limitations including the ignorance of body mass index as a covariate and significant age difference between experimental groups. The small sample size is another limitation of this work.
In conclusion, we showed that Caucasian heavy drinkers in the Russian population carrying PNPLA3 rs738409 CG or CG+GG genotypes or G allele have high risk of liver cirrhosis development, however rs738409 has no effect on the serum levels of IL6, IL10, IL8 or CCL2. PNPLA3 genotyping could be considered as a tool to stratify the risk of ALD and carriers of rs738409 G allele should be carefully addressed for appropriate interventions.AbbreviationsAA alcohol abuse alcohol-related cardiovascular disease alcoholic liver cirrhosis alcoholic liver disease alanine aminotransferase aspartate aminotransferase alkaline phosphatase C-C motif chemokine ligand C-X-C motif chemokine ligand confidential interval granulocyte-macrophage colony-stimulating factor interleukin gamma-glutamyltranspeptidase odds ratio patatin-like phospholipase domain containing tumor necrosis factor
None declared.
Authors’ contributionsAL, AI, OA, IG collected and interpreted clinical data. VB carried out cytokine analyses. DP performed genotyping. DP, VB and AI collected and assembled the data. DP contributed to the statistical analysis of the results, prepared the first draft of the manuscript. SP reviewed the manuscript critically and wrote the final manuscript. NT and ZK conceived, designed and supervised the study. DP is responsible for the integrity of the work as a whole. All authors contributed to and have approved the final manuscript.
Conflict of interestThe authors report no conflicts of interest.