Enzyme production by Aspergillus terreus NCFT 4269.10 was studied under liquid static surface and solid-state fermentation using mustard oil cake as a substrate. The maximum lipase biosynthesis was observed after incubation at 30°C for 96h. Among the domestic oils tested, the maximum lipase biosynthesis was achieved using palm oil. The crude lipase was purified 2.56-fold to electrophoretic homogeneity, with a yield of 8.44%, and the protein had a molecular weight of 46.3kDa as determined by SDS-PAGE. Enzyme characterization confirmed that the purified lipase was most active at pH 6.0, temperature of 50°C, and substrate concentration of 1.5%. The enzyme was thermostable at 60°C for 1h, and the optimum enzyme–substrate reaction time was 30min. Sodium dodecyl sulfate and commercial detergents did not significantly affect lipase activity during 30-min incubation at 30°C. Among the metal ions tested, the maximum lipase activity was attained in the presence of Zn2+, followed by Mg2+ and Fe2+. Lipase activity was not significantly affected in the presence of ethylenediaminetetraacetic acid, sodium lauryl sulfate and Triton X-100. Phenylmethylsulfonyl fluoride (1mM) and the reducing, β-mercaptoethanol significantly inhibited lipase activity. The remarkable stability in the presence of detergents, additives, inhibitors and metal ions makes this lipase unique and a potential candidate for significant biotechnological exploitation.
Lipases (triacylglycerol acylhydrolases, EC 3.1.1.3) belong to the class of serine hydrolases,1 are produced by plants, animals, bacteria and molds2 and contain the consensus sequence G–X1–S–X2–G as the catalytic moiety, where G, X1, S, and X2 are glycine, histidine, serine, and glutamic or aspartic acid, respectively.3 A hydrolase fold and nucleophilic elbow characteristics of lipases are characterized by 3D structures, where the catalytically reactive serine is located in the active site.4 During the last two decades, lipases have gained importance to a certain extent over proteases and amylases, particularly in the area of organic synthesis.5 A true lipase converts emulsified esters of glycerin and long-chain fatty acids (triolein and tripalmitin) into respective polar lipids. Lipase reactions take place at the interface between the aqueous and oil phases due to opposite polarity between the enzymes (hydrophobic) and their substrates (lipophilic).6 In addition, lipases are the supplest biocatalysts and perform a wide range of bioconversion reactions, such as hydrolysis, inter-esterification, esterification, alcoholysis, acidolysis and aminolysis.7 Lipases can act on a variety of substrates, including natural oils and synthetic triglycerides. A number of other low- and high-molecular-weight esters, thiol esters, amides, polyol/polyacid esters, etc., are accepted as substrates by this unique group of enzymes. The inimitable distinctiveness of lipases includes substrate specificity, stereospecificity, regiospecificity, chiral selectivity8 and a capability to catalyze a heterogeneous reaction at the interface of water-soluble and water-insoluble systems. The affinity toward solvents and the ability to catalyze the reverse reaction of synthesis just as efficiently confers on these enzymes an extensive application potential that is literally unlimited. Lipases are the crucial class of enzymes for food technology applications in the fat and oil, dairy, pharmaceutical, and bakery industries, biopolymer synthesis, and biodiesel production,9 as well as for the generation of maltose- and lactose-like sugar fatty acid esters.10 Other applications include treatment of fat-containing waste effluents, enantiopure synthesis of pharmaceuticals and nutraceutical agents, production of cosmetics, leather, detergent formulations, and perfumery, as a biosensor in diagnostics,11 treatment of malignant tumors and detection of target DNA sequences,12 inorganic chemical processing, synthesis of biosurfactants, in agrochemical industry, and paper manufacture. Detergent enzymes alone make up 32% of the total lipase sales.13
A search for these enzymes continues, and solid-state fermentation (SSF) is preferred because of its simplicity, low capital investment, lower levels of catabolite repression and end-product inhibition, low wastewater output, better product recovery and high quality production.14 Furthermore, it is a unique approach for fermentation when the crude fermented matter/broth is used chiefly as the source of enzyme.15 Filamentous fungi are especially suitable for solid-state fermentation because of their hyphal mode of fungal growth and their good tolerance to low water activity and high osmotic pressure conditions, which makes them competitive over other natural microflora for bioconversion of solid substrates.16
In view of the potential applications, the present work aimed at production, purification and characterization of a lipase from a fungal isolate of Aspergillus terreus grown on agro substrates under both submerged fermentation (SmF) and SSF conditions.
Materials and methodsAgro-wastes and chemicalsMustard oil cake (MoC) used as a fermentative substrate was purchased from a local market in Bhubaneswar, Odisha, India. The substrate was transported to the laboratory in a sterilized new steel container and then oven-dried at 60°C for 48h. The sample was powdered and kept in a sterile container until required. Olive oil was obtained from local retail outlets in Bhubaneswar, Odisha, India. Analytical reagent grade chemicals used in the present study were purchased from Hi-Media Ltd., SRL Pvt. Ltd., and Merck India Ltd., Mumbai, India.
Microorganism and inoculumTo conduct this study, a lipase-producing 7-day-old potato dextrose agar slant culture of A. terreus NCFT 4269.1017 was used for all experiments at an initial cell density of 1×107cellsmL−1.18
Fermentation for lipase production and enzyme recoveryFor this study, sterilized and cooled fermentation broth medium (with powdered MoC) was inoculated with 1×107cellsmL−1 from a 7-day-old culture and incubated at 30±1°C under static conditions [liquid static surface fermentation (LSSF)]. After 96h of incubation, the fermented sample was processed and analyzed for lipase activity. Similarly, for SSF the powdered MoC was mixed with 8mL of minimal salt solution to adjust the moisture content up to 80% and autoclaved using a standard protocol for 15min at 121°C and a 15psi pressure. This sterilized fermentation medium was inoculated aseptically with an optimum number of spores and incubated for 4 days at 30±1°C with intermittent observation. At the end of both fermentation processes, LSSF and SSF19 were used for enzyme extraction from the fermented media.
Effect of inducers on lipase productionThe effect of substrate-related compounds on lipase production was determined by supplementing the basal medium with natural oils (sunflower, coconut, palm, sage, almond, mustard, ghee, castor, olive and sesame) at a concentration of 1.0% (v/v).
Purification of lipaseA crude culture filtrate (∼500mL) was purified by precipitation with 80% of ammonium sulfate [(NH4)2S04] with constant stirring on a magnetic stirrer at 4°C up to 24h, followed by size-exclusion column chromatography using Sephadex G-100.20 Fractions of 2mL each were collected for total protein determination. Fractions showing maximum absorption at 750nm were collected and evaluated for their enzyme activity. Enzyme-positive fractions with higher enzyme activity were combined together, lyophilized and stored at −20°C for further characterization.
Assay methodsTotal protein determination and lipase assayThe total protein contents of the crude and purified enzyme extracts were determined by the method of Lowry et al.21 using bovine serum albumin as the standard. Lipase activity was determined by the method described by Mustranta.22 One unit was defined as the amount of the enzyme required for the release of 1μmol of fatty acid per min under the assay conditions. Enzyme activity was expressed as units per mL of the enzyme extract.
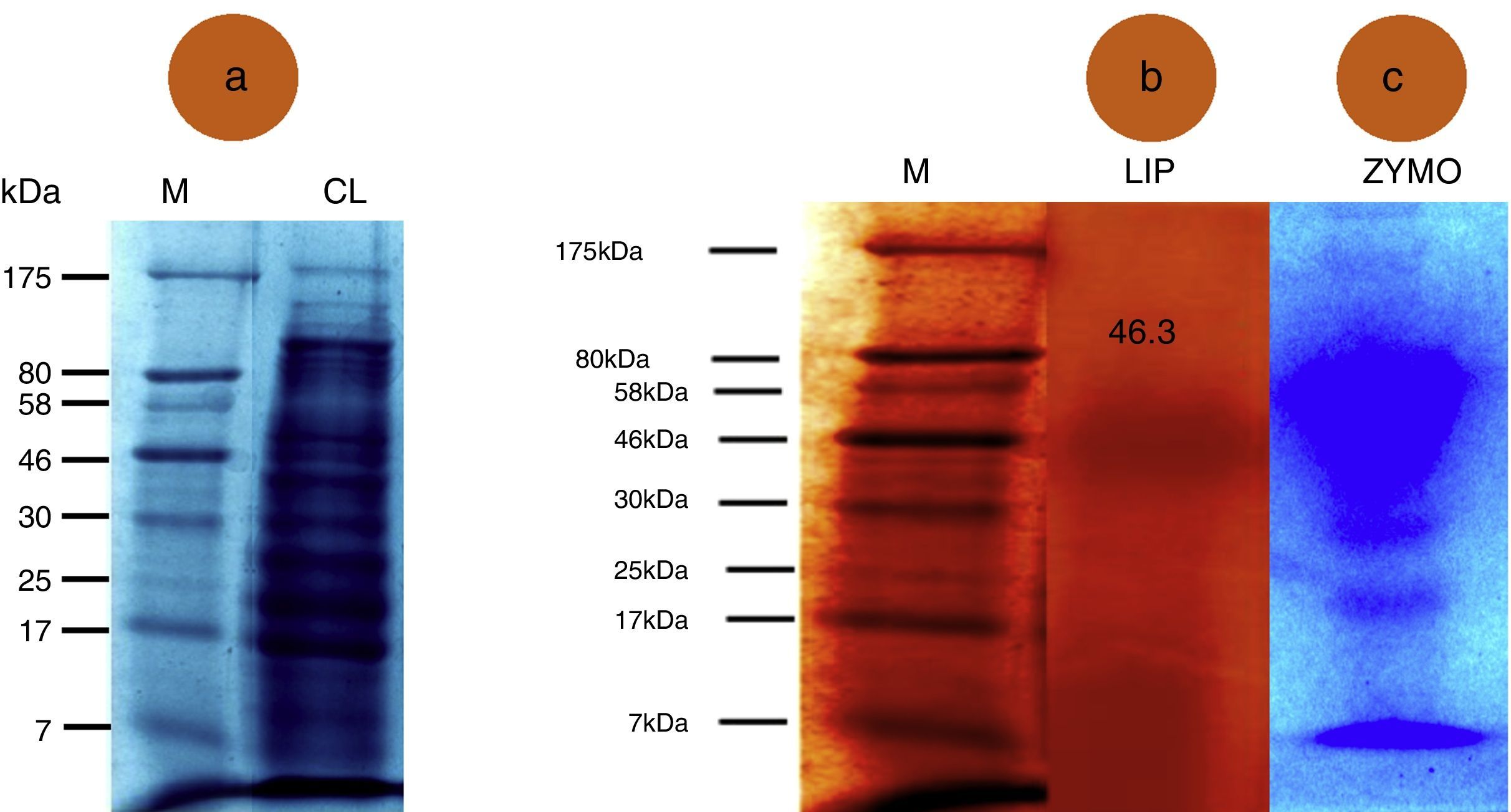
SDS-PAGE and zymographic analysisThe samples (crude and purified lipase) were resolved by electrophoresis in 10% SDS-PAGE gels for protein molecular weight determination.23 Relative positions of the bands were analyzed using a Bio-Rad gel documentation system. Molecular weight protein markers ranging from 7 to 175kDa (Bangalore Genei Ltd.) were used for SDS-PAGE.
Zymographic analysis of the lipase was performed as described by Prim et al.24 in a 10% non-denaturing polyacrylamide gel in a discontinuous buffer system, with olive oil added as the substrate prior to the addition of ammonium persulfate and polymerization. Activity bands were observed for a short time (≤1min) under UV illumination in the gel documentation system and photographed.
Enzyme characterizationEffects of pH (3.0–10.0), temperature (30–90°C), substrate concentration (0.25–2.0%), enzyme–substrate reaction time (10–90min), commercial detergents [sodium dodecyl sulfate (SDS), Ariel, Henko, Surf Excel and Rin (0.1%, w/v)], metal ions (K+, Na+, Ca2+, Mn2+, Hg2+, Mg2+, Fe2+, Cu2+, Ag+, Zn2+ and Co2+), surfactant solutions (Triton X-100, Tween-20 and sodium lauryl sulfate (SLS), 0.03%, w/v) and inhibitors [ethylenediaminetetraacetic acid (EDTA), phenylmethylsulfonyl fluoride (PMSF) (0.5%, w/v) and β-mercaptoethanol, 0.5%, v/v] on enzyme activity were evaluated.
All experiments were carried out in triplicates (n=3) and repeated 3 times. Each value is an average of 3 parallel replicates. The±sign and the error bars represent a standard deviation of the mean. For each individual experiment, one-way ANOVA was calculated using the SPSS 16.0 software. Least significant differences were also calculated using Duncan's new multiple range tests.
Results and discussionFor production of secondary metabolites and enzymes, fungi represent a vast array of filamentous saprophytic organisms with a remarkable genetic repertoire. Although they are of very high biotechnological interest, fungi have not been widely investigated yet for lipase production, and thus there is little information available on lipase-producing fungi. Several enzymes have been biosynthesized by Aspergillus species25 and other bacterial and fungal species under SSF conditions, but only a few reports are available on lipase production by A. terreus in a solid-state system and enzyme characterization.
In one of the studies, agar media supplemented with olive oil and a dye were used for the screening and selection of high lipase-producing Aspergillus species17; however, the media were not appropriate for authentic production of the enzymes and did not unambiguously differentiate between cell-bound and extracellular activities. The native isolate of A. terreus was cultured in liquid medium with olive oil (1%, v/v) as a lipase inducer, because lipase activities associated with cells of A. terreus are induced and enhanced by a lipid substrate present in the fermentation medium.
However, olive oil is a costly component of any lipase production medium. Therefore, its replacement with various agro-residual wastes, such as MoC, as alternative substrates can result in cheaper enzyme production and may help to solve pollution problems, otherwise caused by waste disposal. Therefore, in this study MoC was used as a substrate for lipase biosynthesis both in SSF and LSSC,18 and solid-state fermentation was found to be better than submerged fermentation. Wheat bran and gingelly oil cake were also used as substrates by Mala et al.26 in SSF for lipase production by Aspergillus niger MTCC 2594. Lipase production using SSF has a better economic perspective. Similar conclusions were made by Castilho et al.27 while studying lipase production by Penicillium restrictum under SmF and SSF conditions.
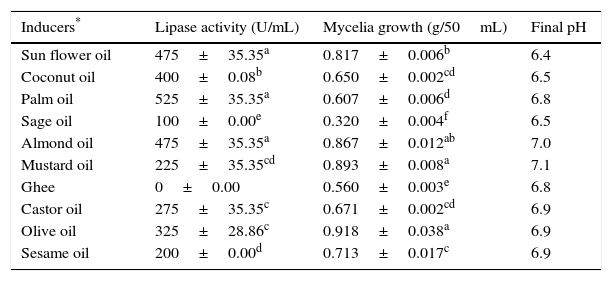
Submerged cultures of A. terreus were carried out in basal medium supplemented with various natural oils (1% v/v) as inducers for the biosynthesis of the extracellular lipase in the medium. Among the oils used in the present study, maximum lipase production was achieved using palm oil (525±35.35U/mL), followed by sunflower (475±35.35U/mL) and almond oils (475±35.35U/mL) (Table 1).
Effect of inducers on lipase production and mycelia growth.
| Inducers* | Lipase activity (U/mL) | Mycelia growth (g/50mL) | Final pH |
|---|---|---|---|
| Sun flower oil | 475±35.35a | 0.817±0.006b | 6.4 |
| Coconut oil | 400±0.08b | 0.650±0.002cd | 6.5 |
| Palm oil | 525±35.35a | 0.607±0.006d | 6.8 |
| Sage oil | 100±0.00e | 0.320±0.004f | 6.5 |
| Almond oil | 475±35.35a | 0.867±0.012ab | 7.0 |
| Mustard oil | 225±35.35cd | 0.893±0.008a | 7.1 |
| Ghee | 0±0.00 | 0.560±0.003e | 6.8 |
| Castor oil | 275±35.35c | 0.671±0.002cd | 6.9 |
| Olive oil | 325±28.86c | 0.918±0.038a | 6.9 |
| Sesame oil | 200±0.00d | 0.713±0.017c | 6.9 |
Inducers were added to the basal medium (Saboraud's Dextrose Broth) at 1% (v/v) concentration and initial pH was adjusted to 6.0 for the whole set up. The static culture experiments were performed for 96h at 30°C. The data represent mean±SD of three replicates. Values in the same column carrying different letters are significantly different between the treatments and control at P≤0.05.
However, the maximum biomass was obtained with olive oil, followed by mustard and almond oils, while significantly reduced activity and biomass production were seen with sage oil. This may be due to the enzyme specificity or to the presence of lipase inhibitors or germicidal agents in sage oil. Lipases are inducible enzymes and require the presence of oil inducers, although the role of the inducers in lipase synthesis and stimulation is poorly understood.28 Natural oils such as soybean, corn, sunflower, olive, palm and cotton seed oils are used as inducers for lipase production and as sole carbon sources in the medium, because the carbon chain moiety of the fatty acid present in the triacylglycerol controls lipase synthesis. A similar finding has been reported for Aspergillus carneus,29 where a considerably higher content of palmitic acid in the oil supported good lipase production.
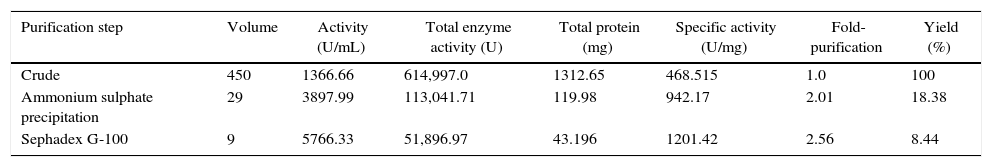
The dialyzed lipase sample contained 113,041.71U of the enzyme with a specific activity of 942.17Umg−1 of protein, representing an 18.38% yield and 2.01-fold purification. The partially purified lipase (dialyzed ammonium sulfate-precipitated fraction) was further fractionated through a Sephadex G-100 column, and fractions 39–45 contained the maximum amounts of the lipase, representing 2.56-fold purification (Table 2). Similarly, lipase of Aspergillus awamori was purified to homogeneity, with 10.6-fold purification, a yield of 18.84% and a specific activity of 1862.2U/mg using ammonium sulfate precipitation, followed by Sephadex G-75 gel filtration chromatography, as reported by Jin-lan et al.30 Lipase from Rhizopus oryzae was purified to homogeneity using ammonium sulphate precipitation, followed by Q-Sepharose chromatography, which enhanced the recovery of purified extracellular lipase up to 8.6-fold.31
Purification summary of isolated lipase from Aspergillus terreus NCFT 4269.10.
| Purification step | Volume | Activity (U/mL) | Total enzyme activity (U) | Total protein (mg) | Specific activity (U/mg) | Fold-purification | Yield (%) |
|---|---|---|---|---|---|---|---|
| Crude | 450 | 1366.66 | 614,997.0 | 1312.65 | 468.515 | 1.0 | 100 |
| Ammonium sulphate precipitation | 29 | 3897.99 | 113,041.71 | 119.98 | 942.17 | 2.01 | 18.38 |
| Sephadex G-100 | 9 | 5766.33 | 51,896.97 | 43.196 | 1201.42 | 2.56 | 8.44 |
SDS-PAGE detected 14 protein bands ranging from 7 to 175kDa in the crude lipase extract (Fig. 1A). The banding pattern was unique representing difference in gene expression. The molecular weight of the purified lipase was 46.3kDa (Fig. 1B). However, when a zymogram was prepared, the lipase sample displayed 4 bands representing the isoenzyme pattern (Fig. 1C). Indeed, A. terreus secrets a mixture of enzymes, among which at least 7 isoenzymes have been reported, which are encoded by a lipase minigene family. Five of the coding genes, namely, lip1 to lip5, from different fungi were sequenced and intensively analyzed, whereas only partial sequencing was performed for lip6 and lip7.32 Recently, a new gene, lipJ08, has been sequenced from Candida rugose.33
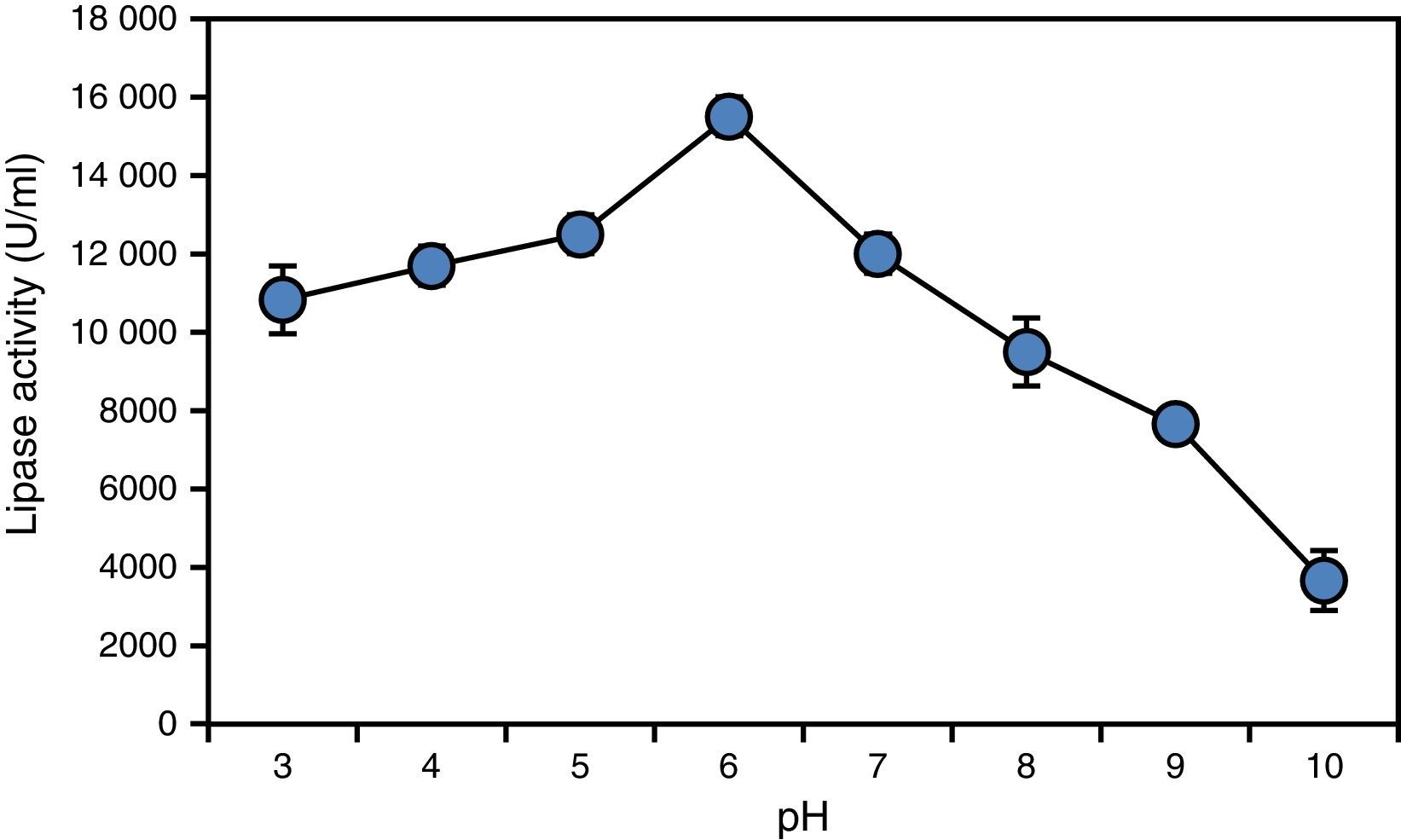
Enzyme characterization revealed that the lipase obtained from A. terreus was significantly active over a broad range of pH, with a pH optimum of 6.0 (Fig. 2). However, the enzyme became unstable at pH values over 9.0 and below 6.0. The same pH optimum was observed for the lipase from C. rugosa.34
The activity and stability levels shown by the lipase from A. terreus at moderately acidic pH values are not common among lipases produced by fungi, which are generally reported to be most active at alkaline and neutral pH values. For instance, Kamini et al.35 reported that the lipase obtained from A. niger exhibited maximum activity at pH 7.0, which is different from our findings. Thus, this lipase showing high activity and stability under moderately acidic conditions deserves to be evaluated for industrial exploitation.
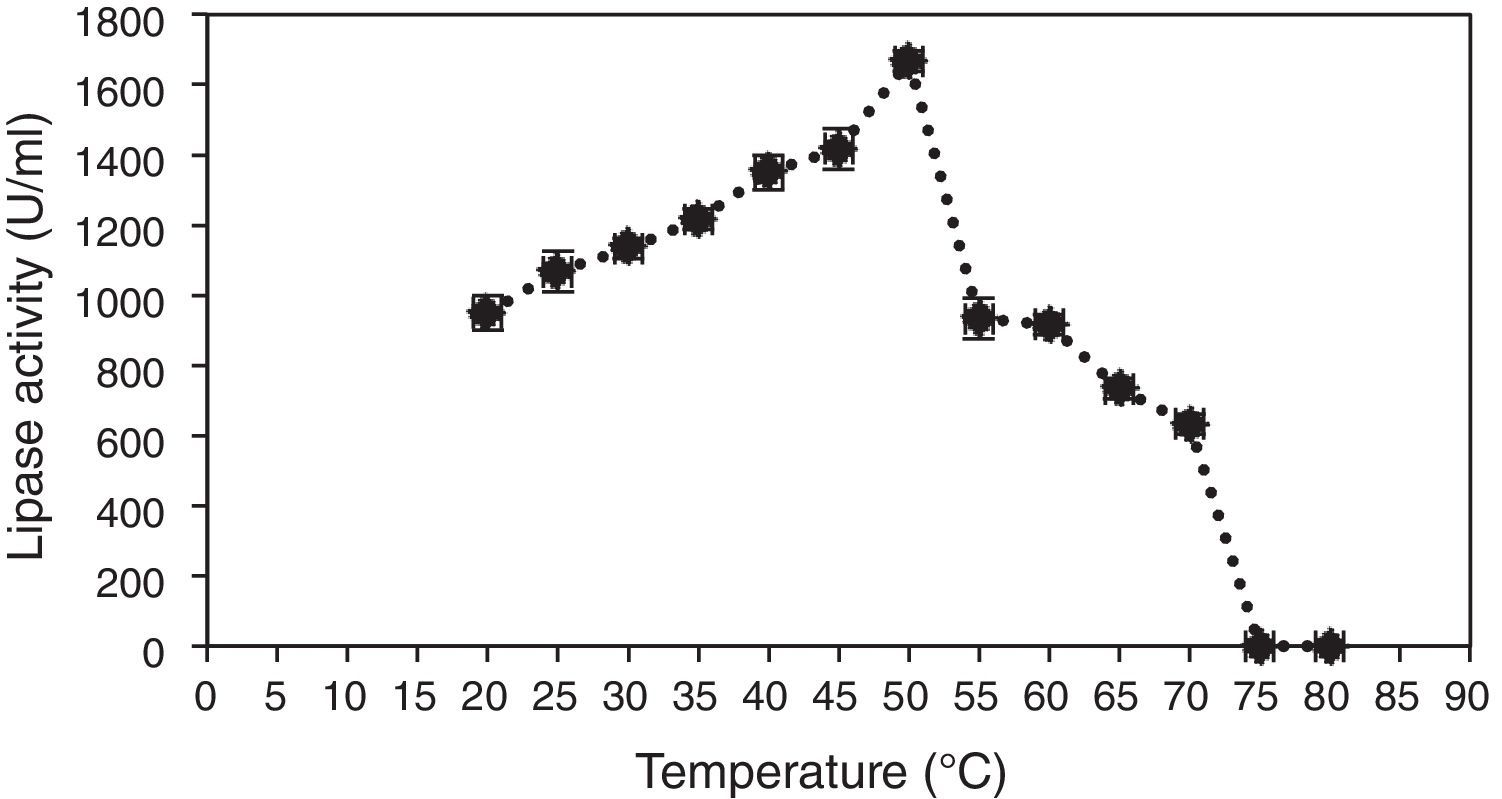
In order to evaluate the effect of temperature, 100μL of lipase was mixed with 0.5mL of 1% olive oil and 0.4mL of 50mM phosphate buffer, pH 7.0, and incubated at temperatures ranging from 20 to 80°C for 30min. Then, activity of the lipase was determined, and the results showed that the enzyme hydrolyzed olive oil in the range of temperatures from 30 to 80°C, with the maximum activity observed at 50°C (Fig. 3).
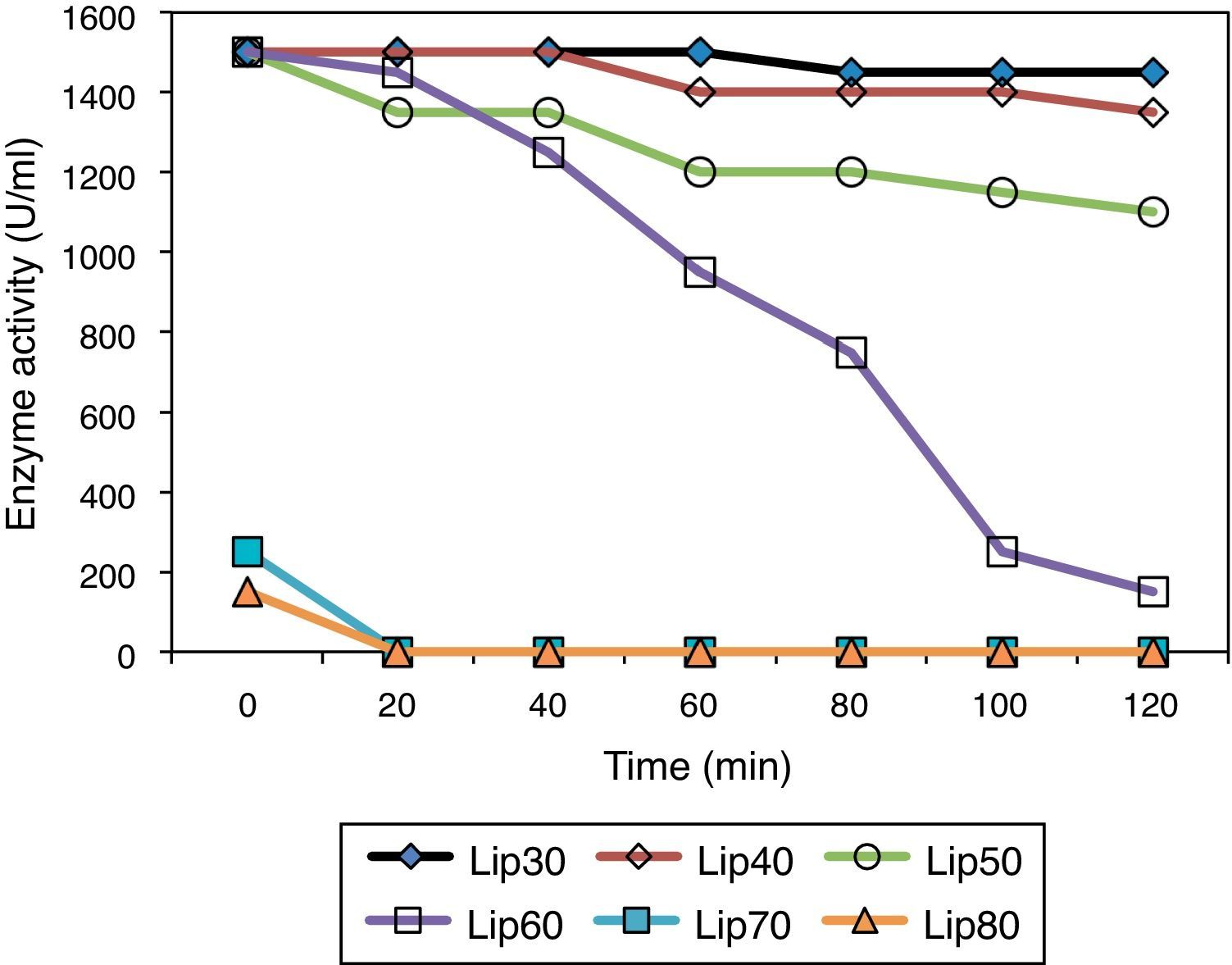
The thermostability of the lipase was also studied, it was found to be highly stable at 60°C for 1h, with more than 80% of its activity preserved (Fig. 4), indicating a high thermal stability of the extracted purified protein. Moreover, the enzyme purified from the SSF system had higher thermal stability compared with that produced by submerged fermentation, indicating a superiority of SSF over the submerged fermentation system.
Although thermostable lipases are rare among fungal and yeast enzymes,36 a temperature-resistant lipase from A. niger has been reported earlier to be active between 40 and 55°C.35 Most lipases from fungi of the genus Penicillium exhibit maximum activities at temperatures in the range from 25 to 45°C.37 One exception is Penicillium aurantiogriseum, which produces a thermostable lipase with maximum activity at 60°C and pH 8.0.36 Thus, the new lipase from A. terreus, showing high activity and stability in the temperature range of 50–60°C, may be appropriate for applications in biocatalytic processes.
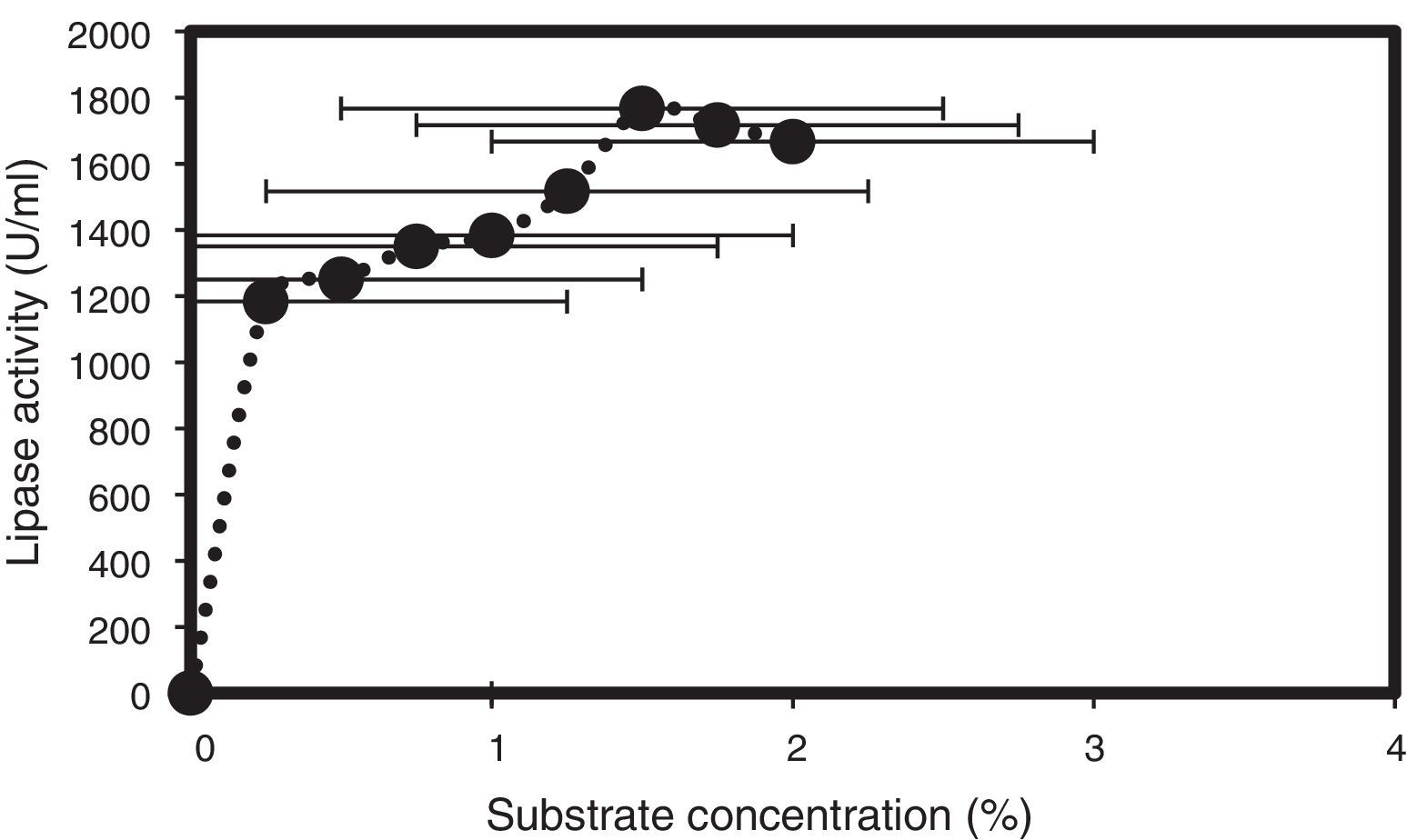
Olive oil was tested at different concentrations (0.25–2.5%) as a substrate for lipase hydrolysis, and the maximum lipase activity was observed at a concentration of 1.5% (v/v) (Fig. 5), even though 1% (v/v) of olive oil was used for the initial screening. Similarly, Gulati et al. (1998) reported that 2.0% (v/v) of neem oil was best for lipase production by A. terreus, and particularly high lipase levels were obtained with cod liver oil, as reported by Esakkiraj et al.38 for Staphylococcus epidermidis.
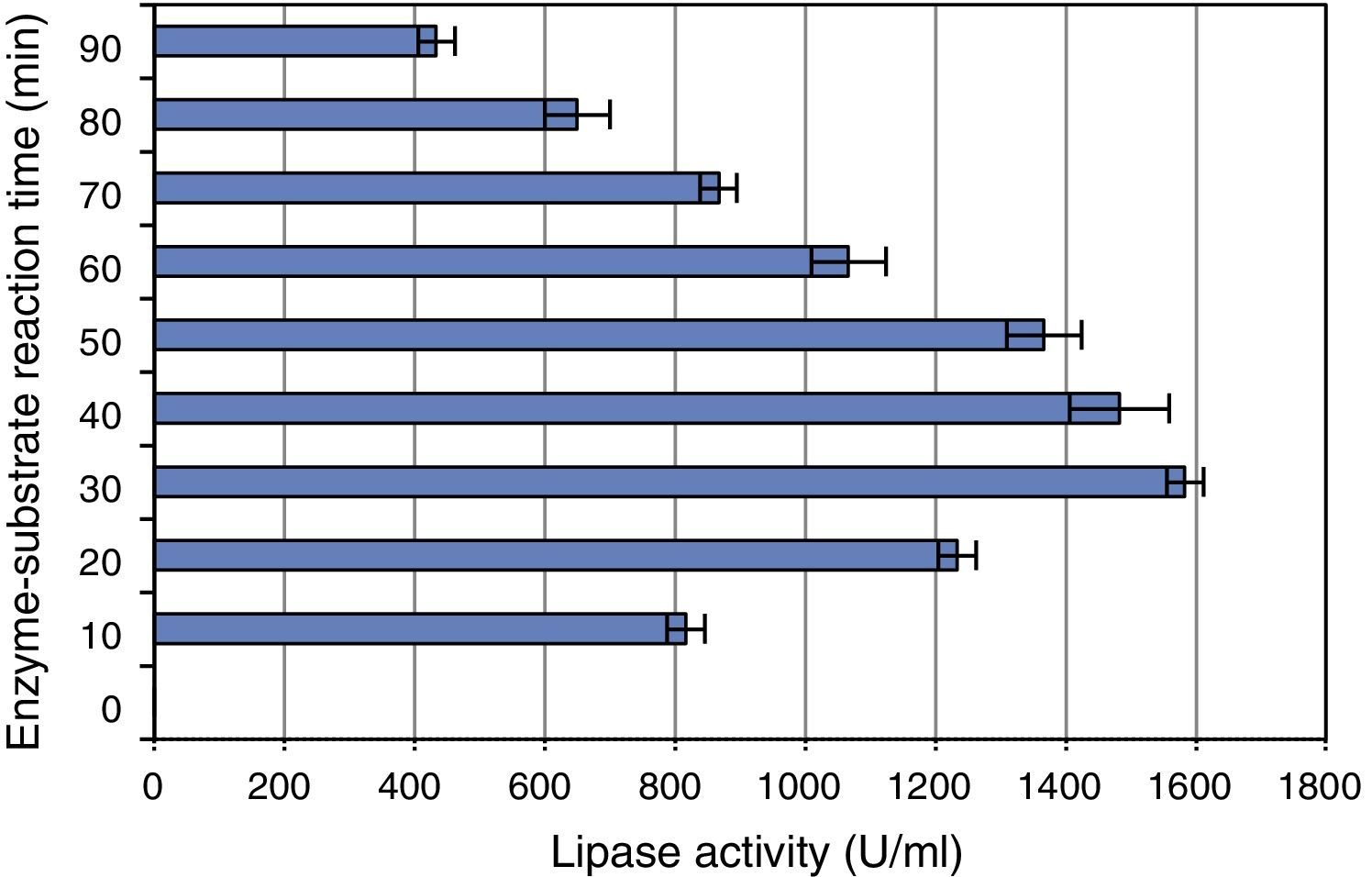
The effect of incubation period (enzyme–substrate reaction time) on the lipase was determined by incubating the substrate and enzyme at 37°C for different periods of time. Activity of the enzyme was determined after incubation for 10, 20, 30, 40, 50, 60, 70, 80 and 90min at the optimum conditions. The maximum activity was obtained after incubation for 30min (Fig. 6), and further increase in the incubation time led to a gradual decrease in enzyme activity.
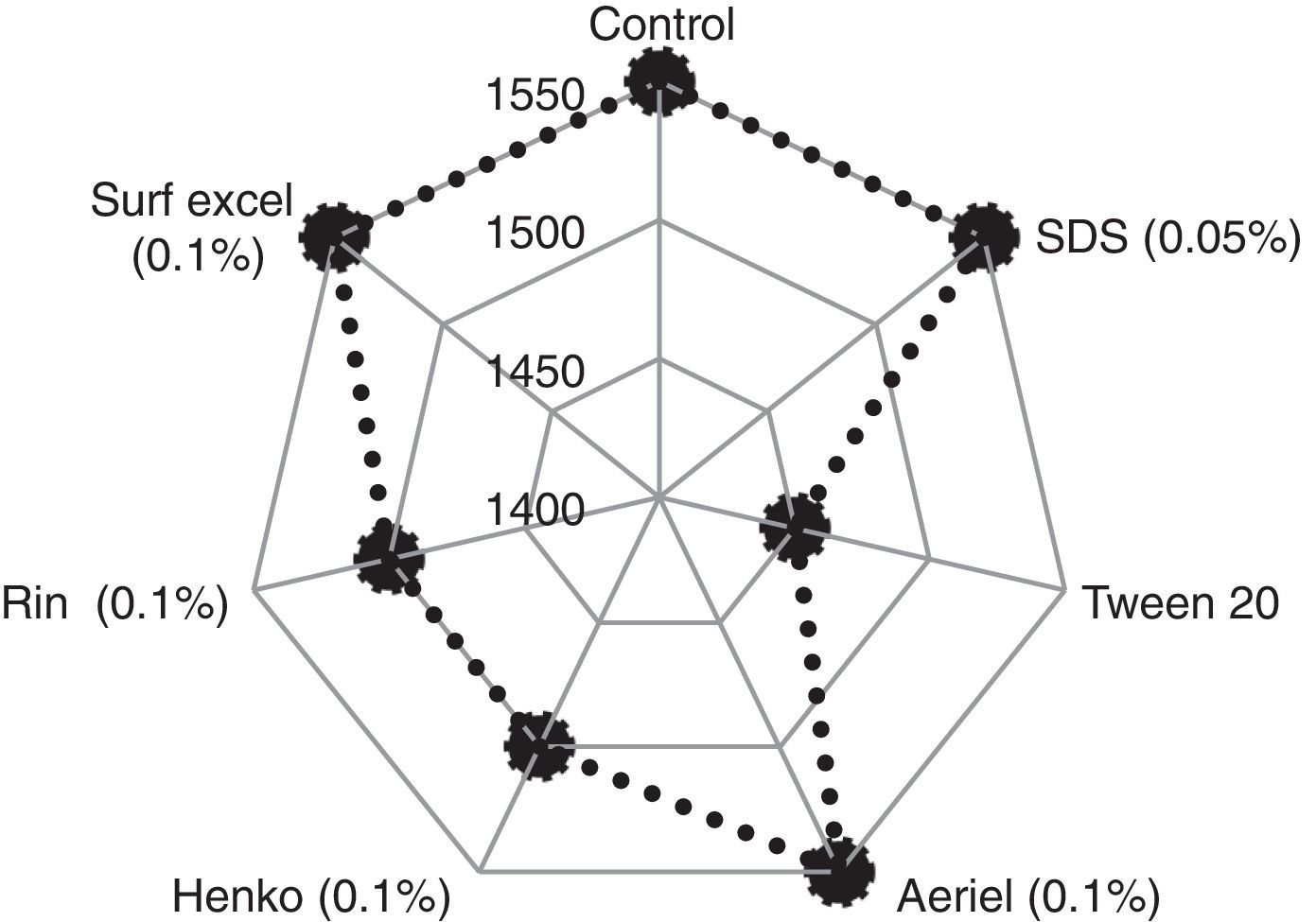
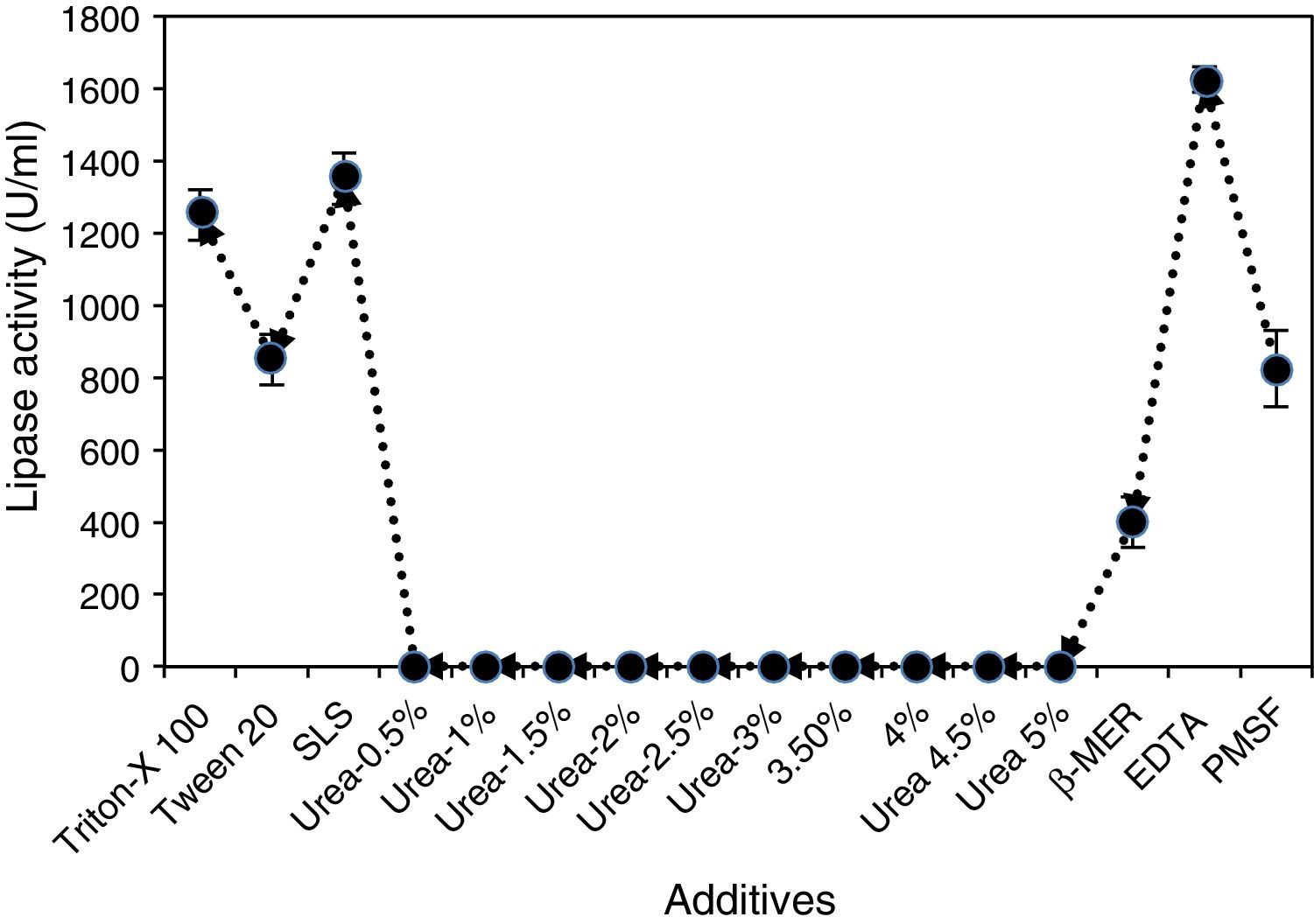
The fungal lipase showed significant stability during a 30-min incubation at 30°C in the presence of all detergents tested at a concentration of 1mg/mL. The enzyme was found to be stable in the presence of Tween 20, the anionic detergent SDS and all commercial detergents, among which SDS and commercial detergents such as Surf Excel and Henko had the least effect on the lipase stability (Fig. 7). Therefore, the lipase may be used in laundry detergents to facilitate the removal of triglyceride soils from human sebum, which are difficult to remove under normal washing conditions.
Similar results were reported for the lipase from A. niger,35 and more than 70% of residual lipase activity was also observed by Gulati et al.39 in the presence of commercial detergents available on international and Indian markets, viz., Surf, Ariel, Nirma, Wheel, and Fena.
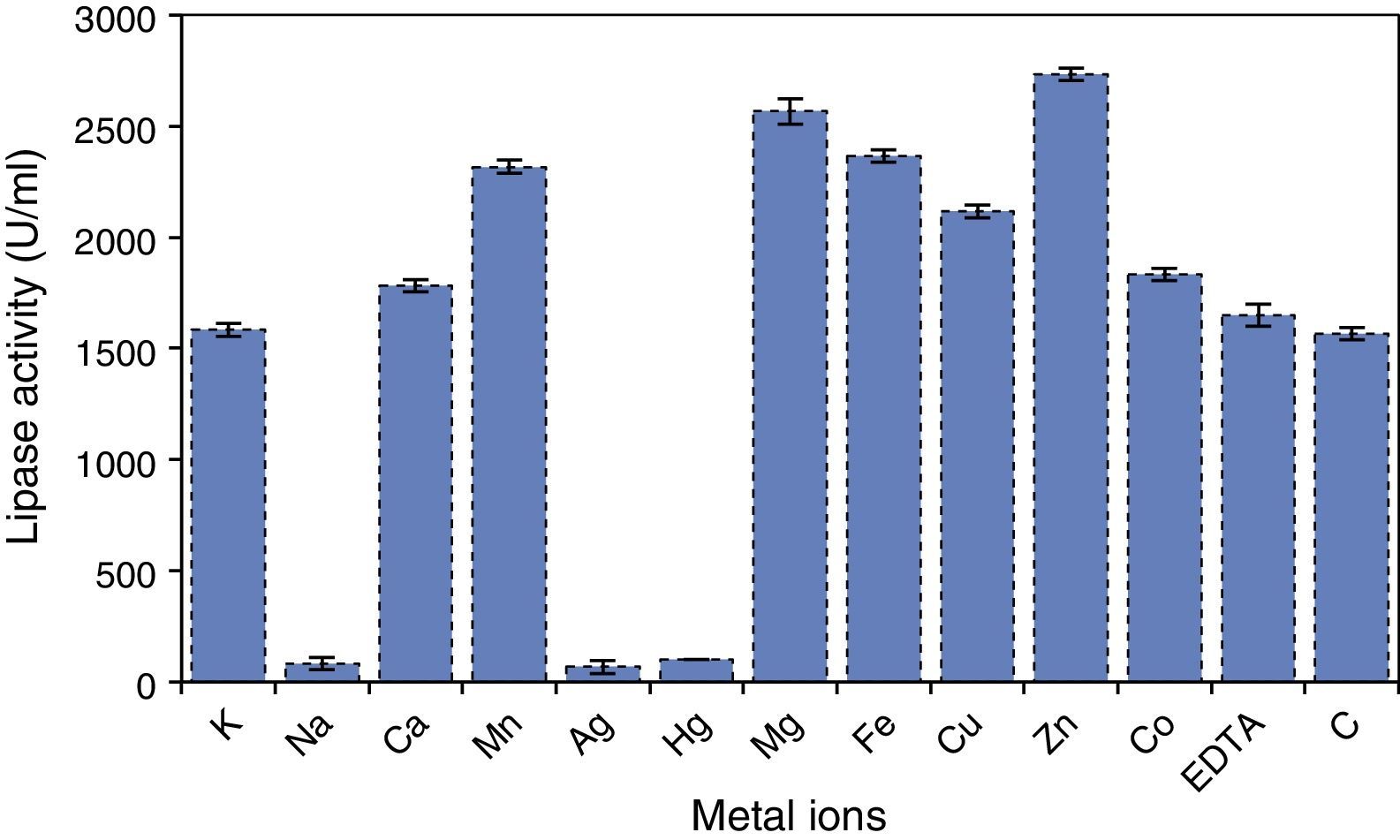
Effects of different metal ions on A. terreus lipase activity and stability were also studied, and the results (Fig. 8) showed that the purified lipase was strongly inhibited by Ag2+ and Hg2+, losing more than 90% of its activity within 30min. This may be due to an alteration in the enzyme conformation, which has been earlier reported for other lipases.36
Most of the metal ions significantly increased enzyme activity, and maximum activity was observed when the enzyme was incubated with Zn2+, followed by Mg2+ and Fe2+. Mn2+ exhibited at par result with Fe2+. The effect of iron on lipase stability was similar to that observed in the case of the lipase from Streptomyces fradiae var. k11.40 Ca2+ ion has been reported to have a positive effect on activity and stability of lipases; however, no noteworthy influence on A. terreus lipase was observed in this study. Lima et al.36 reported Cu2+ as a strong inhibitor of lipase activity, but in this study no strong inhibition of A. terreus lipase activity was observed under the test conditions. The lipase activity and stability was not significantly affected in the presence of EDTA, which suggests a metal-independent nature of A. terreus lipase (Fig. 8). Zhang et al.40 also reported that lipases obtained from Streptomyces rimosus and S. fradiae var. k11 were unaffected by EDTA. The active center of all known lipases contains serine; nevertheless, some lipases are resistant to inactivation by serine-reactive agents. PMSF (1mM) inhibited lipase activity by approximately 30%, possibly suggesting the presence of a hydrophobic lid hindering access to the catalytic site.
Also, the enzyme was not significantly affected in the presence of 0.03% (w/v) SLS, similar to P. aurantiogriseum lipases, which were reported to be stable, with a significant residual activity, in SDS.36 The lipase was not significantly affected in the presence of Triton X-100 and retained 90% of its activity (Fig. 9). The reducing agent β-mercaptoethanol had a remarkable effect on the lipase and reduced its activity by more than 77% when the purified enzyme was treated with β-mercaptoethanol at a concentration of 0.03% (v/v) (Fig. 9).
ConclusionsIn this study, A. terreus was found to be a potent producer of an extracellular lipase. The enzyme showed high thermal stability, activity in a broad range of pH values, with the highest stability at a slightly acidic pH, and a notable stability in the presence of detergents, additives, inhibitors and metal ions, which makes it unique. This new extracellular lipase can be of considerable biotechnological potential. Thus, our results justify further studies to determine possible applications of this enzyme in biocatalysis in organic media. Also, more research is needed to optimize the fermentative process parameters for this strain for enhanced lipase production.
Conflicts of interestThe authors declare no conflicts of interest.
This work was financially supported by UGC-RGNF Scheme, New Delhi, India under Grant No. F.14-2(SC)/2008 (SA-III) dated 31 March 2009. We also thank Prof. Keshab C. Mondal, Head, Department of Microbiology, Vidyasagar University, Midnapore, West Bengal, for allowing to use necessary laboratory facilities to BKS.