Clinical manifestations of SARS-CoV-2 infection include more frequently fever and cough, but complications (such as pneumonia, respiratory distress syndrome, and multiorgan failure) can occur in persons with additional comorbidities. Liver dysfunction is one of the most striking affections among patients suggesting that SARS-CoV-2 may represent a new king of liver aggressor. However, the molecular process underlying this phenomenon is still unclear. In this work, we overview the most recent findings between the molecular biology of the virus, pathogenic mechanisms, and its relationship to liver disease observed in patients.
SARS-CoV-2 is the etiological agent of the disease known as COVID-19, which causes a disease characterized by pneumonia, cough, fever, occasional diarrhea, headache, cardiac injury and in some patients’ liver alterations. COVID-19 has been found to be an extremely complex disease in many severely ill patients. In other infected subjects, an infection is reported to be so severe that it can lead to a disproportionate and mortal reaction of the immune system, known as a “cytokine storm”. All of these factors make COVID-19 highly unpredictable: it is what specialists call a multi-system disease.
Around the world cases of liver dysfunction, denoted by elevated hepatic enzymes in serum such as AST (aspartate aminotransferase) and ALT (alanine transaminase) have been documented among patients infected with SARS-CoV-2. There is still no certainty whether the COVID-19-related liver damage/dysfunction is due mainly to the viral replication per se, drugs toxicity or other coexisting comorbidities.
It is important to understand how liver function can be altered by direct infection with this “respiratory” virus; which mechanisms of viral pathogenesis are involved; to evaluate whether sex-related difference could help to explain why infected men are more predisposed to develop COVID-19-associated liver dysfunction than infected women; to analyze if there is any genetic predisposition related to impaired liver function during the disease; and of course the cross-talk between viral and cellular proteins that mediate this healthy or harmful relationship between the liver and its viral aggressor.
In this paper, we describe a brief overview of the implications for researchers in the field of liver disease of the most recent findings between the molecular biology of the virus, pathogenic mechanisms, and its relationship to liver disease observed in patients.
2Clinical characteristics and liver injury in patients with COVID-19This emerging viral illness is typically characterized by fever, dry cough, myalgia, headache, sore throat, diarrhea and may be aggravated with shortness of breath and respiratory failure [1]. The angiotensin-converting enzyme 2 (ACE2), the functional receptor of the spike glycoprotein of SARS-CoV-2, is widely distributed in the organism. Historically, Hamming et al. reported ACE2 expression in the surface of lung alveolar epithelial cells, enterocytes of the small intestine, arterial and venous endothelial cells and arterial smooth muscle cells [2]. Further transcriptomic and proteomic analyses confirmed their findings and added high ACE2 expression in adipose tissue, bone marrow, duodenum, endometrium, heart, kidney, small intestine, smooth muscle, testis and thyroid [3]. ACE2 is also expressed in liver but in lesser extent. One of the most worrisome complications is the unusual formation of blood clots in many patients with COVID-19, even those who were receiving anticoagulants. Researchers at Mount Sinai, in New York published studies suggesting that clots in the lungs play an important role in the most severe cases of COVID-19 [4]. Regarding the gastrointestinal (GI) tract and liver, over 60% of COVID-19 patients develop GI symptoms such as anorexia, diarrhea, nausea, and vomiting and a significant proportion present with altered liver function tests [5].
COVID-19-associated liver injury is defined as any liver damage that occurs during disease progression and/or COVID-19 treatment in patients with or without a history of previous liver disease. In general, the incidence of increased liver biochemical markers in hospitalized patients with COVID-19, mainly AST and ALT, and slightly elevated bilirubin, varies between 14%–53% of cases [6]. The increase in liver enzymes is observed more frequently in men and is mainly documented in more severe cases [7,8]. AST elevation is the most common abnormality in patients presenting with COVID-19. Decreased albumin levels are associated with severe infection and poor prognosis. Still, there are no reports of acute or subacute liver failure in patients with COVID-19.
The largest cohort study that included 5771 cases of COVID-19 from China showed that 81 (1.4%) had pre-existing chronic liver disease. Lei et al. reported that impaired liver function was related to mortality in COVID-19 patients [9]. Elevated AST was more frequent (39.4%) and significant than the increase of ALT (28.1%) in severe hospitalized patients. Moreover, elevated AST was shown to be associated with highest mortality risk [10,11].
In the study reported by Wang et al. [12], they found that 64 of 156 (41.0%) COVID-19 patients had elevated AST activity. The median levels of ALT were 50 U/L vs. 19 U/L, respectively, AST were 45.5 U/L vs. 24 U/L, respectively in abnormal and normal aminotransferase groups. Liver enzymes abnormality were associated with disease severity, as well as a series of laboratory tests including higher Alveolar-arterial Oxygen Gradient (A-aDO2), higher gamma glutamyl transpeptidase (GGT), lower albumin and lower total protein levels. In addition, they found by ultrastructural examination of coronavirus particles in hepatocytes in COVID-19 cases. SARS-CoV-2 infected hepatocytes displayed conspicuous mitochondrial swelling, endoplasmic reticulum dilatation, and glycogen granule decreased. Histological findings showed apoptosis and binuclear hepatocytes. A report from Gerussi et al. [13], demonstrated that patients under immunosuppressive therapy for autoimmune hepatitis (AIH) developing COVID-19 had a disease course similar to that reported in non- immunosuppressed population.
Taken together, both ultrastructural and histological evidence indicated a typical lesion of viral infection [12]. All these findings by different reports demonstrates that SARS-CoV-2 infection in liver is a crucial cause of hepatic impairment in COVID-19 patients. However, today the cellular and molecular mechanisms that are altered by infection with this coronavirus, the interaction of its proteins with cellular proteins, and consequently, the alteration of cellular metabolism that give rise to systematic alterations and metabolic changes, are still unknown.
3Molecular biology of SARS-Cov-2Coronaviruses are enveloped viruses that contain a positively polarized, unsegmented RNA genome belonging to the Coronaviridae family and the order of Nidovirales, they are distributed in humans and other mammals [14]. The size of the SARS-CoV-2 virions is approximately 50–200 nm in diameter [15–17]. SARS-CoV-2 has a genome that consists of 29,891 nucleotides [18] (encoding 9860 amino acids) and it is composed of a region that encodes for structural proteins and a larger region that encodes two open reading frames (ORF) 1a and 1b, which together encode for the non-structural virus proteins from nsp1 to nsp16 [18,19].The virions have a structural S-spike protein (outer spiky glycoprotein), M-membrane protein (a type III transmembrane glycoprotein), N-nucleocapsid protein (which is within the phospholipid bilayer) and nonstructural proteins, which are encoded by the various genetic loci on the genome [18]. At the center of the virion lies a nucleocapsid composed of the genomic RNA and the nucleocapsid protein [20]. The virus glycoprotein S consists of two subunits: S1 which is at the amino terminus and provides the receptor binding site and S2 which is at the carboxyl terminus, responsible for membrane fusion [21]. The envelope protein E has a role in the assembly and release of the virus [22], as well as protein M which is a type III transmembrane glycoprotein [23,24]. Non-structural proteins have several functions during de viral cycle. For example, nsp 3, 4 and 6 participates in the cellular membrane rearrangements for the replication and transcription complexes [25]. Among the encoded proteins is an RNA-dependent RNA polymerase (RdRp) which is nsp12, and is responsible of the replication and transcription of the virus [26].
The virus enters the cell by endocytosis through the interaction between envelope glycoprotein S with the cell receptor ACE2 and with the participation of the type II transmembrane serine protease (TMPRSS2) [27]. Once it enters the cell, the N protein with viral genome are released within the cytoplasm, then cellular proteases degrade the capsid and the virus genome is left free. Next, the polyprotein containing the viral proteins that are processed to form the replication complex is translated and then the complementary strand of negative sense pre-genomic RNA is synthesized to be used as a template to replicate the positive sense viral genome [28]. Furthermore, the replication and transcription complex will lead to a series of smaller, positive sense subgenomic RNAs, that will translate into viral proteins; this entire process occurs in the cell cytoplasm. The structural proteins synthesized in the endoplasmic reticulum membrane are assembled to form the viral particle, ending the cycle through the release of the viruses by exocytosis [20].
Both, SARS-CoV (2003) and the new SARS-CoV-2 are very similar in structure and pathogenicity, but the major structural protein, S protein, is slightly different between them [29]. Compared to other beta coronaviruses, the presence of a furin-like cleavage site in SARS-CoV-2 enables the S protein priming and facilitates an increase on the efficiency of the spread of SARS-CoV-2 as reported worldwide [30].
4How does the virus select which cells to infect?Viruses can infect only certain species of hosts and only permissive cells within that host. Permissive cells make all the necessary proteins and viral factors to allow virus to replicate. Once a virus gets inside a cell, it hijacks the cellular processes to produce virally encoded proteins that will replicate the virus’s genetic material [31].Viral replication may cause biochemical changes producing cell damage, called cytopathic effects. Like other coronaviruses, SARS-CoV-2 requires cellular receptors to initiate its internalization to the cells that carry these factors [32]. SARS-CoV-2 uses the angiotensin-converting enzyme 2 (ACE2) as a host cell receptor. SARS-CoV-2 spike (S) protein binds ACE2 with significantly high affinity [33,34]. In addition, the main host protease that mediates S-protein activation on primary target cells and initial viral entry is the type II transmembrane serine protease [35]. Other host proteases, such as furin, have also been suggested to promote the pathogenesis of this coronavirus [36,37].
In order to provide insights into the mechanism of SARS-CoV-2 infection, Li et al. [38], analyzed the expression of ACE2 in various normal human tissues using the datasets from the Genotype-Tissue Expression (GTEx) project and The Cancer Genome Atlas (TCGA) program (https://portal.gdc.cancer.gov/). They compared ACE2 expression levels across 31 human tissues, between males and females, and between younger and older persons in these individual tissues. Furthermore, other reports have analyzed the correlations between ACE2 expression levels and immune signature enrichment levels in individual tissues. As reported by Li et al., ACE2 expression levels showed no significant difference between males and females, between younger and older persons, or between Asian and non-Asian races. This finding suggests that the infection risk of SARS-CoV-2 and SARS-CoV may have no significant association with sex, age, or race.
5Is the liver a direct target for SARS-CoV-2?As expected, due to the systemic manifestations of COVID-19, it has been reported that SARS-CoV-2 has an organ tropism beyond the respiratory tract, including the kidneys, liver, heart, and brain, and it is plausible that this organ tropism influences the course of COVID-19 disease and aggravates preexisting conditions. The ACE2 protein is found at high levels in the GI tract as well as the colon, biliary system and liver [2,39]. On the other hand, SARS-CoV-2 RNA shedding in the GI tract has been well-documented [40] supporting its tropism for the GI tract and liver cells, and these may be sites of active viral replication and either direct or indirect tissue injury. Indeed, a large part of the cells distributed in the liver architecture express ACE +/ TMPRSS1 + receptors [38]. The presence of these two host factors in the liver suggests that a direct viral cytopathic effect occurs. Also, in SARS infection, the presence of viral RNA in liver tissue was documented but not as extensively as the new coronavirus.
Data published by Gordon [41] suggest that mitochondrial proteins interact directly with the virus, which helps to understand the potential mechanism by which elevated AST profiles are detected in these patients [42]. Furthermore, in addition to this intracellular effect, the exacerbated inflammatory response in COVID-19 may play a central role, in which high levels of IL-6 have been reported [43], which are involved in both inflammatory and repair responses in liver disease.
More recently [12], identified the clinical and laboratory characteristics of COVID-19 patients with abnormal liver transaminases and they reported that SARS-CoV-2 is able to infect liver cells and cause liver impairment by direct cytopathic effect.
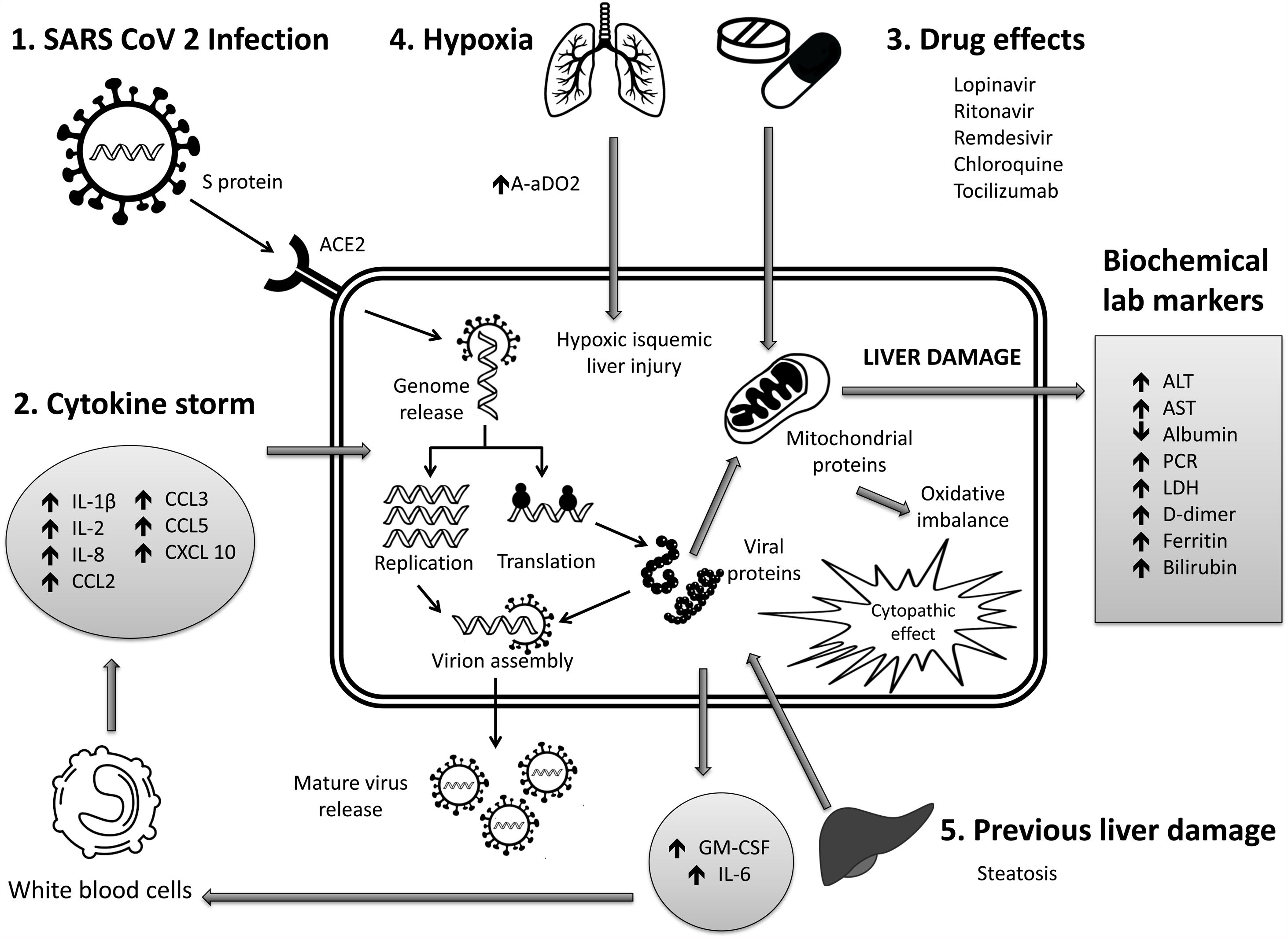
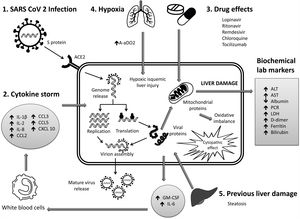
6Mechanisms of liver pathogenicityIf SARS-CoV-2 replication has direct adverse effects on liver function, it is still unknown. Findings in liver biopsy of patients killed by COVID-19 showed moderate micro vesicular steatosis and mild portal and necro-inflammatory activity [44]. This seems to indicate that a direct injury occurred while the infection that could have been directly caused by SARS-CoV-2. Another possibility is that a drug-induced liver injury occurred. To date, there are the following possible mechanisms (Fig. 1).
- 1
Immune damage from exacerbated inflammation in response to SARS-CoV-2 infection: The massive release of cytokines by the immune system in response to the viral infection can result in a cytokine storm and symptoms of sepsis that are the cause of death in 28% of fatal COVID-19 cases [45]. In these cases, uncontrolled inflammation induces multiorgan damage leading including liver failure. Biomarkers of inflammation, such as C-reactive protein (PCR), serum ferritin, LDH, d-dimer, IL-6, IL-2, are have been found to be significantly elevated in critically ill patients with COVID-19 [11,46,47].
- 2
Direct cytopathic effect due to active viral replication in various liver cells: SARS-CoV-2 enters cells through the ACE2 molecule, which is abundantly expressed in the liver and, in particular, in bile epithelial cells [48]. Based on this expression, the liver is a potential target for direct infection with this virus. To understand the pathogenesis of SARS-CoV-2–related liver disease, more studies should be performed for evidence of viral mechanisms of replication in different cell organelles as cytoplasm, endoplasmic reticulum, Golgi apparatus, and lipid-rafts into hepatocytes and liver histology characterization. It is also important to know in cells, their capacity to efficiently produce both infectious and defective non-infective whole virions. There are not yet enough data to know about the viral dynamics in different tissues and associated pathogenesis.
- 3
Anoxia: respiratory failure is one of the main characteristics of COVID-19. Anoxic hypoxic hepatitis is common in patients with severe symptoms [12].
- 4
Drug-induced liver damage (DILI): Initial clinical guidelines recommended antiviral agents for COVID-19, so a variety of drugs have been administered in various studies, such as: lopinavir / ritonavir, remdesivir, chloroquine, tocilizumab, uminefovir, traditional Chinese medicine, so it is important to consider, that they could be potentially hepatotoxic in some patients [49,50].
- 5
Reactivation of pre-existing liver disease: Patients with pre-existing chronic liver disease may be more susceptible to liver damage from SARS-CoV-2. Biological medications such as tocilizumab and baricitinib used to combat the adverse immune reaction may also cause HBV reactivation [51] and induce eventual impairment of liver function (in those patients with HBV). On the other hand, it is still unknown whether SARS-CoV-2 infection exacerbates cholestasis in people with underlying cholestatic liver disease.
- 6
Genetic factors: Genetics may well be one of the determining factors in some patients who become seriously ill with COVID-19, but until now we cannot be sure. It is possible, for example, that the genetic variation that makes an individual more susceptible to high blood pressure or diabetes also makes him more vulnerable to the virus. It will be important to find out what role genetic factors predisposing to liver steatosis have and their sensitivity to severe symptoms of COVID-19. Ji et al., showed that subjects with metabolic-associated fatty liver disease (MAFLD) have a higher risk of COVID-19 severity disease and abnormal liver blood tests than patients without MAFLD [52]. In contrast, Biquard et al [53], demonstrated that MAFLD is not associated with changes in liver expression of genes implicated in SARS-CoV-2 infection. The observed persistence of liver blood test abnormalities reported by Ji et al. is thus likely not explained by increased hepatic SARS-CoV-2 uptake. Still several contradictory reports will help to find the true role of genetic factors in the evolution of this disease.
The scenario is not yet complete, which does not allow us to establish or understand the natural history of the disease and the participation or commitment of the liver in this disease. Certainly, the application of new technological platforms such as single-cell transcriptomic assays, will allow us to quickly know the commitment of each cell type in affected organs and the meaning of viral dynamics in the various affected systems including the liver. However, as we already know from the old hepatotropic viruses, this history is still ongoing. We have much to learn and understand about the virologic characteristics of this emerging RNA virus that will allow us to develop specific antivirals as in the case of HCV and hopefully a vaccine to decrease the impact of this “acute” infection.
Financial SupportFinancial support was providing by Conacyt through the national biobank laboratory project: Consolidation 299077.
Conflict of interestThe authors have no conflicts of interest to declare.
Ethical approvalNot required.