About 250 million people around the world are chronically infected with the hepatitis B virus (HBV). Those people are at risk of developing hepatocellular carcinoma. The HBV genome is organized as a minichromosome in the infected hepatocyte and is associated with histones and non-histone proteins. In recent years, many groups have investigated the transcriptional regulation of HBV mediated by post-translational modifications on the histones associated with the covalently closed circular DNA (cccDNA). Our aim is to investigate the role of the histone variant H3.3.
Materials and methodsAn in vitro HBV replication model system based on the transfection of linear HBV genome monomeric molecules was used. We then either ectopically expressed or reduced the levels of H3.3, and of its histone chaperone HIRA. Viral intermediates were quantified and the level of H3K4me3 using Chromatin immunoprecipitation (ChIP) assay was measured.
ResultsHistone variant H3.3 ectopically expressed in cells assembles into the viral cccDNA, correlating with increasing levels of the active histone mark H3K4me3 and HBV transcription. The opposite results were found upon diminishing H3.3 levels. Furthermore, the assembly of H3.3 into the cccDNA is dependent on the histone chaperone HIRA. Diminishing HIRA levels causes a reduction in the HBV intermediates.
ConclusionsHistone variant H3.3 positively regulates HBV transcription. Importantly, the characterization of the viral chromatin dynamics might allow the discovery of new therapeutic targets to develop drugs for the treatment of chronically-infected HBV patients.
Infection with the hepatitis B virus (HBV) continues to be a serious worldwide health problem, despite the existence of an effective vaccine. Data from the World Health Organization estimate that more than 250 million people around the world are chronically infected with HBV. Complications of chronic HBV infection include cirrhosis and hepatocellular carcinoma [1,2].
HBV is a small, enveloped DNA virus and a prototypic member of the Hepadnaviridae family. The HBV genome is a partially double-stranded circular DNA molecule of 3.2 kb, surrounded by the viral capsid. The infection starts with the binding of HBV to its cellular receptor, the NTCP protein that localizes in the hepatocyte surface [3]. In the nucleus, the viral genome repairs to generate the covalently closed circular DNA (cccDNA) molecule that serves as a template for viral transcription [4]. The cccDNA organizes as a minichromosome associated with histones and non-histone proteins [5,6], and its transcription depends on the cellular transcriptional machinery. In recent years, many groups have characterized how post-translational modifications on the histones associated with the cccDNA regulate viral transcription [7–16]. Histone acetylation and methylation of the lysine 4 of the histone H3 (H3K4me) correlate with active viral transcription, whereas histone hypoacetylation, H3K9me and H4R3me correlate with viral repression. Consistently, chromatin-modifying enzymes that impose marks on histone proteins are recruited to cccDNA [7,9,13,14,16].
The incorporation of histone variants to chromatin is another way of regulating chromatin function. These are non-canonical (non-allelic) variants of histones, characterized by differences in few amino acids, and expressed at low levels compared to the canonical ones. They have specific expression, localization, and developmental pattern expression [17–19]. The canonical histones H3 are H3.1 and H3.2, which express coupled to the DNA synthesis and incorporated into the chromatin during DNA replication by the histone chaperone CAF1 [20]. The most common H3 variant is H3.3, differing in only five amino acids compared to H3.1. This variant is expressed all through the cell cycle, and it is assembled into chromatin independently of DNA replication. H3.3 enriches at actively transcribed genes, promoters, and regulatory elements, where it is assembled by the histone chaperone HIRA [20]. Consistently, histone H3.3 from different species is enriched in post-translational modifications associated with transcriptional activation [21–23]. However, H3.3 also accumulates at pericentric heterochromatin and telomeres [24–27], and is also required for silencing endogenous retroviral elements in mouse embryonic stem cells [28,29]. The histone chaperone Daxx/ATRX assembles H3.3 at these loci [28,29]. Therefore, H3.3 is involved in gene transcription and silencing. The diverse H3.3 roles are most likely linked to how it assembles into chromatin at specific genomic regions. Here, we investigated the function of the histone variant H3.3 in the HBV transcription. We found that H3.3 is assembled into the cccDNA by the histone chaperone HIRA and that this assembly correlates with increased levels of the active mark H3K4me and the activation of HBV transcription.
2Materials and methods2.1Reagents and primersThe following antibodies were used: anti-Flag beads (Sigma Aldrich A2220), β-actin (Millipore 04-1116), Flag (Sigma Aldrich F3165-2MG), histone H3 (Abcam ab1791), H3K4me3 (Abcam ab8580), HIRA (Abcam ab20655). The following plasmids were used: pOZ-H3.3-Flag, pcDNA3-HA-FLAG. The following primers were used: primer cccDNA, amplify the region between 1662–1923 (261 bp product) of Hepatitis B virus strain F1b, complete genome (GenBank: KM233681.1) [30], Forward 5′-ACT CTT GGA CTT TCA GGA AGG-3, Reverse 5′-TCT TTA TAA GGG TCA ATG TCC AT-3′; primer core promoter region, amplify the area between 1677–1790 (113 bp product) of Hepatitis B virus strain F1b, complete genome (GenBank: KM233681.1) [30], Forward 5′-GGA AGG TCA ATG ACC TGG ATC-3′, Reverse 5′-ATG CCT ACA GCC TCC TAA TAC-3′; primer PreS1 promoter region, amplify the area between 2697–2801 (104 bp product) of Hepatitis B virus strain F1b, complete genome (GenBank: KM233681.1) [30], Forward 5′-CCC TAT TAT CCT GAT AAC GTG G-3′, Reverse 5′-GCT ACG TGT GGA TTC TCT CTT-3′; primer X promoter region, amplify the area between 1219–1290 (71 bp product) of Hepatitis B virus strain F1b, complete genome (GenBank: KM233681.1) [30], Forward 5′-ATT GGC CAT CAG CGC ATG CG-3′, Reverse 5′-AGC TGC AAG GAG TTC CGC AGT-3′; primer H3 F3A: Forward 5′-GCA AGA GTG CGC CCT CTA CTG-3′, Reverse 5′- GGC CTC ACT TGC CTC CTG CAA A-3′; primer H3 F3B: Forward 5′-GTG GCG CTT CGA GAG ATT C-3′, Reverse 5′-GCG AGC CAA CTG GAT GTC TT-3′; primer GAPDH: Forward 5′-AGA AGG CTG GGG CTC ATT TG-3′, Reverse 5′-AGG GGC CAT CCA GAC TCT TC-3′; primer HIRA: Forward 5′–AAG GAG GCC ATG TGT CTG TC-3′, Reverse 5′-CCC CAC CAC TGT CAC TTC AT–3′.
2.2Cells and cell cultureThe Huh-7 human hepatocarcinoma cell line was grown in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% fetal bovine serum (FBS), 100U/mL penicillin (Hyclone), 100ug/mL streptomycin, and 2 mM glutamine. The cells were incubated at 37 °C and 5% CO2. To harvest the cells, they were trypsinized and collected in completed DMEM.
2.3HBV DNA, plasmids, and si/shRNA transfectionsHuh7 cells were seeded at a density of 5 × 105 and transfected after 24 h. For shRNA treatment, 3 μg of either pLL3.7 empty vector as control or with the H3.3 target sequence [31] were transfected using Lipofectamine 2000 48 h before HBV transfection, according to the manufacturer's instructions. For H3.3-Flag overexpression, 5 × 105 Huh7 cells were transfected with Lipofectamine 2000 48 h before HBV genome transfection. For siRNA treatment, 10 nM of either control siRNA (Silencer negative control #1 siRNA, Ambion, Life Technologies) or human HIRA siRNA (sc-43836, Santa Cruz Biotechnology) were transfected with Lipofectamine 2000 48 h before HBV transfection. siRNA products from Santa Cruz Biotechnology consist of three to five 19–25 nt siRNAs target-specific designed to knockdown gene expression. Treated or untreated Huh7 cells were transfected with 1.3 µg of full-length HBV genotype F genome obtained as described previously [12,13], using Lipofectamine 2000 (Invitrogen). The transfection efficiency of Huh7 was determined by flow cytometry with BD FACSDIVA™ Software (BD Bioscience).
2.4cDNA analysisRNA was obtained using the RNeasy® Mini Kit (Qiagen). cDNA was synthesized using the GoScript™ Reverse Transcriptase System (Promega) and quantified by qPCR. The data obtained were analyzed by the ΔΔCt method and plotted in Graph Pad Prism 6.
2.5Purification and analysis of HBV cytoplasmic intermediates and cccDNAHBV intermediates were purified as described previously [12,13]. In brief, nucleic acids from 10% of the harvested cells were purified by treating cells for 1 h with input lysis buffer (10 mM Tris, 5 mM EDTA, 0,5% SDS), then with proteinase K, phenol–chloroform (1:1) extraction and ethanol precipitation. The rest of the cells were incubated with lysis buffer (10 mM Tris pH7.5, 50 mM NaCl, 0,5 % Nonidet P-40, 1 mM EDTA) and centrifuged at 2400 x g for 10 min. The supernatant, containing cytoplasmic and viral particles, was treated with DNase I, proteinase K, and DNA purified by phenol-chloroform (1:1) extraction and ethanol precipitation. The pellet, containing the cccDNA, was incubated with lysis buffer (100 mM NaOH, 6% SDS) for 30 min. Sodium acetate was added to a final concentration of 600 mM and centrifuged at 9600 × g for 20 min. The cccDNA was purified from the supernatant by phenol-chloroform (1:1) extraction and precipitated with ethanol. HBV intermediates were examined by real-time PCR. The values were presented as change folds and plotted with the Graph Pad Prism 6 software.
2.6cccDNA chromatin immunoprecipitation assaysThe assay was performed as previously described [12,13], with minor modifications. Precleared samples (2 µg) were taken as input control for ChIP reactions. KAPA SYBR FAST (KAPA Biosystems) was used to quantify the immunoprecipitated DNA, according to the manufacturer's instructions. Immunoprecipitated DNA was quantified by creating a line of best fit from a standard curve using serial dilutions of Input DNA. PCR was carried out for 40 cycles with 95 °C for 10 s (melting), 59 °C for 5 s (annealing) and 72 °C for 5 s (extension).
2.7Cell cycle analysisTreated Huh7 cells were fixed with 70% cold ethanol and stored overnight at −20 °C. Cells were then washed with PBS and incubated with RNaseA for 1 h at 37 °C. Cells were washed with PBS and DNA stained with propidium iodide. Samples were analyzed by flow cytometry with BD FACSDIVA™ Software (BD Bioscience), and cell cycle examined with FlowJovX.0.7 (tree Star).
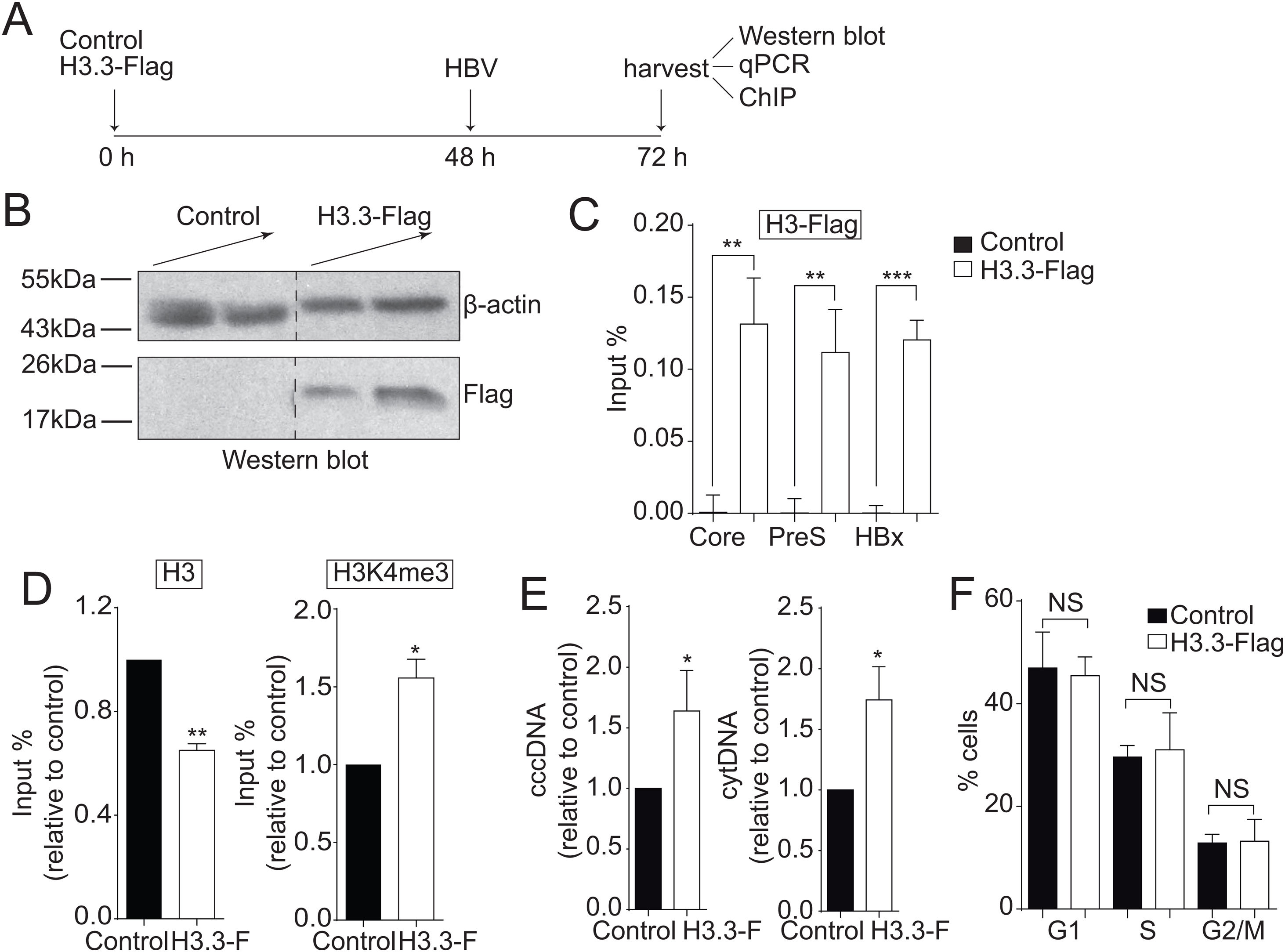
3Results3.1The histone variant H3.3 associates to the cccDNA and activates HBV transcriptionWe investigated the role of H3.3 on HBV transcription. However, there are no suitable antibodies available to differentiate between H3.1 and H3.3 variants. Therefore, we exogenously expressed Flag-tagged histone H3.3 so we could purify H3.3 by Flag-immunoprecipitation (Fig. 1A) [22]. Overexpression of histones is harmful to the cell; therefore, we expressed the exogenous H3.3 gene under the regulation of a retroviral promoter, allowing a low expression of H3.3, representing about 8% of the endogenous variant [22]. The transfection efficiency was about 30% (data not shown). As shown in Fig. 1B, we detected H3.3 with Flag antibodies. We then examined whether histone H3.3 associated with the cccDNA by Flag-ChIP (Chromatin immunoprecipitation) analysis. We found that the variant H3.3 associates to the three viral promoters analyzed: core, PreS, and HBx (Fig. 1C). We then examined if the association of H3.3 to the cccDNA results in changes in the transcriptional state of the viral cccDNA. We previously showed that the methylation of the lysine 4 of the histone H3 (H3K4me3) correlates with an active cccDNA chromatin state [13]. Thus, we performed ChIP analysis and observed that H3K4me3 is enriched in the viral core promoter upon expression of H3.3-Flag compared to control cells (Fig. 1D, right). We also observed reduced levels of the endogenous histone H3 at the viral core promoter upon expression of H3.3-Flag (Fig. 1D, left), consistent with the fact that actively transcribed genes have nucleosome-free promoters [32]. Besides, we found that both cytoplasmic viral intermediates significantly increased upon expression of H3.3-Flag (Fig. 1E). We found no changes in the cell cycle upon expression of H3.3-Flag (Fig. 1F). We thus concluded that the histone H3.3 associates to the HBV cccDNA and activates HBV transcription.
The histone variant H3.3 associates to the cccDNA and activates HBV transcription.
(A) Scheme illustrating H3.3-Flag expression and detection of the HBV viral intermediates. (B) Western blot analysis of H3.3-Flag expression. Seventy-two hours after transfection, 15 and 30 µg of total cell extracts derived from transfection were separated on 10% SDS-PAGE and Western blotted for Flag. On the left, the migration of molecular size markers is shown. (C) H3.3 association to viral promoters was assayed by ChIP analysis. Immunoprecipitated DNA was quantified by qPCR using specific primers for core, PreS and HBx promoters. The results are expressed as % of input. The standard deviation was obtained from three PCR reactions and the graph is representative of three independent experiments. (D) Covalent post-translational modifications on histone H3 were determined by ChIP analysis using the specific antibodies: H3 (left) and H3K4me3 (right). Immunoprecipitated DNA was quantified by qPCR using specific primers for core promoter. The results are expressed as fold changes of % Input with respect to the control. The standard deviation was obtained from three PCR reactions and the graphs are representative of two independent experiments. (E) The HBV replicative intermediates cytoplasmic viral core particles (cytDNA, right) and cccDNA (left) were determined 72 h post-transfection of the H3.3-Flag construct. The value obtained in Control and in H3.3-Flag was divided by the Control value, thus the results are expressed relative to the Control. The standard deviation was obtained from four independent experiments. (F) Cell cycle profile of Huh7 cells expressing H3.3-Flag. The standard deviation was obtained from three independent experiments. *: p < 0.05, **: p < 0.01, ***: p < 0.001, Student´s t-test.
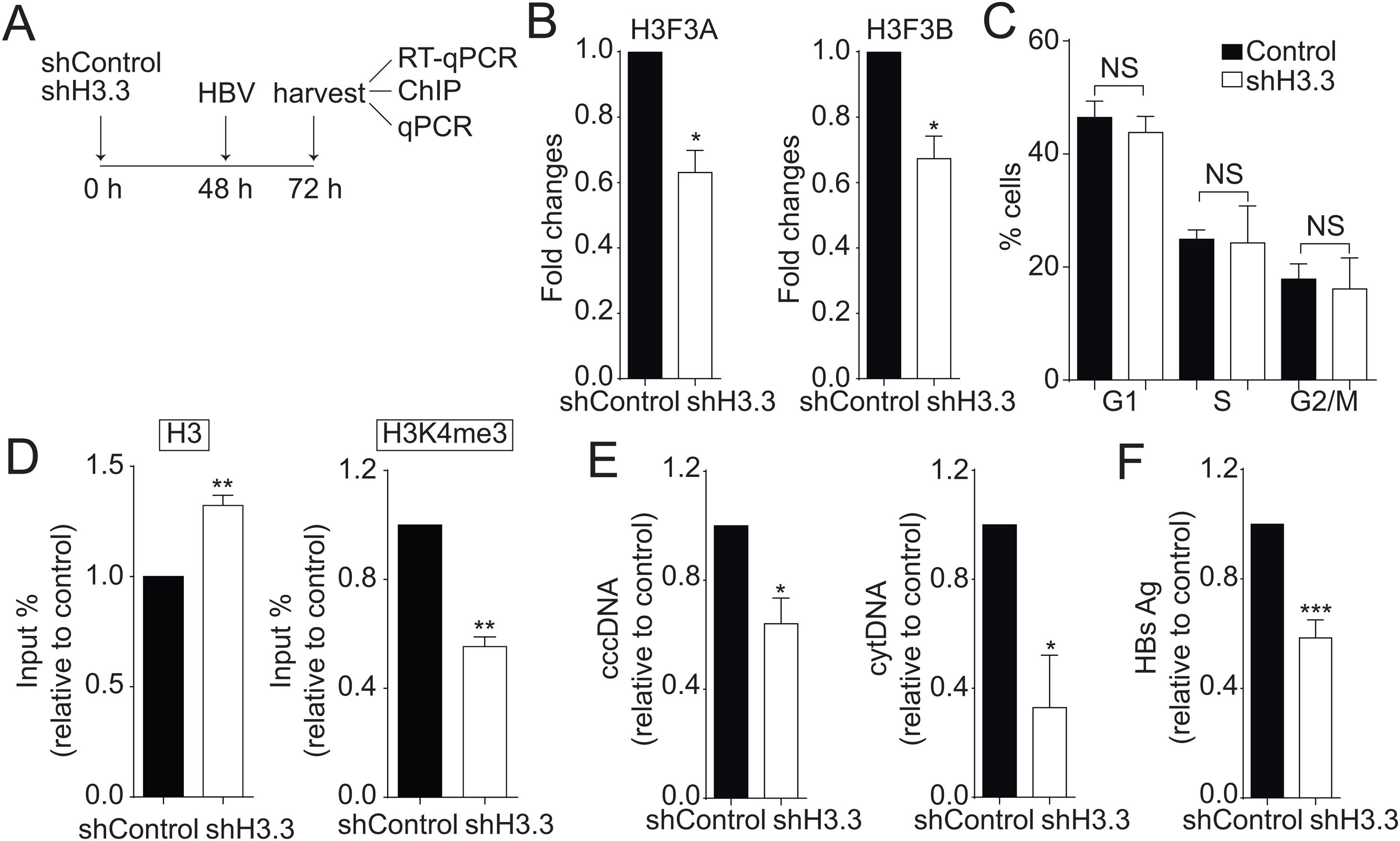
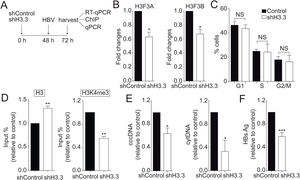
We then decided to diminish the levels of the H3.3 variant. As illustrated in Fig. 2A, we utilized shRNA against H3.3 (shH3.3). The transfection efficiency was about 50% (data not shown). Two genes encode histone H3.3, H3 F3A, and H3 F3B [19], and we reduced the expression of both mRNAs (Fig. 2B). We found no changes in the cell cycle upon treatment with shH3.3 (Fig. 2C). The H3.3 knocked-down correlated with a significant reduction in the H3K4me3 levels, with a concomitant increase in the amount of histone H3 present on the viral core promoter (Fig. 2D). Consistently, upon H3.3 knockdown, both viral intermediates significantly decreased (Fig. 2E). We also examined the HBs viral antigen, used as a marker of acute infection, showing that shH3.3 gave rise to a reduction on the HBs levels (Fig. 2F). Taken together, the data indicate that H3.3 binds to the cccDNA and positively regulates HBV transcription by activating the cccDNA chromatin state.
The histone variant H3.3 histone is necessary to regulate HBV transcription.
(A) Scheme illustrating the shH3.3 experiment and detection of the HBV viral intermediates. (B) Levels of mRNA on samples derived from shControl or shH3.3 treated Huh7 cells measured by real-time PCR. The graph shows mRNA levels expressed relative to the shControl. Each expression level was normalized to that of GAPDH. The standard deviation was obtained from three independent experiments. (C) Cell cycle profile of Huh7 cells treated with shH3.3. The standard deviation was obtained from three independent experiments. (D) Covalent post-translational modifications on histone H3 were determined by ChIP analysis using the specific antibodies: H3 (left) and H3K4me3 (right). Immunoprecipitated DNA was quantified by qPCR using specific primers for core promoter. The results are expressed as fold changes of % Input with respect to the control. The standard deviation was obtained from three PCR reactions and the graphs are representative of two independent experiments. (E) The HBV replicative intermediates cytoplasmic viral core particles (cytDNA, right) and cccDNA (left) were determined 72 h post-transfection of the shH3.3 construct. The value obtained in shControl and in shH3.3 was divided by the shControl value, thus the results are expressed relative to the Control. The standard deviation was obtained from three independent experiments. (F) The quantity of the viral antigen HBsAg in the supernatant of the transfected cells was determined by ELISA, using specific antibodies. The result is shown as fold changes with respect to the control. The standard deviation was obtained from three independent experiments. *: p < 0.05, **: p < 0.01, Student’s t-test.
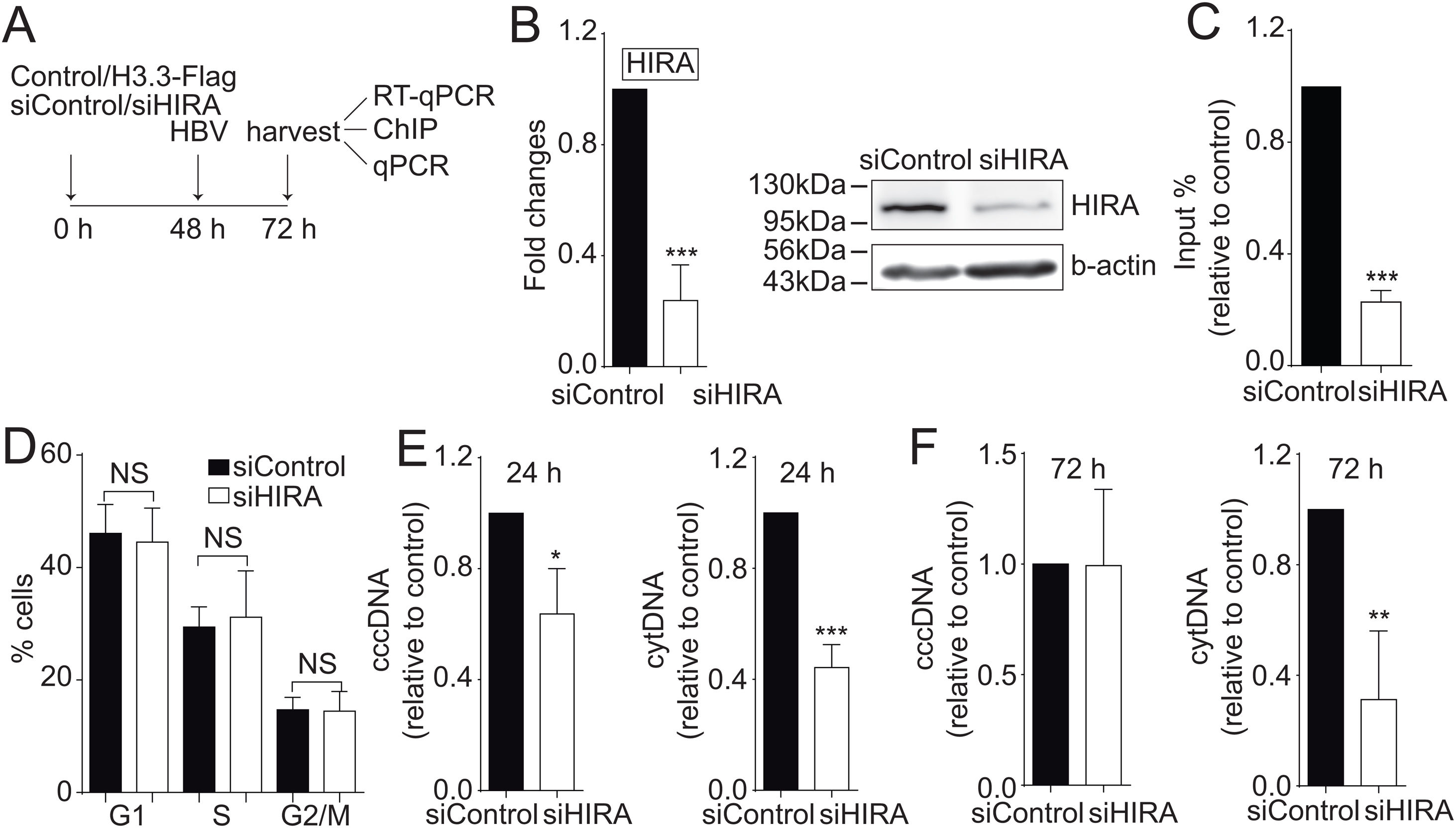
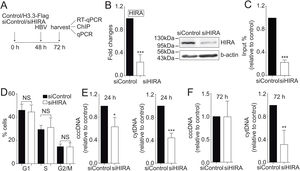
We finally investigated the molecular mechanism by which the H3.3 histone variant associates to the viral HBV cccDNA. Two histone chaperones assemble H3.3 to the cellular chromatin: HIRA and Daxx/ATRX [19]. We decided to focus on the chaperone HIRA because it is involved in the assembly of H3.3 to euchromatic regions of the DNA [20]. Upon knocking down HIRA with a commercial siHIRA (Fig. 3A and B), we observed a reduction in the amount of H3.3-Flag associated to the viral core promoter (Fig. 3C), indicating that HIRA participates in the assembly of H3.3 to the viral cccDNA. We found no changes in the cell cycle upon treatment with siHIRA (Fig. 3D). Consistently, in the 24 or 72 h HIRA knocked-down conditions, both viral intermediates significantly decreased (Fig. 3E and F, respectively). Taken together, we conclude that H3.3 is assembled into the HBV cccDNA by the histone chaperone HIRA, one of the steps that contribute to establishing a transcriptionally active cccDNA chromatin state.
The histone chaperone HIRA assembles the histone H3.3 into the HBV cccDNA.
(A) Scheme illustrating the siHIRA and detection of the HBV viral intermediates. (B) Levels of mRNA (left) and protein (right) on samples derived from siControl and siHIRA treated Huh7 cells. The graph shows mRNA levels expressed relative to the siControl. The expression level was normalized to that of GAPDH. The standard deviation was obtained from three independent experiments. (C) H3.3 association to viral promoters was assayed by ChIP analysis under siControl and siHIRA conditions. Flag-immunoprecipitated DNA was quantified by qPCR using specific primers for core promoter. The results are expressed as fold changes of % Input with respect to the control. The standard deviation was obtained from three PCR reactions and the graph is representative of two independent experiments. (D) Cell cycle profile of Huh7 cells treated with siHIRA. The standard deviation was obtained from three independent experiments. (E–F) The HBV intermediates cytoplasmic viral core particles (cytDNA, right) and cccDNA (left) were determined 24 (E) and 72 (F) h post-transfection of the siHIRA, as indicated. The value obtained in siControl and in siHIRA was divided by the siControl value, thus the results are expressed relative to the Control. The standard deviation was obtained from three independent experiments. *: p < 0.05, **: p < 0.01, ***: p < 0.001, Student´s t-test.
The HBV cccDNA molecule is the source of persistence in chronically infected HBV patients. Given that therapeutic treatments for chronically infected patients are inefficient, the characterization of the molecular mechanisms by which HBV regulates its transcription is highly relevant, as it can shed light onto new therapeutic targets [33]. The HBV genome associates with cellular histones and utilizes cellular machineries to regulate its transcription in a chromatin context. Several studies have contributed to the understanding of the molecular mechanisms that regulates HBV transcription in a chromatin context, mainly focusing on histone post-translational modifications and DNA methylation [7–16,34].
Much effort has been spent in understanding the role that histone variants have in the regulation of chromatin dynamics. In the viral context, one report showed that the variants H3.1 and H3.3 are incorporated into the human cytomegalovirus (HCMV) chromatin, and this incorporation occurs independently of DNA replication [35]. Time-course experiments with the herpes virus, on the other hand, have shown that H3.3 is assembled into HSV-1 chromatin before the canonical variant H3.1, which is assembled into the viral chromatin when HSV-1 DNA replicates [36,37]. This observation is consistent with the fact that H3.1 is incorporated into the cellular chromatin coupled to DNA replication [20]. The incorporation of H3.3 to HSV-1 is dependent on the chaperone HIRA [36]. Further experiments should address whether the differences observed in HSV-1 and HCMV viruses regarding the dependency of DNA replication and the incorporation of H3.1 into the viral chromatin is due to intrinsic viral differences. We showed that the histone variant H3.3 is deposited into the HBV cccDNA by the histone chaperone HIRA, and that this association correlates with increased levels of the active mark H3K4me3 and with the activation of HBV transcription.
In this study, we utilized the HBV Genotype F, not commonly used in research. This genotype is present in Central and South America and our previous studies showed that it is prevalent in Chile [38]. We also showed that when compared to HBV Genotype A, the most commonly genotype used in research, genotype A replicated more than genotype F and viral transcriptional activities in both genotypes correlated with their cccDNA chromatin state [13]. Thus, our study extends research into one of the less characterized genotypes of the human hepatitis B virus.
5ConclusionsThis study demonstrated that, in addition to histone post-translational modifications and DNA methylation, histone variants, particularly histone H3.3, contribute to regulate in vitro the HBV cccDNA functional state. Further studies are required to confirm the functionality of these machineries in an in vivo model, as well as to investigate how long changes on the HBV chromatin dynamics last. Thus, the characterization of chromatin regulators that play roles in viral chromatin dynamics might reveal new therapeutic targets to develop drugs for the treatment of chronically infected HBV patients.List of abbreviationsHBV hepatitis B virus covalently closed circular DNA Sodium taurocholate cotransporting polypeptide chromatin immunoprecipitation Histone Regulator A herpes simplex virus-1
Not applicable
Consent for publicationNot applicable
Availability of data and materialsThe datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Conflict of interestThe authors have no conflicts of interest to declare.
Authors’ contributionsFAA, RAV, and AL conceived and designed the experiments; FAA, DG, MVG, JLG, RAV, and AL performed experiments and analyzed data; FAA, RAV and AL wrote the manuscript; and all authors read and approved the final manuscript.
Laboratory members for critical suggestions.