Introduction and aim. HAVCR1 protein is the cellular receptor for hepatitis A virus (HAV). Genetic polymorphism in this gene may alter the outcome of HAV infection. In a previous study, a 6-amino acid insertion (157insMTTTVP) in HAVCR1 gene was associated with more severe disease. We decided to investigate this association further.
Material and methods. We sequenced exon 4 of the HAVCR1 gene in patients with clinical hepatitis A attending our institution, and a group of healthy controls in a disease-endemic setting in India. Frequencies of different haplotypes of a genomic region with two overlapping insertion-deletion polymorphisms (indels; rs141023871 and rs139041445) were compared between patients and controls, as well as between patients with and without a severe form of disease (liver failure).
Results. The gene had three haplotypes in the region of interest - a short form, an intermediate-form with a 5-amino acid 157insMTTVP insertion and a long-form with a 6-amino acid 157insMTTTVP insertion. The allele frequency (29/150 [19%] vs. 43/146 [29%]; p = ns) and haplotype frequency (29/75 [39%] vs. 39/73 [53%]; p = ns) of the 157insMTTTVP variant were similar in hepatitis A patients and healthy controls (30%). Further, the allele frequency (12/58 [21%] vs. 17/92 [18%]; p = ns) and haplotype frequency (12/29 [41%] vs.17/46 [37%]; p = ns) of the longest variant were also similar in patients with severe and mild disease.
Discussion. In the study population, the 157insMTTTVP variant of HAVCR1 gene was not associated with more severe outcome of HAV infection. Further studies in other populations around the world are needed to assess the relation of this genetic variation with disease outcome.
Hepatitis A virus (HAV) is a hepatotropic virus with 27-nm sized virions that contain a 7.5-Kb long, single-stranded RNA genome of positive polarity.1 It is excreted in the feces of infected persons and is transmitted by the enteric route. The infection is distributed worldwide, though the risk of transmission varies widely across countries, depending on socioeconomic status.2 The severity of clinical disease following HAV infection depends heavily on the age at infection.3 Thus, infection during early childhood is most often asymptomatic. By contrast, infection in older children or adults more often leads to icteric illness, and to fulminant liver failure and death. Infection provides life-long immunity against reinfection.
The association of risk of HAV infection with hygiene and sanitation, the dependence of disease severity on age at infection, and the induction of lifelong immunity following HAV infection, result in varying epidemiologic patterns of this disease in different geographical areas.2 Thus, in low-income countries, HAV infection is highly endemic with a high transmission rate, leading to nearly universal exposure during early childhood and a high seropositivity rate by adolescence, albeit with little disease. By contrast, in high-income countries with low endemicity, HAV transmission is infrequent, leading to low rates of seroprevalence as well as disease. A third pattern, with intermediate endemicity, is seen in developing countries that are transitioning to better hygiene; falling transmission in these areas has led to an increase in average age at exposure to HAV, with a paradoxical increase in disease rates and mortality.
Hepatitis A virus cellular receptor 1 (HAVCR1), a protein expressed on human hepatocyte membrane, has been identified as the cellular receptor for HAV.4 This protein, also known as T-cell immunoglobulin and mucin domain 1 (TIM-1) or kidney injury molecule 1 (KIM-1) protein, has additional physiological roles, including in the induction of cellular immunity.5
The gene for HAVCR1 is located on chromosome 5q33.3, and shows marked genetic polymorphism.6 Being the receptor for HAV, variations in it have been proposed to affect an individual’s susceptibility to HAV infection. Recently, in Argentina, a 6-amino acid insertion at residue 157 of the HAVCR1 protein (termed 157insMTTTVP) was shown to be associated with an increased risk of severe hepatitis A.7 This association is believed to be mediated by a more efficient binding of HAV by the longer HAVCR1 variant on hepatocytes, and by a higher cytotoxicity against HAV-infected cells of natural killer T cells that carry this variant HAVCR1 protein.7
However, in the Argentine study, the statistical measure of association between severe hepatitis A disease and presence of HAVCR1 157insMTTTVP polymorphism was marginal. Thus, there is a need for further studies on this relationship, particularly from other geographical regions with different epidemiologic patterns of this disease. We, therefore, undertook such a study in India, where HAV infection is highly endemic.
Material and MethodsSubjectsBetween May 2013 and July 2015, we enrolled children with acute viral hepatitis A (AVH-A) attending the outpatient or inpatient services of the Department of Pediatric Gastroenterology at our institution. The diagnosis of AVH-A was based on suggestive clinical picture, elevated serum transaminase levels (> 3-fold the upper limit normal) and positive IgM anti-HAV [Mini Vidas; bioMérieux, Marcy l’Etoile, France]). Persons with clinical or laboratory findings suggesting another concomitant disease or chronic liver disease were excluded.
Patients were examined for the presence of hepatic encephalopathy (HE). HE, if present, was graded using the standard criteria in children aged > 3 years.8 For younger children, a set of modified criteria, which have been recommended for this age group, were used.9 In addition, prothrombin time-international normalized ratio (PT-INR) was measured after parenteral administration of one dose of vitamin K. Children with AVH-A who fulfilled the criteria laid down by the Pediatric Acute Liver Failure Study Group,9 i.e. if HE was present and PT-INR was > 1.5, or if the PT-INR was > 2.0 even in the absence of HE, were classified as having acute liver failure (ALF-A); the remaining children were considered to have uncomplicated AVH-A.
In addition, left-over blood specimens from a group of healthy adult blood donors were used as controls.
HAVCR-1 genotypingGenomic DNA was extracted from whole blood collected in EDTA using the phenol-chloroform method, and its purity and concentration were determined using Nanodrop spectrophotometer 1000 (Thermo Fisher Scientific, Waltham, MA, USA). A 441-basepair region of HAVCR-1 gene was amplified using specific primers (forward: 5’-TCT GTC ATT GGT GTG CTA GGG-3’ and reverse: 5’-TGG TCC GCA GCT CCT CAT TA-3’; Figure 1). Each PCR reaction (20 µL) contained 40 ng of DNA template, 1X Invitrogen buffer, 1.5 mM MgCl2, 0.2 µM dNTPs, 5 pmol of each primer and 2 units of TaqDNA polymerase (Invitrogen, Waltham, MA, USA). Cycling conditions were: denaturation at 94 °C for 5 min, 36 cycles, each consisting of denaturation at 94 °C for 30 sec, annealing at 63 °C for 30 sec and extension at 72 °C for 90 sec, and a final extension at 72 °C for 7 min. The amplicons were purified using PureLink PCR purification kit (Invitrogen, Thermo Fisher), and sequenced in both directions using the Big Dye Terminator v3.1 (Life Technologies, Thermo Fisher) chemistry and an ABI 3130 genetic analyzer.
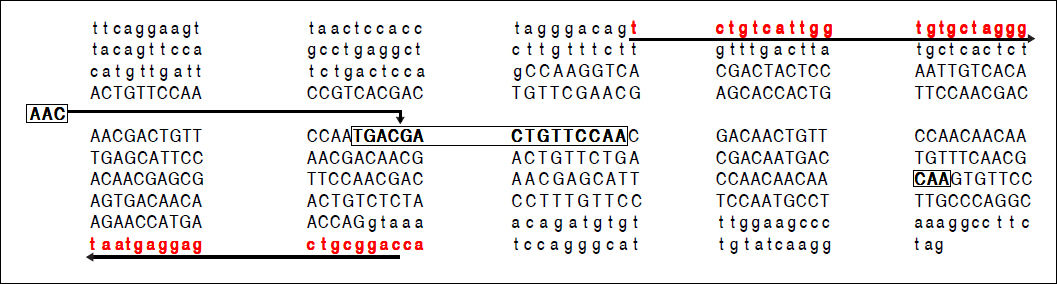
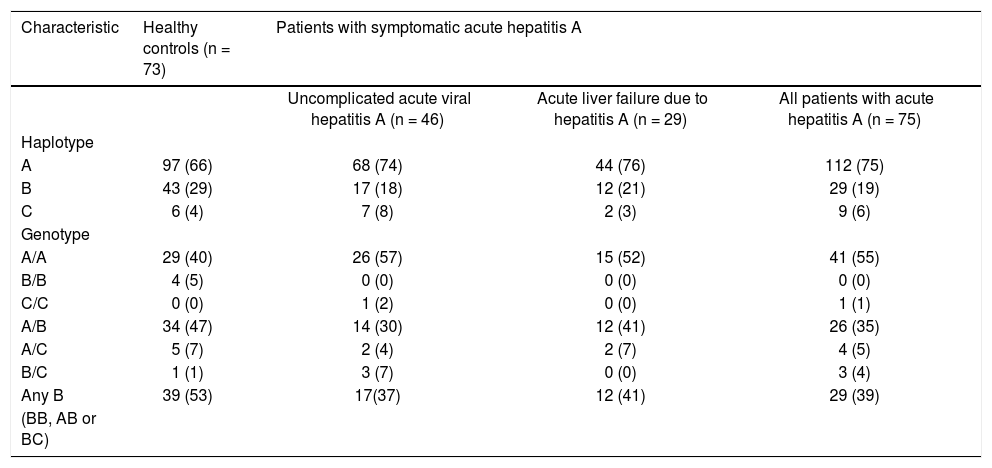
A part of the reference sequence (NG_017001.1; GenBank database) of HAVCR1 gene showing exon 4 (upper case letters) with parts of surrounding introns (lower case letters). The locations of primers used for amplification of the exon 4 are shown using solid underlined arrows. The large box in the sequence shows a 15-base (TGA CGA CTG TTC CAA) ins/del polymorphism (rs141023871). Within this region is located a 3-base (AAC) ins/del polymorphism (rs139041445) shown in the margin. The smaller box in the main sequence shows a 3-base (CAA) ins/del polymorphism (rs45439103).
The sequencing electropherograms were analyzed using FinchTV (http://www.geospiza.com/Products/finchtv. shtml). The sequences were further analyzed by aligning these to the reference genomic sequence of HAVCR1 gene (NG_017001.1; NCBI nucleotide database), using BioEdit software version 7 (www.mbio.ncsu.edu/bioedit/bioedit. html), to identify nucleotide substitutions and insertion/ deletions (indels).10 In particular, two previously-reported, overlapping indels in exon 4 of the HAVCR1 gene (Figure 1), namely p.Met158_Pro162del (rs141023871) and p.Thr160_Val161insThr (rs139041445), were mapped for each subject. The sequences were also analyzed for the presence of another 3-nucleotide indel (p.Thr200del; rs45439103). Frequencies of various alleles among controls were tested for Hardy-Weinberg equilibrium. The allele and genotype frequencies were compared between groups using the χ2 test.
The study was approved by our institution’s Ethics Committee. Informed consent was obtained from each study subject or from one of the parents.
ResultsStudy subjectsA total of 75 children with AVH-A were enrolled, of whom 46 had uncomplicated AVH-A and 29 had ALF-A. Specimens from 73 blood donors were studied as controls. Clinical and laboratory details of the study patients are shown in table 1.
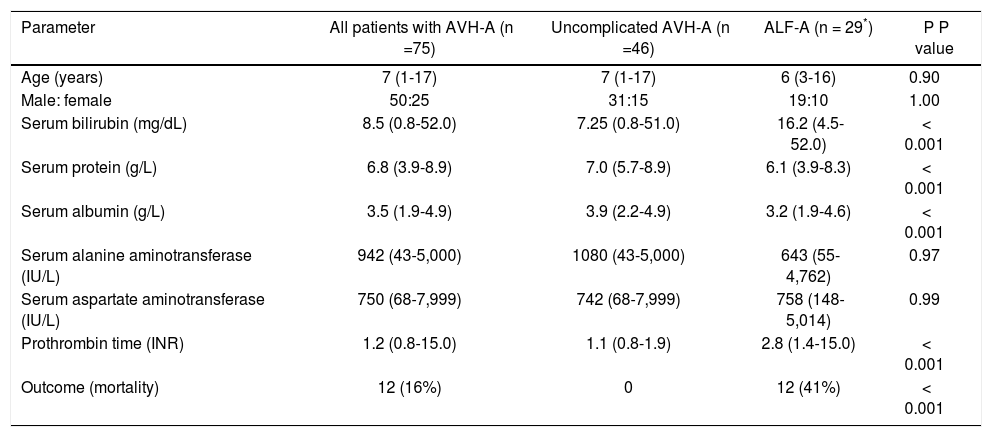
Clinical and laboratory characteristics of study subjects.
| Parameter | All patients with AVH-A (n =75) | Uncomplicated AVH-A (n =46) | ALF-A (n = 29*) | P P value |
|---|---|---|---|---|
| Age (years) | 7 (1-17) | 7 (1-17) | 6 (3-16) | 0.90 |
| Male: female | 50:25 | 31:15 | 19:10 | 1.00 |
| Serum bilirubin (mg/dL) | 8.5 (0.8-52.0) | 7.25 (0.8-51.0) | 16.2 (4.5-52.0) | < 0.001 |
| Serum protein (g/L) | 6.8 (3.9-8.9) | 7.0 (5.7-8.9) | 6.1 (3.9-8.3) | < 0.001 |
| Serum albumin (g/L) | 3.5 (1.9-4.9) | 3.9 (2.2-4.9) | 3.2 (1.9-4.6) | < 0.001 |
| Serum alanine aminotransferase (IU/L) | 942 (43-5,000) | 1080 (43-5,000) | 643 (55-4,762) | 0.97 |
| Serum aspartate aminotransferase (IU/L) | 750 (68-7,999) | 742 (68-7,999) | 758 (148-5,014) | 0.99 |
| Prothrombin time (INR) | 1.2 (0.8-15.0) | 1.1 (0.8-1.9) | 2.8 (1.4-15.0) | < 0.001 |
| Outcome (mortality) | 12 (16%) | 0 | 12 (41%) | < 0.001 |
The data are shown as median (range) or number (percent). AVH-A: acute viral hepatitis A (uncomplicated). ALF-A: acute liver failure due to hepatitis A. INR: international normalized ratio.
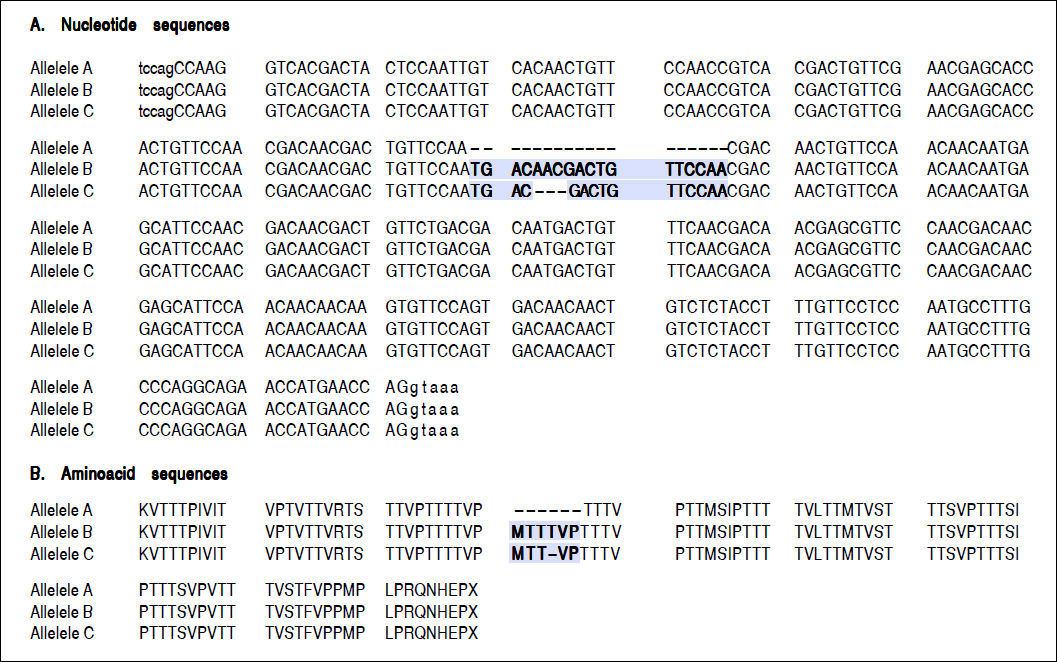
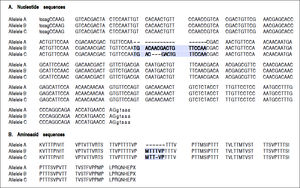
For the region of interest with two overlapping indels, three haplotypes were observed (Figures 2 and 3). These three haplotypes, in the decreasing order of their frequencies, were:
- •
rs141023871_del and rs139041445_del (the shortest variant).
- •
rs141023871_ins and rs139041445_ins (the longest variant), with a 18 nucleotide insertion (corresponding to a 6-amino acid insertion - 157insMTTTVP).
- •
rs141023871_ins and rs139041445_del (corresponding to NCBI reference sequence, NG_017001.1), with a 15-nucleotide insertion (5 amino acid – 157insMT-TVP) as compared to the shortest variant.
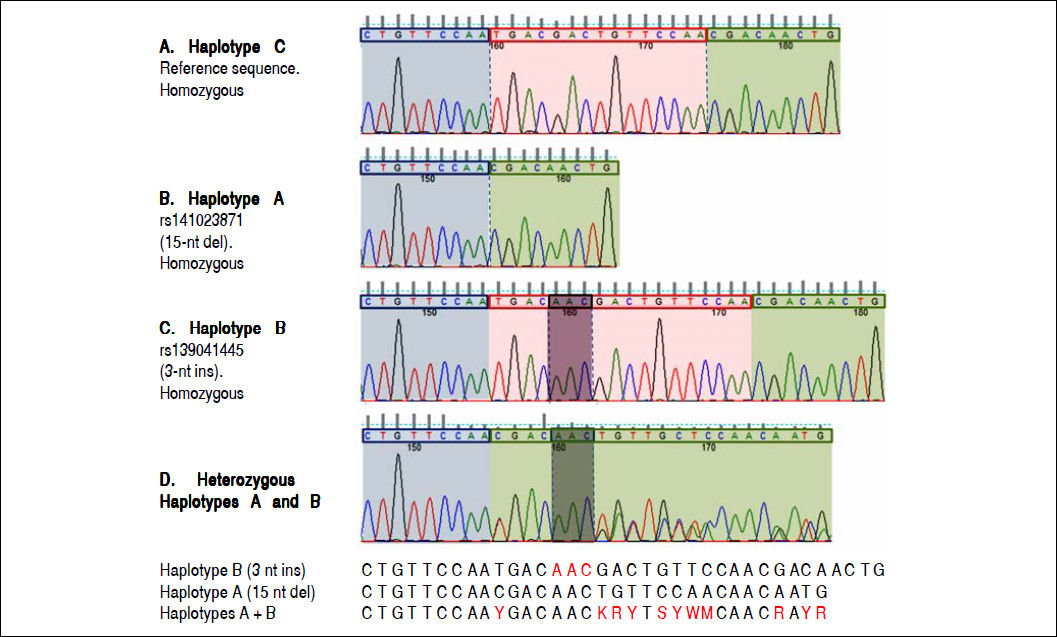
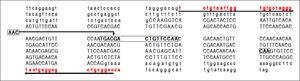
Electropherograms showing the results of sequencing for the region of interest (the region of 157insMTTTVP) in exon 4 of the HAVCR1 gene in persons who were homozygous for the three haplotypes (Panel A: homozygous for reference sequence [haplotype C]; Panel B: homozygous for a 15-nucleotide (nt) deletion [haplotype A]; Panel C: homozygous for a 3-nt insertion [haplotype B]). Panel D shows the electropherogram for a subject who was heterozygous for haplotypes A and B; the panel below this electropherogram shows the sequences for the two haplotypes and the expected merged sequence (the ambiguous codes are represented as per the notation laid down by the International Union of Pure and Applied Chemistry).
These haplotypes are referred to subsequently as haplotypes A, B and C, respectively.
The allele frequencies of the three haplotypes in patients with symptomatic AVH-A were similar to those in the control subjects (Table 2). In particular, the allele frequencies of allele B, the allele of interest, were similar in patients and controls (29/150 [19%] vs. 43/146 [29%]). Also, the combined frequencies of genotypes with 157insMTT-TVP on either one or both alleles (BB, AB or BC genotype) were similar in patients and controls (29/75 [39%] vs. 39/73 [53%]).
Genotype and haplotype frequencies in different subjects groups.
| Characteristic | Healthy controls (n = 73) | Patients with symptomatic acute hepatitis A | ||
|---|---|---|---|---|
| Uncomplicated acute viral hepatitis A (n = 46) | Acute liver failure due to hepatitis A (n = 29) | All patients with acute hepatitis A (n = 75) | ||
| Haplotype | ||||
| A | 97 (66) | 68 (74) | 44 (76) | 112 (75) |
| B | 43 (29) | 17 (18) | 12 (21) | 29 (19) |
| C | 6 (4) | 7 (8) | 2 (3) | 9 (6) |
| Genotype | ||||
| A/A | 29 (40) | 26 (57) | 15 (52) | 41 (55) |
| B/B | 4 (5) | 0 (0) | 0 (0) | 0 (0) |
| C/C | 0 (0) | 1 (2) | 0 (0) | 1 (1) |
| A/B | 34 (47) | 14 (30) | 12 (41) | 26 (35) |
| A/C | 5 (7) | 2 (4) | 2 (7) | 4 (5) |
| B/C | 1 (1) | 3 (7) | 0 (0) | 3 (4) |
| Any B | 39 (53) | 17(37) | 12 (41) | 29 (39) |
| (BB, AB or BC) | ||||
The data are shown as number (percent). A, B and C represent three alternative haplotypes as follows: A: rs141023871 Del/rs139041445 Del (15 bases shorter than the reference sequence, NG_017001.1) – corresponds to the absence of 157insMTTTVP. B: rs141023871Ins/rs139041445Ins (3 bases longer than the reference sequences) - corresponds to a 6-amino acid 157insMTTTVP insertion. C: rs141023871Ins / rs139041445 Del (reference sequence) – corresponds to a 5-amino acid 157insMTTVP insertion.
Furthermore, the patients with ALF-A and those with uncomplicated AVH-A had similar allele frequencies of allele B (12/58 [21%] vs. 17/92 [18%]; p = ns) and composite frequencies of genotypes that contain allele B (12/29 [41%] vs. 17/46 [37%]; p = ns).
rs45439103 polymorphismThis 3-nucletide deletion polymorphism was strongly associated with the rs139041445 polymorphism, i.e. it was observed only in persons with the 157insMTTTVP insertion allele (allele B) at the latter locus. The allelic frequency of this deletion was similar in patients with hepatitis A (19/150 [13%]) and the control subjects (32/146 [22%]; p = ns).
DiscussionIn this study, we assessed the distribution of different haplotypes of a part of exon 4 of the HAVCR1 gene, which included two overlapping indel polymorphisms, among patients with AVH-A as compared to healthy controls in the Indian population, as also between patients with AVH-A of different severities. It revealed that allele and genotype frequencies of the haplotype with a 6-amino acid MTTTVP insertion, previously implicated in the causation of severe hepatitis A, was comparable among patients with AVH-A and healthy controls, as well as among patients with severe and mild forms of AVH-A.
The HAVCR1 protein belongs to the T-cell immunoglobulin mucin (TIM) protein family, which contains several type I cell-surface transmembrane glycoproteins.5,11 It contains an immunoglobulin-like domain, a heavily-glycosylated mucin domain, a single transmembrane domain and a short C-terminal cytoplasmic tail. It is initially expressed by all CD4+ T cells; however, during differentiation, its expression on TH1 and TH17 cells declines whereas that on TH2 cells remains high.
HAVCR1 is expressed on hepatocyte membrane and serves as the cellular receptor for HAV.12–14 In particular, its mucin-like domain plays a role in the uncoating and internalization of the virus. It also appears to serve as the cellular receptor for Ebolavirus, flaviviruses, alphaviruses and arenaviruses.15–17
The HAVCR1 protein, in particular exon 4, is highly polymorphic.6 The 157insMTTTVP polymorphism, located in this exon,18 is located in the extracellular mucin-like region of the protein, which is important for uncoating of HAV before its entry into the cell.19 An insertion at this site may be expected to affect the binding and internalization of HAV into the hepatocytes, and hence influence the clinical outcome of HAV infection.
The association of 157insMTTTVP polymorphism with human disease was first studied in atopic conditions in which HAVCR1 gene was suspected to play a role. This was done using a cross-sectional study, in which 375 individuals were evaluated for history of atopy, serologic evidence of HAV infection and the presence of this insertion.18 In this study, HAV seropositivity was found to protect against atopy in individuals with the 157insMTTTVP variant, but not in those without this insertion. This suggested the existence of an interaction between HAV infection and HAVCR1 genotype that contributed to the occurrence of atopic diseases, and provided a possible basis for the hygiene hypothesis. This interaction was explained through a hypothesis that immune cells with the longer variant of HAVCR1 protein bind HAV better; thus, HAV infection in persons with this variant could alter the Th1/Th2 balance and thus protect against atopy. However, subsequent studies on this issue have shown conflicting results, with an association being observed in some population groups20,21 but not in some others.22,23 Interestingly, the relationship between the insertion and atopy has varied even within studies done in the Chinese population.21,23 In addition, several studies have also looked at the association of this polymorphism with autoimmune disorders with conflicting results. Thus, while two single-nucleotide polymorphisms in this gene were associated with susceptibility to rheumatoid arthritis in one study,24 no association of polymorphisms in this gene was found with systemic lupus erythematosus in another study.25 In Thailand, persons with HAVCR1 exon 4 haplotypes with lower expression levels of TIM1 were found to have a delayed progression of human immunodeficiency virus infection to acquired immunodeficiency syndrome.26
If a genetic variation in the HAVCR1 protein affects its binding to HAV, one may expect it to affect the risk and outcome of HAV infection. This was examined by Kim, et al., who looked for the presence of 157insMTTTVP allele in 30 Argentine patients with ALF-A and 102 controls who had HAV antibodies but no history of liver disease7 and found the insertion allele to be significantly associated (22/60 [37%] vs. 57/204 [28%]; p = 0.037) with HAV-induced liver disease. They further showed that the variant HAVCR1 protein with this 6-amino acid insertion had more efficient in vitro binding to HAV than the protein without this insertion. Furthermore, human natural killer T cells that expressed the longer HAVCR1 protein showed greater cytotoxic activity against HAV-infected liver cells than those expressing its shorter form. These findings appeared to provide a pathogenetic explanation for the greater disease severity among persons with the MTTTVP insertion haplotype. However, though the frequency of the insertion allele in that study was significantly higher in persons with disease than in controls, the absolute difference (37% vs. 28%) was small. We may add here that our study had a much larger number of cases with AVH-A (n = 75) than this the previous study (n = 30). No other study has yet assessed the relationship of the 157insMTTTVP insertion with the severity of hepatitis A.
By contrast, in our study, frequency of the 157insMTT-TVP insertion in patients with AVH-A was similar to that in healthy controls. If anything, the frequency of this insertion was somewhat lower in our patient group than in the control group, as opposed to the higher frequency in patients in the previous study. This suggests that this polymorphism did not have a role in determining the severity of liver injury in our population. Further, we also compared the frequency of this allele in patients with ALF-A and those with uncomplicated AVH-A, and found these to be similar; the previous study did not do such a comparison. Thus, our findings indicate that this polymorphism may not have a role in determining the severity of HAV disease in at least some settings. This is somewhat similar to the association between this insertion and the occurrence of atopy, which has also been found in some populations but not in the others.
What could be the possible reasons for the differences between the results obtained in our and the Argentine studies? First, the two populations have different ethnic origins. Despite the similar frequencies of the HAVCR1 157insMTTTVP haplotype in the two populations, being 28% in the Argentine study and 30% in our population, the two populations would be expected to have several other genetic differences, which could have influenced the relationship between the presence of the 157insMTTTVP insertion and disease severity.
Second, the HAV strains prevalent in India and Argentina could be different. Thus, differences in amino acid sequences, and structures of proteins in viral strains prevalent in the two regions could influence the association of severity of HAV disease with HAVCR1 alleles in these sites. We are unable to comment on this since we did not assess the viral characteristics. However, all HAV genotypes are known to share one highly conserved antigenic neutralization site and have a single serotype with cross-protection across genotypes,27 making this explanation less likely.
It may be pertinent to point out a limitation of our study. Subjects enrolled as controls were not tested for IgG anti-HAV, a marker of prior HAV infection. However, previously published data28 and our unpublished experience show that > 95% of adult blood donors in this area are positive for this antibody.
Overall, our study indicates that there may be an association between genomic variations in the HAVCR1 gene and severity of disease associated with HAV infection in some settings but not in others. This suggests that further studies in various populations are needed to confirm or refute this association, and to determine the factors that influence the presence or absence of a relationship. These studies should also help us better understand the factors which influence the severity of hepatitis A disease, and possibly methods to prevent and treat such severe disease.
Abbreviations- •
ALF-A: acute liver failure due to hepatitis A.
- •
AVH-A: acute viral hepatitis A.
- •
HAV: hepatitis A virus.
- •
HAVCR1: Hepatitis A virus cellular receptor 1.
- •
HE: hepatic encephalopathy.
- •
INR: international normalized ratio.
- •
KIM-1: kidney injury molecule 1.
- •
PT-INR: prothrombin time-international normalized ratio.
- •
TIM: T-cell immunoglobulin mucin.
- •
TIM-1: T-cell immunoglobulin and mucin domain 1.
None of the authors has any conflict of interest.
Research Involving Human Participants and/or AnimalsThe study was approved by the Ethics Committee of Sanjay Gandhi Postgraduate Institute of Medical Sciences, Lucknow, India. Funding information is not applicable.
Informed ConsentInformed consent was obtained from each study subject or from one of the parents.
Author ContributionsMercilena Benjamin: Study design and laboratory work. Shikha Agnihotry: Analysis of sequencing data. Anshu Srivastava: Finalization of study design, and clinical work-up of subjects. Rishi Bolia: Enrolment and clinical work-up of subjects: SK Yachha: Clinical work-up of subjects. Rakesh Aggarwal: Conception and design of the study, conduct, and supervision of laboratory work, analysis of data including those related to sequence data, writing the manuscript.
In addition, all the authors participated in critical revision of the manuscript drafts and approved the final version of the manuscript. RA takes overall responsibility for the work.
AcknowledgmentsThe Bioinformatics Center at Sanjay Gandhi Postgraduate Institute of Medical Sciences, Lucknow is funded by Indian Council of Medical Research (ICMR), New Delhi. Mercilena Benjamin and Shikha Agnihotry were funded by ICMR during this work.





![Electropherograms showing the results of sequencing for the region of interest (the region of 157insMTTTVP) in exon 4 of the HAVCR1 gene in persons who were homozygous for the three haplotypes (Panel A: homozygous for reference sequence [haplotype C]; Panel B: homozygous for a 15-nucleotide (nt) deletion [haplotype A]; Panel C: homozygous for a 3-nt insertion [haplotype B]). Panel D shows the electropherogram for a subject who was heterozygous for haplotypes A and B; the panel below this electropherogram shows the sequences for the two haplotypes and the expected merged sequence (the ambiguous codes are represented as per the notation laid down by the International Union of Pure and Applied Chemistry). Electropherograms showing the results of sequencing for the region of interest (the region of 157insMTTTVP) in exon 4 of the HAVCR1 gene in persons who were homozygous for the three haplotypes (Panel A: homozygous for reference sequence [haplotype C]; Panel B: homozygous for a 15-nucleotide (nt) deletion [haplotype A]; Panel C: homozygous for a 3-nt insertion [haplotype B]). Panel D shows the electropherogram for a subject who was heterozygous for haplotypes A and B; the panel below this electropherogram shows the sequences for the two haplotypes and the expected merged sequence (the ambiguous codes are represented as per the notation laid down by the International Union of Pure and Applied Chemistry).](https://static.elsevier.es/multimedia/16652681/0000001700000004/v1_201906150923/S1665268119304788/v1_201906150923/en/main.assets/thumbnail/gr3.jpeg?xkr=ue/ImdikoIMrsJoerZ+w96p5LBcBpyJTqfwgorxm+Ow=)




