Coronavirus disease 2019 (COVID-19) has brought great challenges to global public health. However, a comprehensive analysis of the relationship between liver biochemical parameters and COVID-19 mortality is quite limited.
MethodsWe searched the following electronic databases: PubMed, Embase, Cochrane Library, Web of Science, Scopus, Wanfang and China National Knowledge Infrastructure database until May 5, 2020. STATA software was used for the statistical analyses.
ResultsA total of 25 studies involving 5971 COVID-19 patients were included in our analysis. Compared with non-survivors, survivors had lower levels of aspartate aminotransferase (AST) (weighted mean difference [WMD]=−16.71U/L, 95%CI=[−21.03,−12.40], P<0.001), alanine transaminase (ALT) (WMD=−5.20U/L, 95%CI=[−8.00,−2.41], P<0.001), total bilirubin (TBIL) (WMD=4.40μmol/L, 95%CI=[−5.11,−3.70], P<0.001) and lactic dehydrogenase (LDH) (WMD=−252.44U/L, 95%CI=[−289.57,−215.30], P<0.001), and higher albumin (ALB) level (WMD=4.47g/L, 95%CI=[3.47,5.47], P<0.001). Besides, survivors had lower proportions of these abnormally increased parameters (AST: OR=0.25, 95%CI=[0.15,0.41], P<0.001; ALT: OR=0.49, 95%CI=[0.37,0.64], P<0.001; TBIL: (OR=0.20, 95%CI=[0.12,0.34], P<0.001; LDH, OR=0.09, 95%CI=[0.06,0.14], P<0.001), and lower proportion of abnormally decreased ALB (OR=0.16, 95%CI=[0.07,0.38], P<0.001). Meta-analysis based on standard mean difference and sensitivity analysis did not change the conclusions. Egger test did not detect the presence of publication bias.
ConclusionsLiver biochemical parameters were strongly correlated with COVID-19 mortality. Measurement of these liver biochemical parameters might assist clinicians to evaluate the prognosis of COVID-19.
coronavirus disease 2019
severe acute respiratory syndrome coronavirus 2
confidence interval
weighted mean difference
aspartate aminotransferase
lactic dehydrogenase
alanine transaminase
total bilirubin
albumin
globulin
gamma-glutamyl transpeptidase
direct bilirubin
alkaline phosphatase
ALB-to-GLB ratio
interquartile range
Newcastle–Ottawa Scale
standard mean difference
odd risk
angiotensin-converting enzyme 2
transmembrane serine protease 2
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
The outbreak and spread of coronavirus disease 2019 (COVID-19) since December, 2019, has brought great challenges to global public health [1]. The pathogen has been identified as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which could be transmitted from person to person through close contact, respiratory droplets and aerosol [2,3]. As of October 11, 2020, more than 37 million confirmed cases have been reported including over 1 million deaths [4]. As cold season approaches, the greatest increase (34%) in cases has been reported in the European Region and a substantial rise in deaths has been documented in the African Region reported in the previous week [4]. Worse still, the mortality rate is increasing with no sign of a turning point in many countries. Given this, the identification of clinical risk factors for COVID-9 mortality is imperative to facilitate more aggressive management to reduce fatal outcomes.
Liver biochemical parameters could be used to reflect the extent of liver damage in clinical practice [5]. A previous study demonstrated that almost one half of COVID-19 patients experienced liver damage [6]. SARS-CoV-2 was reported to enter the host cells via binding to angiotensin-converting enzyme 2 (ACE2), followed by its priming by transmembrane serine protease 2 (TMPRSS2) [7]. Pirola and colleagues revealed that ACE2 and TMPRSS2 were expressed in cholangiocytes and hepatocytes, suggesting the possibility that SARS-CoV-2 may cause direct liver damage by viral cytopathic effect [8]. Moreover, there are also some other possible mechanisms involved including immune mediated damage, hypoxic hepatitis and drug-induced liver damage [9]. Numerous studies have demonstrated the association of COVID-19 severity with liver injury [10,11], and some meta-analyses suggested that abnormal liver chemistry could be used as prognostic markers for the severity of COVID-19 [12,13]. However, a comprehensive analysis of the relationship between liver biochemical parameters and COVID-19 mortality is quite limited. In the current systematic review and meta-analysis, we aimed to provide an overview of the association between liver biochemical parameters and COVID-19 mortality.
2Methods2.1Search strategyOur systematic review and meta-analysis was performed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [14]. We searched the following electronic databases from their inception to May 5, 2020: PubMed, Embase, Cochrane Library, Web of Science, Scopus, Wanfang and China National Knowledge Infrastructure database. The following combined search terms were used: (“Coronavirus disease 2019” OR “Coronavirus 2019” OR “COVID-19” OR “COVID19” OR “Severe acute respiratory syndrome coronavirus 2” OR “SARS-CoV-2” OR “nCoV-2019” OR “2019-nCoV” OR “Novel coronavirus”) AND (“Mortality” OR “Death” OR “Dead” OR “Fatality” OR “Non-survival” OR “Non-survivors” OR “Non-survivor” OR “Prognosis” OR “Deceased”). We did not apply any restriction on language or study design. We also manually reviewed the reference lists of all included studies, major reviews and meta-analyses for further potentially relevant articles [15]. The identifier of systematic review registration was PROSPERO CRD42020184995.
2.2Study selectionStudies reporting the liver biochemical parameters in COVID-19 patients were included if they met the following criteria: (1) patients were diagnosed as COVID-19 and could be divided into survivors group or non-survivors group; (2) continuous levels or dichotomous outcomes of liver biochemical parameters were available between survivors and non-survivors group. Liver biochemical parameters included AST, alanine transaminase (ALT), total bilirubin (TBIL), albumin (ALB), LDH, globulin (GLB), gamma-glutamyl transpeptidase (GGT), direct bilirubin (DBIL), total protein, alkaline phosphatase (ALP), ALB-to-GLB ratio (A/G) and prealbumin. If there were two or more studies from the same authors or institutions, only the study with the largest sample size was chosen. To be specific, two studies [16,17] were excluded because they are from same authors and had smaller sample size, compared with these two references [18,19]. Studies were excluded if patients were asymptomatic carriers and did not fulfill the inclusion criteria.
2.3Data extraction and quality assessmentAll the records from the initial search were exported to bibliographic files and then imported into Endnote X9 to exclude any duplicate and irrelevant studies. The following data were extracted: first authors, publication date, country of origin, grouping situation, cases, age, sex and continuous levels or dichotomous outcome of liver biochemical parameters in survivors and non-survivors groups. Stratified data or interquartile range (IQR) were converted to mean (SD) based on the mathematical formulas for meta-analysis [20,21]. Newcastle-Ottawa Scale (NOS) was used for quality assessments of all eligible studies. Studies with NOS stars lower than 7 were regarded as studies with inferior quality and therefore excluded. Two authors (GD and FZ) independently selected and evaluated the included articles. Disagreements were resolved by discussion.
2.4Statistical analysisAll the statistical analyses were carried out by STATA (Version 12.0; STATA Corporation, College Station, TX, USA) software. Weighted mean difference (WMD) with 95% confidence intervals (95% CI) was calculated for the continuous levels of liver biochemical parameters in survivors and non-survivors groups. Standard mean difference (SMD) was used to explore the consistence of the conclusion. Odd risk (OR) with 95% CI was calculated for the dichotomous outcomes of liver biochemical parameters in survivors and non-survivors groups. In the case of heterogeneity among studies (I2>50% or P<0.1), random-effect model was used and would give a more conservative estimate of the 95% CI. In other cases, fixed-effect model was adopted (I2≤50% and P≥0.1) [22]. Sensitivity analysis was performed by omitting one study each time through influence analysis to assess the stability of results. Egger test was used to evaluate publication bias and the Duval and Tweedie trim-and-fill method was implemented to adjust for this bias [15]. P<0.05 was considered statistically significant.
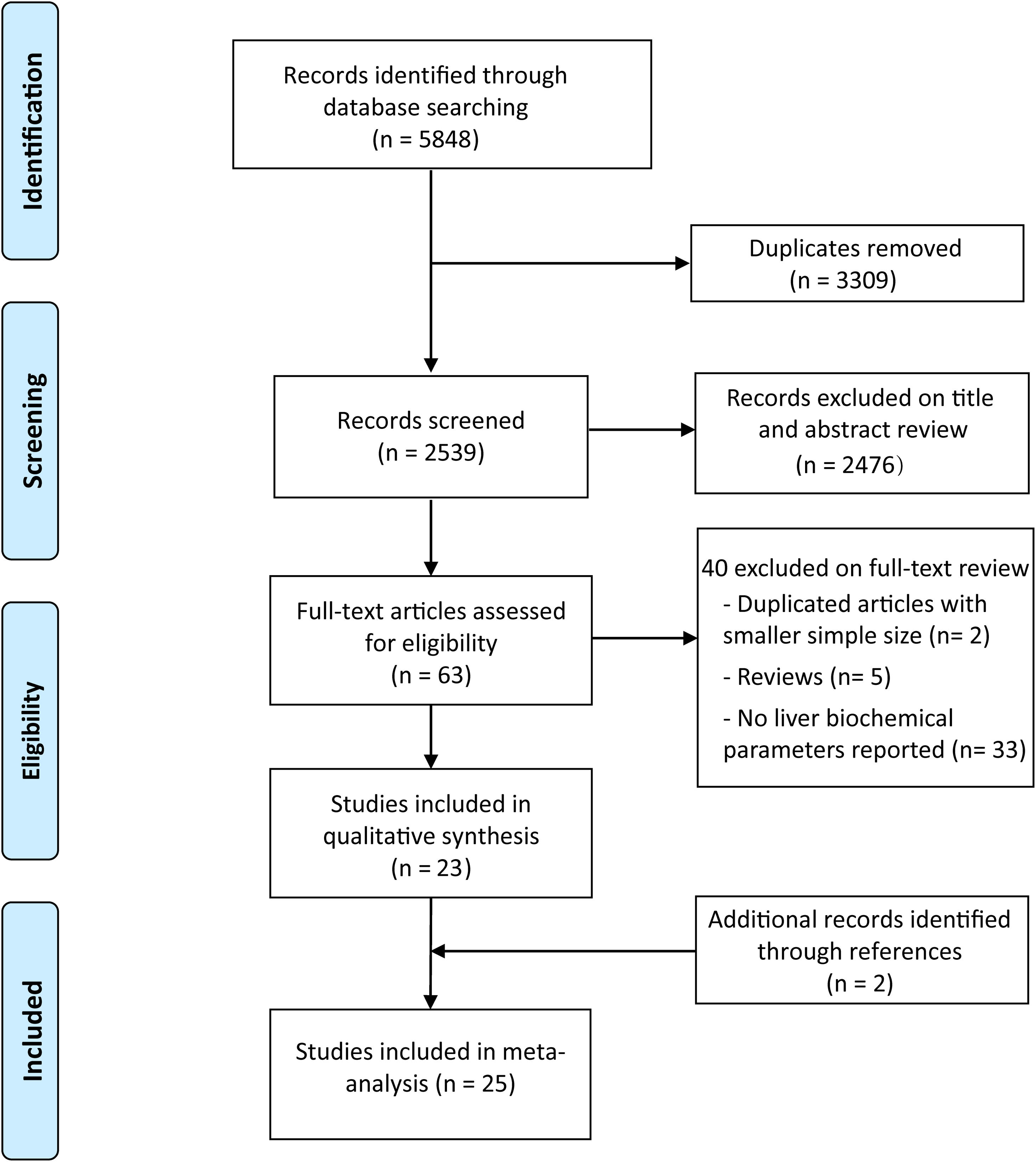
3Results3.1Literature search and studies characteristicsA total of 5848 records were initially identified by our searchers. After exclusion of duplicates, 2539 studies remained. After review of the titles and abstracts, 63 studies were considered to meet the criteria for full-text scanning. After removal of reviews, duplicate articles with smaller sample size, and studies which did not report liver biochemical parameters, 23 studies met the inclusion criteria [18,19,23–43]. Additionally, two studies were enrolled after review of the reference lists of all included studies and major review and meta-analyses [44,45]. Finally, 25 studies involving 5971 COVID-19 patients were included in our analysis [18,19,23–45] (Fig. 1).
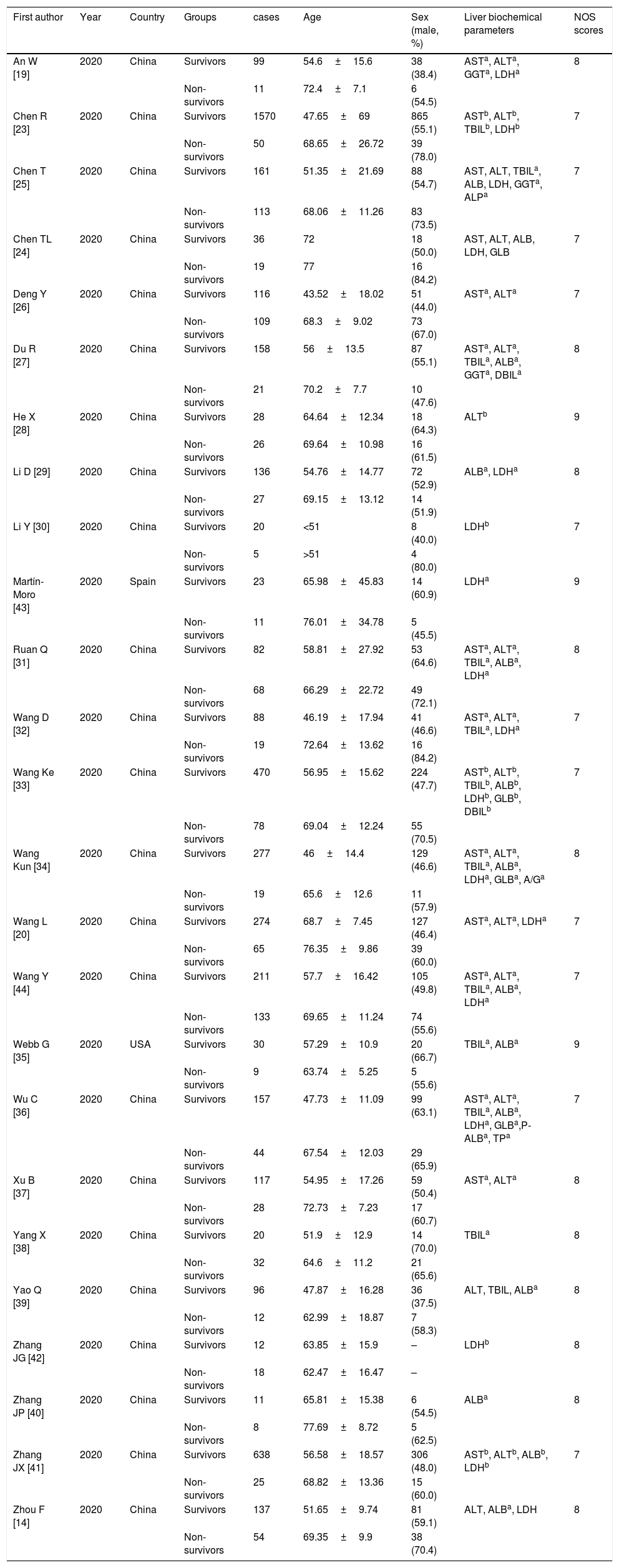
The main characteristics of eligible studies were presented in Table 1. All these studies were published in 2020 and could be divided into non-survivors group and survivors group. Among them, 23 studies were from China [18,19,23–34,36–43,45], one from the United States [35] and one from Spain [44]. Among the 23 studies from China, 20 studies were written in English and three in Chinese [18,28,42]. In general, patients in non-survivors group were older than those in survivors group except for Zhang's study [42], and most of studies showed more male patients in non-survivors than survivors group. All studies were deemed of high quality with 7 or more NOS scores and details were shown in Table A.1.
Characteristics of eligible studies.
| First author | Year | Country | Groups | cases | Age | Sex (male, %) | Liver biochemical parameters | NOS scores |
|---|---|---|---|---|---|---|---|---|
| An W [19] | 2020 | China | Survivors | 99 | 54.6±15.6 | 38 (38.4) | ASTa, ALTa, GGTa, LDHa | 8 |
| Non-survivors | 11 | 72.4±7.1 | 6 (54.5) | |||||
| Chen R [23] | 2020 | China | Survivors | 1570 | 47.65±69 | 865 (55.1) | ASTb, ALTb, TBILb, LDHb | 7 |
| Non-survivors | 50 | 68.65±26.72 | 39 (78.0) | |||||
| Chen T [25] | 2020 | China | Survivors | 161 | 51.35±21.69 | 88 (54.7) | AST, ALT, TBILa, ALB, LDH, GGTa, ALPa | 7 |
| Non-survivors | 113 | 68.06±11.26 | 83 (73.5) | |||||
| Chen TL [24] | 2020 | China | Survivors | 36 | 72 | 18 (50.0) | AST, ALT, ALB, LDH, GLB | 7 |
| Non-survivors | 19 | 77 | 16 (84.2) | |||||
| Deng Y [26] | 2020 | China | Survivors | 116 | 43.52±18.02 | 51 (44.0) | ASTa, ALTa | 7 |
| Non-survivors | 109 | 68.3±9.02 | 73 (67.0) | |||||
| Du R [27] | 2020 | China | Survivors | 158 | 56±13.5 | 87 (55.1) | ASTa, ALTa, TBILa, ALBa, GGTa, DBILa | 8 |
| Non-survivors | 21 | 70.2±7.7 | 10 (47.6) | |||||
| He X [28] | 2020 | China | Survivors | 28 | 64.64±12.34 | 18 (64.3) | ALTb | 9 |
| Non-survivors | 26 | 69.64±10.98 | 16 (61.5) | |||||
| Li D [29] | 2020 | China | Survivors | 136 | 54.76±14.77 | 72 (52.9) | ALBa, LDHa | 8 |
| Non-survivors | 27 | 69.15±13.12 | 14 (51.9) | |||||
| Li Y [30] | 2020 | China | Survivors | 20 | <51 | 8 (40.0) | LDHb | 7 |
| Non-survivors | 5 | >51 | 4 (80.0) | |||||
| Martín-Moro [43] | 2020 | Spain | Survivors | 23 | 65.98±45.83 | 14 (60.9) | LDHa | 9 |
| Non-survivors | 11 | 76.01±34.78 | 5 (45.5) | |||||
| Ruan Q [31] | 2020 | China | Survivors | 82 | 58.81±27.92 | 53 (64.6) | ASTa, ALTa, TBILa, ALBa, LDHa | 8 |
| Non-survivors | 68 | 66.29±22.72 | 49 (72.1) | |||||
| Wang D [32] | 2020 | China | Survivors | 88 | 46.19±17.94 | 41 (46.6) | ASTa, ALTa, TBILa, LDHa | 7 |
| Non-survivors | 19 | 72.64±13.62 | 16 (84.2) | |||||
| Wang Ke [33] | 2020 | China | Survivors | 470 | 56.95±15.62 | 224 (47.7) | ASTb, ALTb, TBILb, ALBb, LDHb, GLBb, DBILb | 7 |
| Non-survivors | 78 | 69.04±12.24 | 55 (70.5) | |||||
| Wang Kun [34] | 2020 | China | Survivors | 277 | 46±14.4 | 129 (46.6) | ASTa, ALTa, TBILa, ALBa, LDHa, GLBa, A/Ga | 8 |
| Non-survivors | 19 | 65.6±12.6 | 11 (57.9) | |||||
| Wang L [20] | 2020 | China | Survivors | 274 | 68.7±7.45 | 127 (46.4) | ASTa, ALTa, LDHa | 7 |
| Non-survivors | 65 | 76.35±9.86 | 39 (60.0) | |||||
| Wang Y [44] | 2020 | China | Survivors | 211 | 57.7±16.42 | 105 (49.8) | ASTa, ALTa, TBILa, ALBa, LDHa | 7 |
| Non-survivors | 133 | 69.65±11.24 | 74 (55.6) | |||||
| Webb G [35] | 2020 | USA | Survivors | 30 | 57.29±10.9 | 20 (66.7) | TBILa, ALBa | 9 |
| Non-survivors | 9 | 63.74±5.25 | 5 (55.6) | |||||
| Wu C [36] | 2020 | China | Survivors | 157 | 47.73±11.09 | 99 (63.1) | ASTa, ALTa, TBILa, ALBa, LDHa, GLBa,P-ALBa, TPa | 7 |
| Non-survivors | 44 | 67.54±12.03 | 29 (65.9) | |||||
| Xu B [37] | 2020 | China | Survivors | 117 | 54.95±17.26 | 59 (50.4) | ASTa, ALTa | 8 |
| Non-survivors | 28 | 72.73±7.23 | 17 (60.7) | |||||
| Yang X [38] | 2020 | China | Survivors | 20 | 51.9±12.9 | 14 (70.0) | TBILa | 8 |
| Non-survivors | 32 | 64.6±11.2 | 21 (65.6) | |||||
| Yao Q [39] | 2020 | China | Survivors | 96 | 47.87±16.28 | 36 (37.5) | ALT, TBIL, ALBa | 8 |
| Non-survivors | 12 | 62.99±18.87 | 7 (58.3) | |||||
| Zhang JG [42] | 2020 | China | Survivors | 12 | 63.85±15.9 | – | LDHb | 8 |
| Non-survivors | 18 | 62.47±16.47 | – | |||||
| Zhang JP [40] | 2020 | China | Survivors | 11 | 65.81±15.38 | 6 (54.5) | ALBa | 8 |
| Non-survivors | 8 | 77.69±8.72 | 5 (62.5) | |||||
| Zhang JX [41] | 2020 | China | Survivors | 638 | 56.58±18.57 | 306 (48.0) | ASTb, ALTb, ALBb, LDHb | 7 |
| Non-survivors | 25 | 68.82±13.36 | 15 (60.0) | |||||
| Zhou F [14] | 2020 | China | Survivors | 137 | 51.65±9.74 | 81 (59.1) | ALT, ALBa, LDH | 8 |
| Non-survivors | 54 | 69.35±9.9 | 38 (70.4) |
NOS, Newcastle–Ottawa Scale; AST, aspartate aminotransferase; ALT, alanine transaminase; TBIL, total bilirubin; ALB, albumin; LDH, lactic dehydrogenase; GLB, globulin; GGT, gamma-glutamyl transpeptidase; ALP, alkaline phosphatase; DBIL, direct bilirubin; A/G, ALB-to-GLB ratio; P-ALB, prealbumin.
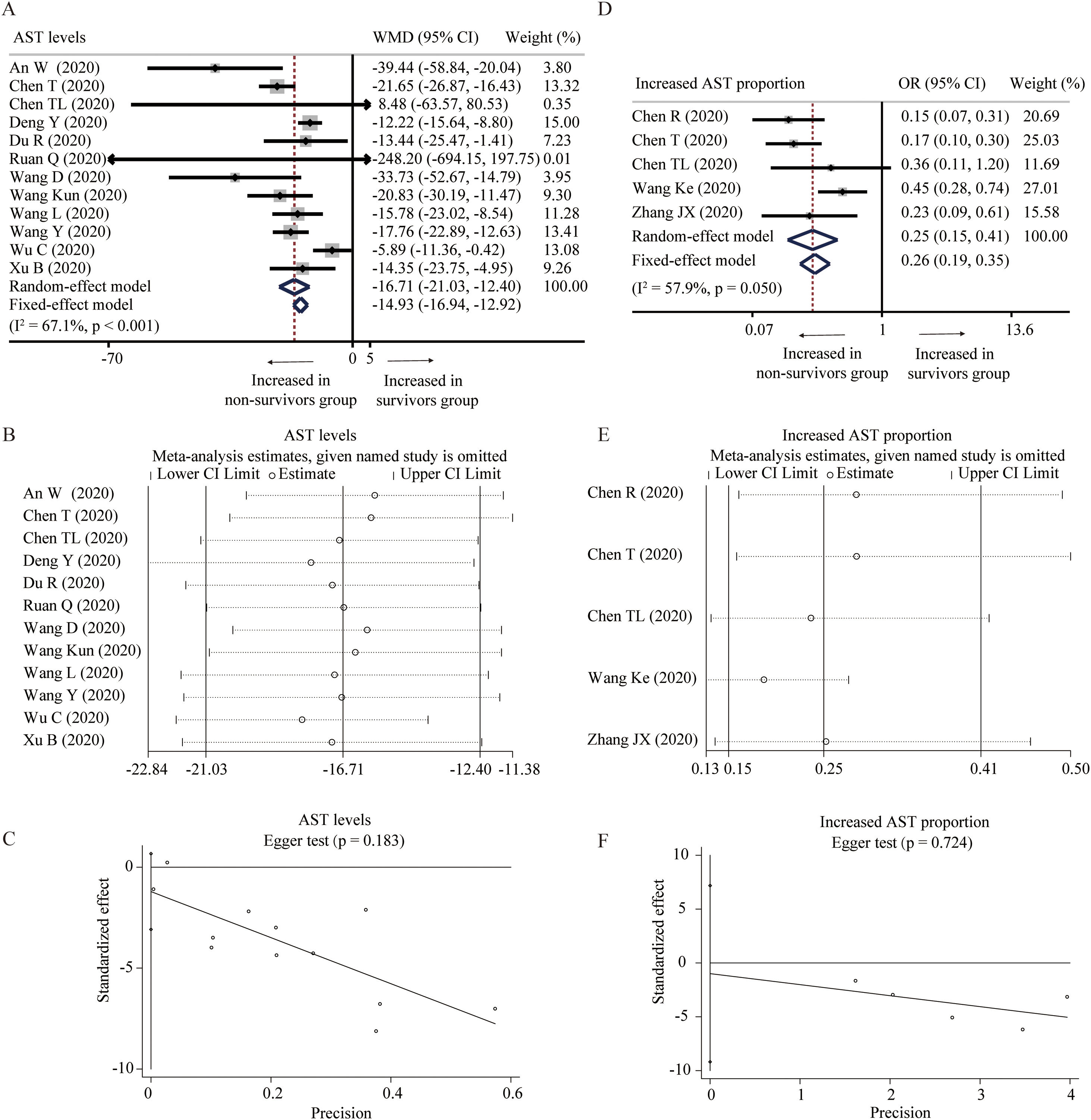
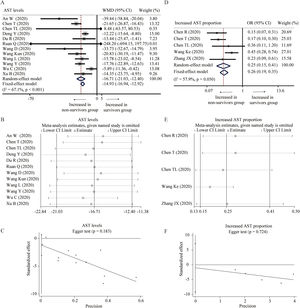
Twelve studies compared the levels of AST in non-survivors and survivors groups [18,19,24–27,31,32,34,36,37,45]. Random-effect model suggested that survivors had lower level of AST than non-survivors (WMD=−16.71U/L, 95% CI=[−21.03,−12.40], P<0.001) with significant heterogeneity (I2=67.1%, P<0.001) (Fig. 2A). Random-effect model based on SMD increased the heterogeneity and did not change the conclusion (Table 2). Sensitivity analysis also arrived at a similar conclusion by omitting one study each time (Fig. 2B). Egger test did not observe the publication bias (Fig. 2C). Moreover, five studies reported the proportion of increased AST in these two groups [23–25,33,41]. With heterogeneity (I2=57.9%, P=0.050), random-effect model showed that survivors had lower proportion of increased AST than non-survivors (OR=0.25, 95% CI=[0.15,0.41], P<0.001) (Fig. 2D). Similar results were found through fixed-effect model and sensitivity analysis, and no publication bias was detected by Egger test (Fig. 2D and F).
Forest plot, sensitivity analyses and publication bias assessment of AST. (A–C) Forest plot (A), sensitivity analyses (B) and Egger test (C) of continuous levels of AST between survivors and non-survivors. (D–F) Forest plot (D), sensitivity analyses (E) and Egger test (F) of the proportion of abnormally increased AST between survivors and non-survivors.
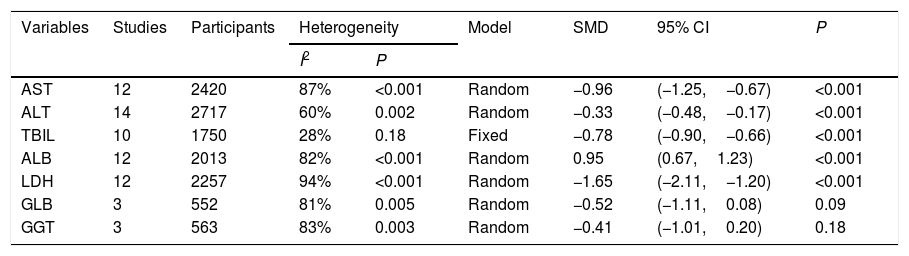
The results of meta-analysis based on standard mean difference (SMD).
| Variables | Studies | Participants | Heterogeneity | Model | SMD | 95% CI | P | |
|---|---|---|---|---|---|---|---|---|
| I2 | P | |||||||
| AST | 12 | 2420 | 87% | <0.001 | Random | −0.96 | (−1.25,−0.67) | <0.001 |
| ALT | 14 | 2717 | 60% | 0.002 | Random | −0.33 | (−0.48,−0.17) | <0.001 |
| TBIL | 10 | 1750 | 28% | 0.18 | Fixed | −0.78 | (−0.90,−0.66) | <0.001 |
| ALB | 12 | 2013 | 82% | <0.001 | Random | 0.95 | (0.67,1.23) | <0.001 |
| LDH | 12 | 2257 | 94% | <0.001 | Random | −1.65 | (−2.11,−1.20) | <0.001 |
| GLB | 3 | 552 | 81% | 0.005 | Random | −0.52 | (−1.11,0.08) | 0.09 |
| GGT | 3 | 563 | 83% | 0.003 | Random | −0.41 | (−1.01,0.20) | 0.18 |
AST, aspartate aminotransferase; ALT, alanine transaminase; TBIL, total bilirubin; ALB, albumin; LDH, lactic dehydrogenase; GLB, globulin;
GGT, gamma-glutamyl transpeptidase.
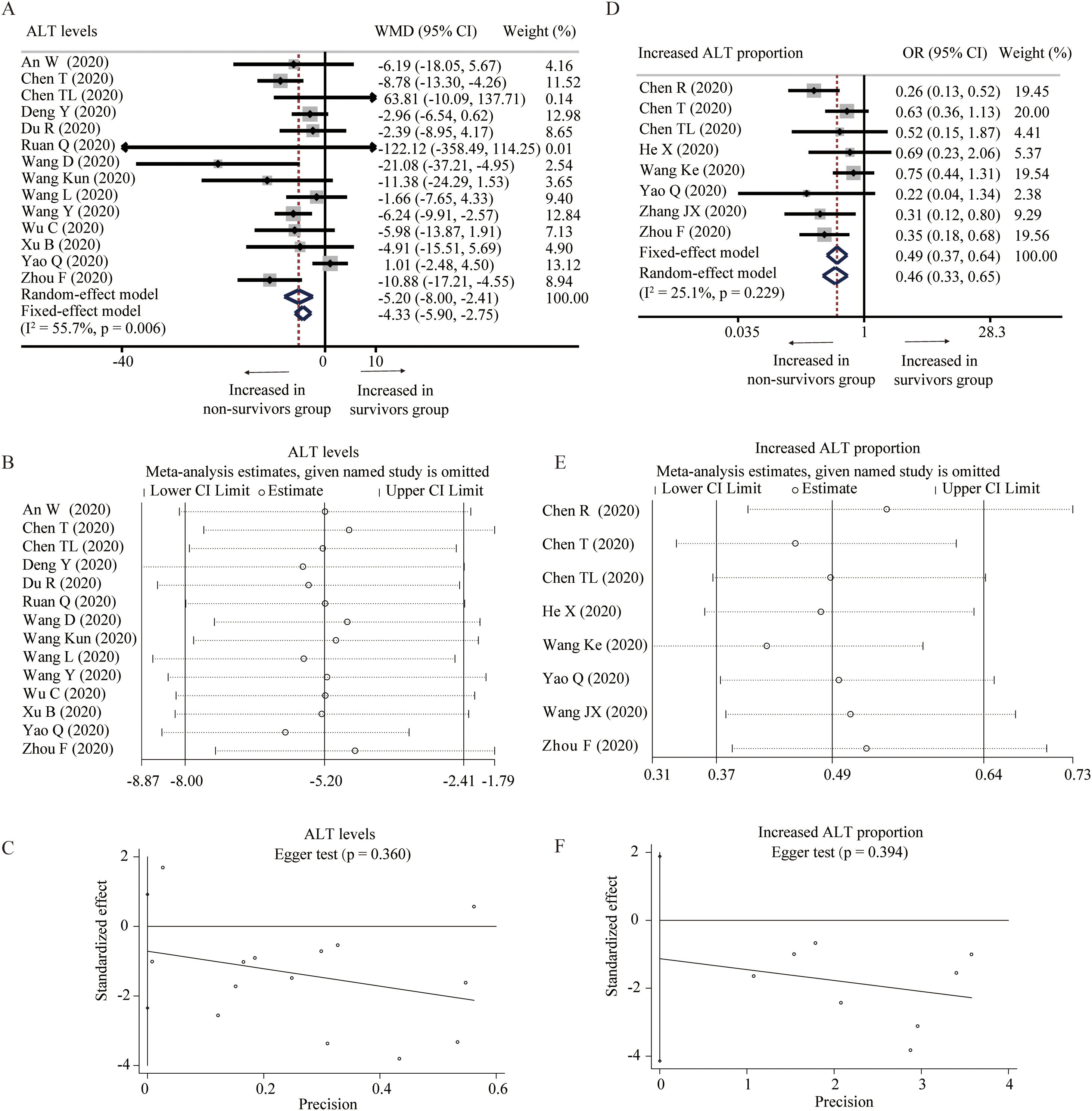
Fourteen studies provided the levels of ALT between survivors and non-survivors group [18,19,24–27,31,32,34,36,37,39,43,45]. With heterogeneity (I2=55.7%, P=0.006), random-effect model was used and showed that survivors had lower level of ALT than non-survivors (WMD=−5.20U/L, 95% CI=[−8.00,−2.41], P<0.001) (Fig. 3A). Random-effect model based on SMD and sensitivity analysis did not change the conclusion, and Egger test did not detect the presence of publication bias (Table 2 and Fig. 3B and C). Eight studies described the proportion of increased ALT in the two groups [23–25,28,33,39,41,43]. The heterogeneity test showed low heterogeneity among these studies (I2=25.1%, P=0.229), and fixed-effect model was used for the meta-analysis. The results demonstrated that survivors had lower proportion of increased ALT than non-survivors (OR=0.49, 95% CI=[0.37,0.64], P<0.001) without publication bias, and sensitivity analysis also did not change the conclusion (Fig. 3D–F).
Forest plot, sensitivity analyses and publication bias assessment of ALT. (A–C) Forest plot (A), sensitivity analyses (B) and Egger test (C) of continuous levels of ALT between survivors and non-survivors. (D–F) Forest plot (D), sensitivity analyses (E) and Egger test (F) of the proportion of abnormally increased ALT between survivors and non-survivors.
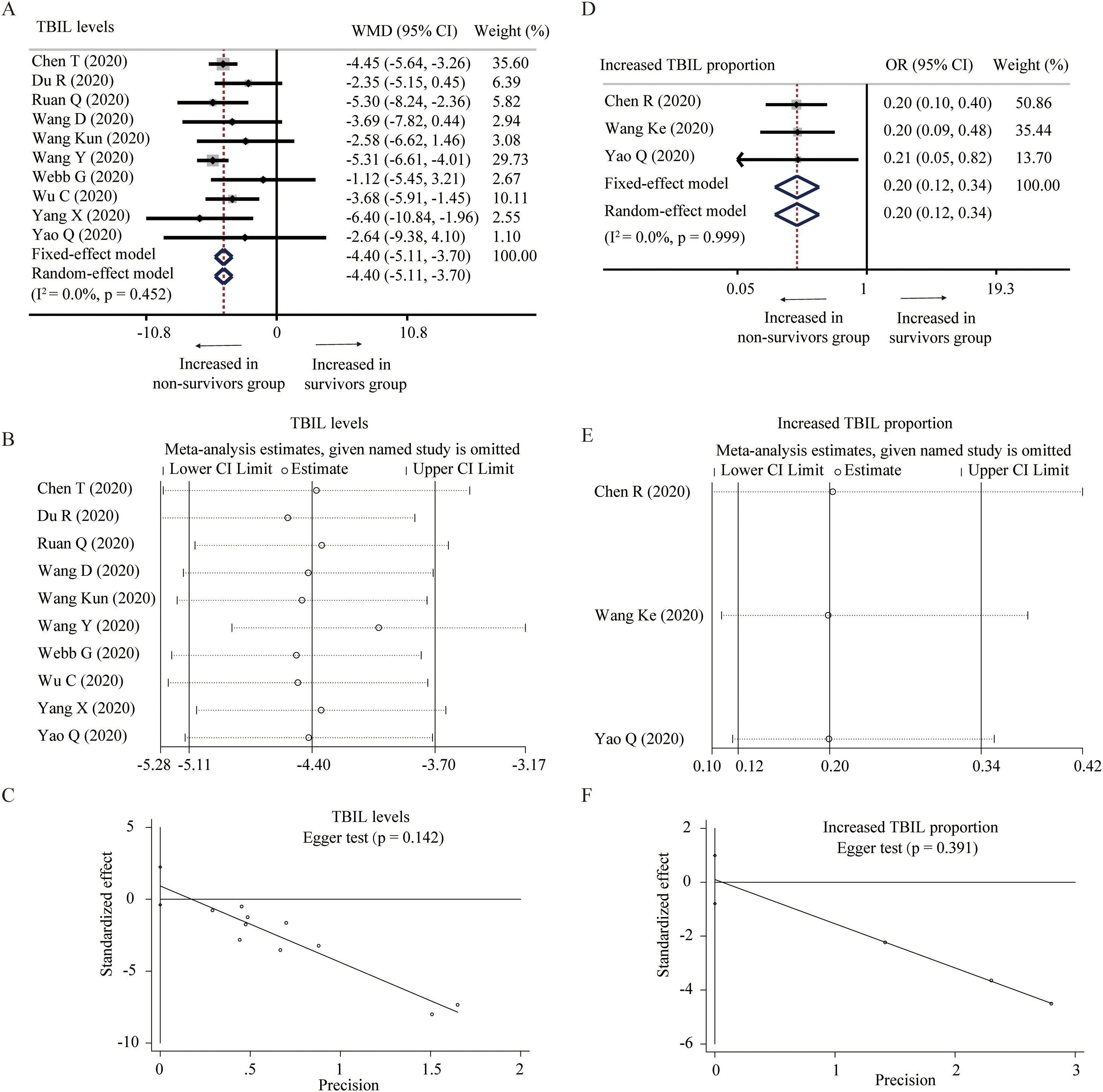
Ten studies described the level of TBIL between survivors and non-survivors group [25,27,31,32,34–36,38,39,45]. No significant heterogeneity was found among these studies and a fixed-model effect was selected (I2=0%, P=0.452). The results showed that survivors had lower level of TBIL than non-survivors without the presence of publication bias (WMD=−4.40μmol/L, 95% CI=[−5.11,−3.70], P<0.001). Fixed-effect model based on SMD, random-effect model, and sensitivity analysis did not change the conclusion (Table 2 and Fig. 4A–C). Additionally, three studies reported the proportion of increased TBIL [23,33,39]. Fixed-effect model showed lower proportion of increased TBIL in survivors than non-survivors (OR=0.20, 95% CI=[0.12,0.34], P<0.001) without detecting the heterogeneity (I2=0%, P=0.999) and publication bias. The conclusion was consistent through sensitivity analysis (Fig. 4D–F).
Forest plot, sensitivity analyses and publication bias assessment of TBIL. (A–C) Forest plot (A), sensitivity analyses (B) and Egger test (C) of continuous levels of TBIL between survivors and non-survivors. (D–F) Forest plot (D), sensitivity analyses (E) and Egger test (F) of the proportion of abnormally increased TBIL between survivors and non-survivors.
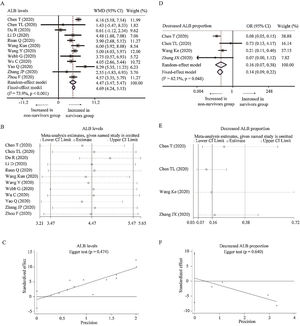
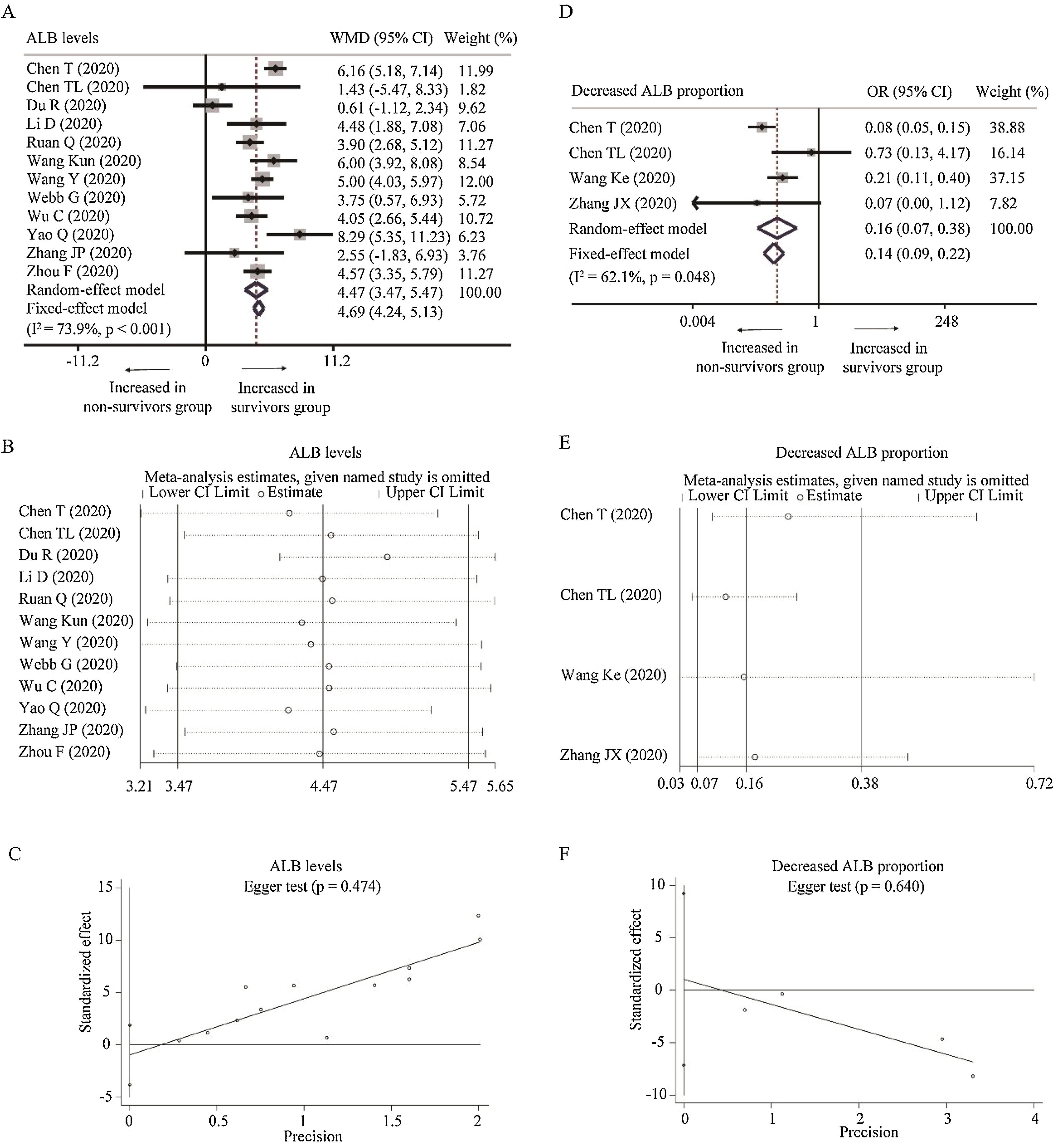
Twelve studies reported the relationship between the level of ALB with COVID-19 mortality [24,25,27,29,31,34–36,39,40,43,45]. With heterogeneity (I2=73.9%, P<0.001), random-effect model was used and suggested that survivors had higher level of ALB than non-survivors (WMD=4.47g/L, 95% CI=[3.47,5.47], P<0.001) with the absence of publication bias. The conclusion was not changed by random-effect model based on SMD or sensitivity analysis (Table 2 and Fig. A.1A–C). What is more, as for the proportion of decreased ALB, random-effect model showed that survivors had lower proportion of decreased ALB than non-survivors (OR=0.16, 95% CI=[0.07, 0.38], P<0.001) with heterogeneity (I2=62.1%, P=0.048). Sensitivity analysis did not change the conclusion, and Egger test did not show the presence of publication bias (Fig. A.1D–F).
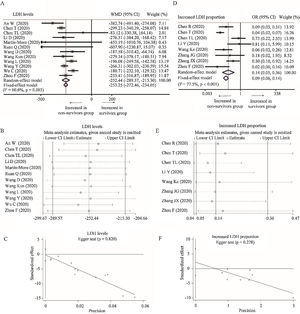
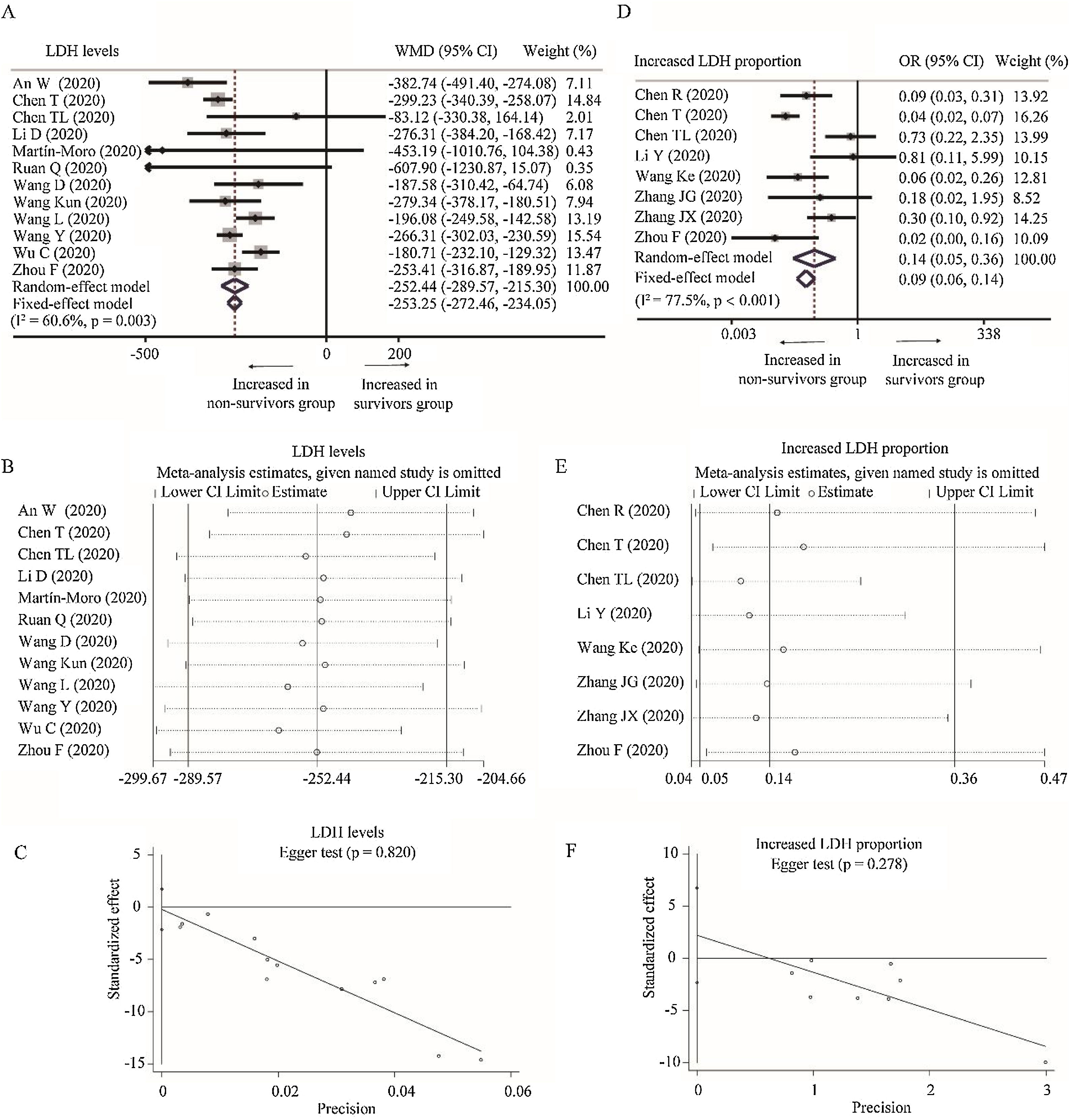
Twelve studies depicted the level of LDH between survivors and non-survivors group [18,19,24,25,29,31,32,34,36,43–45]. Considering the heterogeneity among studies (I2=60.6%, P=0.003), random-effect model was used and suggested that survivors had lower level of LDH than non-survivors (WMD=−252.44U/L, 95% CI=[−289.57,−215.30], P<0.001). The conclusion did not change through random-effect model based on SMD or sensitivity analysis, and no publication bias was detected (Table 2 and Fig. A.2A–C). Besides, eight studies reported the proportion of increased LDH between these two groups [23–25,30,33,41–43]. With heterogeneity (I2=77.5%, P<0.001), random-effect model revealed lower proportion of increased LDH in survivors than non-survivors group (OR=0.09, 95% CI=[0.06,0.14], P<0.001). Sensitivity analysis did not change the conclusion and Egger test did not detect the presence of publication bias (Fig. A.2D–F).
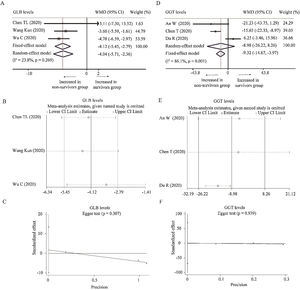
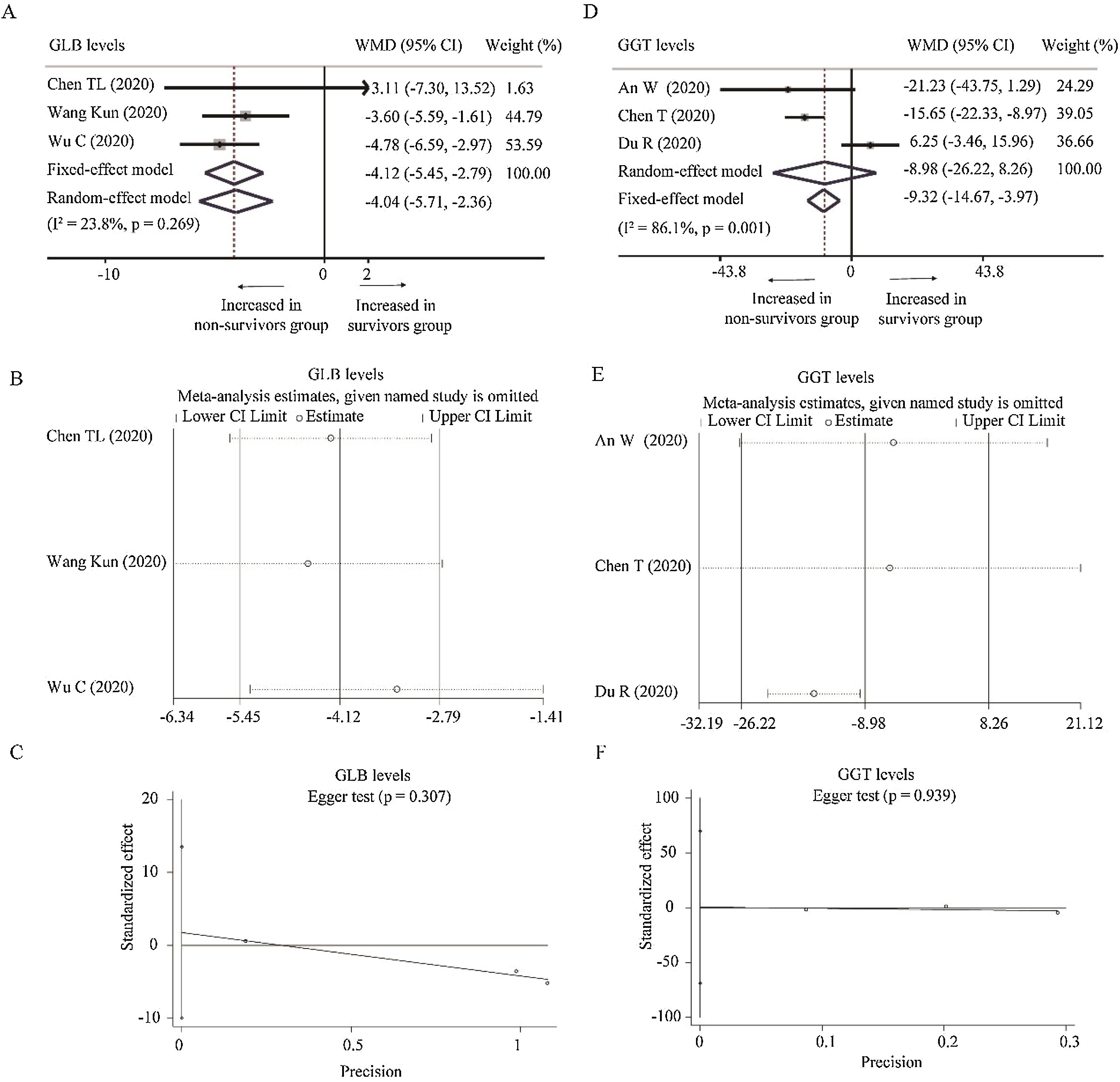
Three studies reported the levels of GLB [24,34,36] and GGT [18,25,27] between survivors and non-survivors group, respectively. With low heterogeneity (I2=23.8%, P=0.269), fixed-effect model showed that survivors had lower level of GLB than non-survivors (WMD=−4.12g/L, 95% CI=[−5.45,−2.79, P<0.001). Random-effect model based on SMD changed the conclusion while the heterogeneity became larger, suggesting that using WMD to analyze the pooled data was more recommended. Moreover, sensitivity analysis did not change the conclusion, and Egger test did not observe the publication bias (Table 2 and Fig. A.3A–C). As for GGT, random-effect model did not find the difference between survivors and non-survivors group (WMD=−8.98U/L, 95% CI=[−26.22,8.26], P=0.310) with great heterogeneity (I2=86.1%, P=0.001). When deleting Du et al.’s study [27], the heterogeneity disappeared (I2=0%, P=0.640) and fixed model changed the results (WMD=−16.10U/L, 95% CI=[−22.51,−9.69], P<0.001). No publication bias was observed among these studies (Fig. A.3D–F).
Additionally, one study on the level of DBIL [27], one study on the proportion of increased DBIL [33], one study on the level of total protein [36], one study on the level of ALP [25], one study on the level of A/G [34], one study on the level of prealbumin [36] and two study on the proportion of increased GLB [24,33], were not included in our meta-analysis due to their inadequate data.
4DiscussionCOVID-19 is rapidly expanding around the world while there is still no specific medication until now [24]. To reduce the COVID-19-related death, numerous studies focus on the risk factor for COVID-19 mortality. Liver biochemical parameters such as AST and LDH have been identified as predictors for the prognosis of COVID-19 patients in some studies [1,46], while their prognostic values in other studies are not statistically significant [47,48]. A comprehensive evaluation of the association between liver biochemical parameters and COVID-19 mortality is needed to identify those COVID-19 patients at high risk and finally to prevent fatal outcomes.
In the systematic review and meta-analysis, 25 studies including 5971 COVID-19 patients were enrolled in our analysis. We found that patients in survivors group had lower levels of AST, ALT, TBIL, LDH, GLB and GGT, and lower proportion of these abnormally increased parameters, compared with those in non-survivors group. Moreover, survivors had higher level of ALB and lower proportion of abnormally decreased ALB than non-survivors. Sensitivity analyses indicated that the results were not influenced by excluding any one specific study except GGT between survivors and non-survivors groups. Furthermore, meta-analysis based on SMD did not change the conclusion except GLB. Notably, Egger test did not detect the presence of publication bias in all of these comparisons. These findings suggested that patients in non-survivors group were more likely to suffer from liver damage than those in survivors group, and liver biochemical parameters could be used to predict the COVID-19 mortality.
COVID-19-related liver damage could be traced back to the early report about 99 cases in Wuhan, China. Chen and his colleagues found that 28% patients had abnormally elevated ALT, 35% patients had abnormally elevated AST, and 18% patients had abnormally elevated TBIL. More seriously, one patient had severely increased liver damage (ALT 7590U/L, AST 1445U/L) [49]. Moreover, Guan et al. revealed that 22.2%, 21.3% and 10.5% patients had abnormally elevated AST, ALT and TBIL, separately. Also, compared with non-severe patients and patients without presence of composite primary end point, severe patients and patients with presence of composite primary end point had higher proportion of abnormally increased AST, ALT and TBIL [50]. Furthermore, Chen et al. suggested that abnormally elevated AST could be used to predict the overall survival of COVID-19 through multivariate cox regression analysis [1]. Besides, Ji and his colleague demonstrated that the risk of severe COVID-19 was increasing with LDH level going up [51]. To conclude, our findings further validated the above reports and highlighted the prognostic value of liver biochemical parameters for COVID-19 mortality.
There still exists debates on the mechanism of COVID-19-related liver damage. One possible mechanism is that SARS-CoV-2 could directly cause liver damage. Recent studies revealed that TMPRSS2 could cleave and activate the spike protein of SARS-CoV-2 and allow the virus to release fusion peptides for membrane fusion, suggesting that co-expression of ACE2 and TMPRSS2 might be important for the entry of SARS-CoV-2 to host cells [7]. Interestingly, the expressions of ACE2 and TMPRSS2 have been detected in cholangiocytes and hepatocytes, which provide a possibility that SARS-CoV-2 directly causes liver damage [8]. Moreover, antipyretic agents including acetaminophen were used to treat COVID-19 patients, and these drugs could cause significant liver damage and even liver failure [52]. Some antiviral drugs such as oseltamivir and lopinavir also had some hepatotoxic effects. Besides, virus infection could induce multiple proinflammatory signals through Toll-like receptor and activation of killer T lymphocytes. The necrosis and apoptosis of host cell caused by T lymphocytes could further amplify the inflammatory signals [53]. Inflammatory storm or systemic inflammatory response syndrome caused by SAR-CoV-2 infection was strongly associated with multiple organ injuries including liver damage. Additionally, COVID-19-related complications such as respiratory distress syndrome and multiple organ failure could lead to hypoxia and shock, causing hepatic ischemia and hypoxia-reperfusion dysfunction [54]. In summary, COVID-19 could directly and indirectly cause liver damage. Our systematic review and meta-analysis based on all the published paper validated the existence of liver damage, providing a solid basis on the mechanism research of liver damage.
Admittedly, our study had some limitations. Firstly, previous studies have identified some clinical risk factors for the poor prognosis of COVID-19, including older age, male sex, BMI, and the presence of comorbidities [55–57]. It is possible that our conclusions were influenced by these confounding factors due to an inherent limitation of all meta-analyses of observational data. Second, we could not exclude the impact of treatment on these outcomes because some patients might receive hepatotoxic drugs. Third, there exists noticeable heterogeneity in most of the comparisons. Though different effect models, sensitivity analysis and pooled data based on SMD were used for the meta-analysis, yet the heterogeneity could not be eliminated completely. Besides, due to insufficient data of DBIL, total protein, ALP, A/G, we could not evaluate their prognostic value for COVID-19 mortality. Also, we could not determine the cutoff of these liver biochemical parameters to stratify high-risk and low-risk COVID-19 patients owing to limited data. Finally, it is underpowered to investigate the underlying mechanism of COVID-19-related liver damage.
5ConclusionsIn this study, liver biochemical parameters were strongly correlated with COVID-19 mortality. Non-survivors had higher levels of AST, ALT, TBIL, LDH, GGT and GLB, higher proportion of these abnormally increased parameters, lower ALB level and lower proportion of abnormally decreased ALB than survivors. Measurement of these liver biochemical parameters might assist clinicians to evaluate the prognosis of COVID-19.
Author's contributionFZ, YD, and GD were responsible for study design, literature screening and manuscript writing. LY, YD, YW, and YY collected the data. FZ, YD, and GD analyzed the data. JZ, YD, and BC revised the manuscript. All authors edited and approved the final version of the manuscript.
FundingThis study was funded by grants from the China Postdoctoral Science Foundation (No. 2020M682594).
Conflict of interestThe authors declare that they have no conflict of interest.
Forest plot, sensitivity analyses and publication bias assessment of ALB. (A–C) Forest plot (A), sensitivity analyses (B) and Egger test (C) of continuous levels of ALB between survivors and non-survivors. (D–F) Forest plot (D), sensitivity analyses (E) and Egger test (F) of the proportion of abnormally decreased ALB between survivors and non-survivors.
Forest plot, sensitivity analyses and publication bias assessment of LDH. (A–C) Forest plot (A), sensitivity analyses (B) and Egger test (C) of continuous levels of LDH between survivors and non-survivors. (D–F) Forest plot (D), sensitivity analyses (E) and Egger test (F) of the proportion of abnormally increased LDH between survivors and non-survivors.
Forest plot, sensitivity analyses and publication bias assessment of GLB and GGT. (A–C) Forest plot (A), sensitivity analysis (B) and Egger test (C) of continuous levels of GLB between survivors and non-survivors. (D–F) Forest plot (D), sensitivity analysis (E) and Egger test (C) of continuous levels of GGT between survivors and non-survivors.
Methodological quality of enrolled studies based on Newcastle–Ottawa Scale (NOS).
| Included studies | Year | Is the definition adequate? | Representativeness of the cases | Selection of controls | Definition of controls | Comparability of both groups | Ascertainment of diagnosis | Same ascertainment method for both groups | Nonresponse rate | Total scores |
|---|---|---|---|---|---|---|---|---|---|---|
| An W | 2020 | 8 | ||||||||
| Chen R | 2020 | – | 7 | |||||||
| Chen T | 2020 | – | 7 | |||||||
| Chen TL | 2020 | – | 7 | |||||||
| Deng Y | 2020 | – | 7 | |||||||
| Du R | 2020 | 8 | ||||||||
| He X | 2020 | 9 | ||||||||
| Li D | 2020 | 8 | ||||||||
| Li Y | 2020 | – | 7 | |||||||
| Martín-Moro | 2020 | 9 | ||||||||
| Ruan Q | 2020 | 8 | ||||||||
| Wang D | 2020 | – | 7 | |||||||
| Wang Ke | 2020 | – | 7 | |||||||
| Wang Kun | 2020 | 8 | ||||||||
| Wang L | 2020 | – | 7 | |||||||
| Wang Y | 2020 | – | 7 | |||||||
| Webb G | 2020 | 9 | ||||||||
| Wu C | 2020 | – | 7 | |||||||
| Xu B | 2020 | 8 | ||||||||
| Yang X | 2020 | 8 | ||||||||
| Yao Q | 2020 | 8 | ||||||||
| Zhang JG | 2020 | 8 | ||||||||
| Zhang JP | 2020 | 8 | ||||||||
| Zhang JX | 2020 | – | 7 | |||||||
| Zhou F | 2020 | 8 |








 AST. (A–C) Forest plot (A), sensitivity analyses (B) and Egger test (C) of continuous levels of
AST. (A–C) Forest plot (A), sensitivity analyses (B) and Egger test (C) of continuous levels of 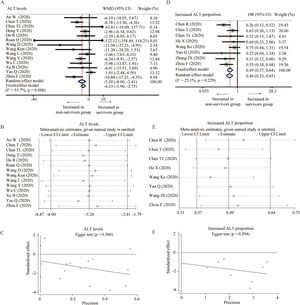 ALT. (A–C) Forest plot (A), sensitivity analyses (B) and Egger test (C) of continuous levels of
ALT. (A–C) Forest plot (A), sensitivity analyses (B) and Egger test (C) of continuous levels of 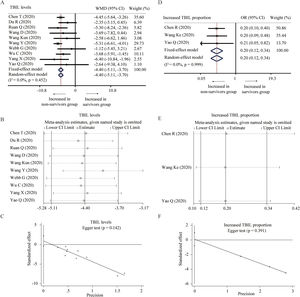 TBIL. (A–C) Forest plot (A), sensitivity analyses (B) and Egger test (C) of continuous levels of
TBIL. (A–C) Forest plot (A), sensitivity analyses (B) and Egger test (C) of continuous levels of