Enterobacter cloacae and E. aerogenes have been increasingly reported as important opportunistic pathogens. In this study, a high prevalence of multi-drug resistant isolates from Brazil, harboring several β-lactamase encoding genes was found. Several virulence genes were observed in E. aerogenes, contrasting with the E. cloacae isolates which presented none.
Bacteria belonging to the Enterobacter genus are gram-negative facultative anaerobes and widely distributed in nature. In the last decades, species of the genus Enterobacter have aroused greater concern, since they are increasingly associated with nosocomial infections, especially in immunocompromised patients.1Enterobacter species are members of ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter species) which are described as the leading cause of nosocomial infections throughout the world.2,3 Among the species, E. aerogenes and E. cloacae stand out as opportunistic pathogens, especially in patients on mechanical ventilation.4
Enterobacter sp., including E. aerogenes and E. cloacae easily acquire numerous genetic mobile elements containing resistance and virulence genes, which strongly contribute to the increased pathogenicity of these bacteria. These species are extended spectrum β-lactamase producers and posses intrinsic resistance to ampicillin, amoxicillin, first-generation cephalosporins and cefoxitin, due to the production of constitutive AmpC β-lactamase. Thus Enterobacter sp. may develop antimicrobial resistance during treatment, limiting therapeutic options.3,4
Although studies have been pointing to the increasing prevalence of E. cloacae and. E. aerogenes as opportunistic pathogens, frequently associated with multidrug-resistance, little is known about their virulence mechanisms.1 Therefore, the aim of this study was to characterize E. aerogenes and E. cloacae clinical isolates from Brazil regarding the genetic relationships, antimicrobial susceptibility profile, and the presence of virulence and β-lactamase encoding genes.
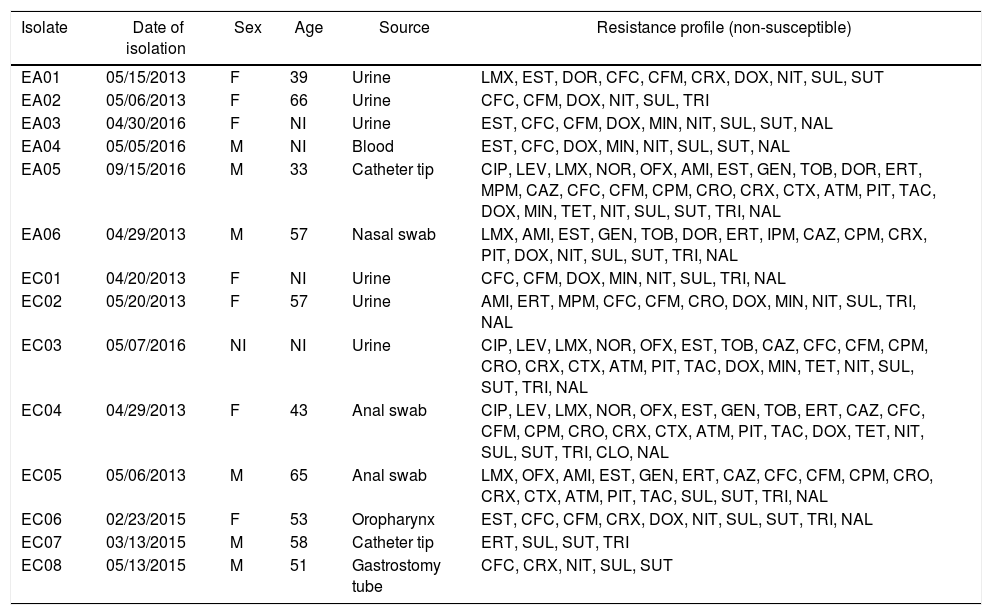
In this study a total of 14 Enterobacter isolates were included, being eight E. cloacae (EC) and six E. aerogenes (EA). Among the isolates, seven were obtained from male patients, six from female patients and one was not informed. These isolates were obtained between March 2013 to September 2016 from three Brazilian tertiary hospitals located in different regions: the southeast (Ribeirão Preto city, São Paulo state), northeast (Teresina city, Piauí state) and the north region (Manaus city, Amazonas state). These isolates were obtained from different sources such as urine, blood, anal and nasal swabs, catheter tip, oropharynx and gastrostomy tube (Table 1).
General data relating to six E. aerogenes (EA) and eight E. cloacae (EC) isolates from this study.
| Isolate | Date of isolation | Sex | Age | Source | Resistance profile (non-susceptible) |
|---|---|---|---|---|---|
| EA01 | 05/15/2013 | F | 39 | Urine | LMX, EST, DOR, CFC, CFM, CRX, DOX, NIT, SUL, SUT |
| EA02 | 05/06/2013 | F | 66 | Urine | CFC, CFM, DOX, NIT, SUL, TRI |
| EA03 | 04/30/2016 | F | NI | Urine | EST, CFC, CFM, DOX, MIN, NIT, SUL, SUT, NAL |
| EA04 | 05/05/2016 | M | NI | Blood | EST, CFC, DOX, MIN, NIT, SUL, SUT, NAL |
| EA05 | 09/15/2016 | M | 33 | Catheter tip | CIP, LEV, LMX, NOR, OFX, AMI, EST, GEN, TOB, DOR, ERT, MPM, CAZ, CFC, CFM, CPM, CRO, CRX, CTX, ATM, PIT, TAC, DOX, MIN, TET, NIT, SUL, SUT, TRI, NAL |
| EA06 | 04/29/2013 | M | 57 | Nasal swab | LMX, AMI, EST, GEN, TOB, DOR, ERT, IPM, CAZ, CPM, CRX, PIT, DOX, NIT, SUL, SUT, TRI, NAL |
| EC01 | 04/20/2013 | F | NI | Urine | CFC, CFM, DOX, MIN, NIT, SUL, TRI, NAL |
| EC02 | 05/20/2013 | F | 57 | Urine | AMI, ERT, MPM, CFC, CFM, CRO, DOX, MIN, NIT, SUL, TRI, NAL |
| EC03 | 05/07/2016 | NI | NI | Urine | CIP, LEV, LMX, NOR, OFX, EST, TOB, CAZ, CFC, CFM, CPM, CRO, CRX, CTX, ATM, PIT, TAC, DOX, MIN, TET, NIT, SUL, SUT, TRI, NAL |
| EC04 | 04/29/2013 | F | 43 | Anal swab | CIP, LEV, LMX, NOR, OFX, EST, GEN, TOB, ERT, CAZ, CFC, CFM, CPM, CRO, CRX, CTX, ATM, PIT, TAC, DOX, TET, NIT, SUL, SUT, TRI, CLO, NAL |
| EC05 | 05/06/2013 | M | 65 | Anal swab | LMX, OFX, AMI, EST, GEN, ERT, CAZ, CFC, CFM, CPM, CRO, CRX, CTX, ATM, PIT, TAC, SUL, SUT, TRI, NAL |
| EC06 | 02/23/2015 | F | 53 | Oropharynx | EST, CFC, CFM, CRX, DOX, NIT, SUL, SUT, TRI, NAL |
| EC07 | 03/13/2015 | M | 58 | Catheter tip | ERT, SUL, SUT, TRI |
| EC08 | 05/13/2015 | M | 51 | Gastrostomy tube | CFC, CRX, NIT, SUL, SUT |
Note: F, female; M, male, NI, not informed; AMI, amikacin; ATM, aztreonam; CFC, cefaclor; CPM, cefepime, CFM, cefixime; CRO, ceftriaxone; CAZ, ceftazidime; CTX, cefotaxime; CRX, cefuroxime; CIP, ciprofloxacin; CLO, chloramphenicol; DOR, doripenem; DOX, doxycycline; ERT, ertapenem; GEN, gentamicin; IPM, imipenem; LVX, levofloxacin; LMX, lomefloxacin; MPM, meropenem; MIN, minocycline; NAL, nalidixic acid; NIT, nitrofurantoin; NOR, norfloxacin; OFX, ofloxacin; PIT, piperacillin-tazobactam; EST, streptomycin; SUL, sulphonamide; TET, tetracycline; TAC, ticarcillin-clavulanate; TRI, trimethoprim; TOB, tobramycin; SUT, trimethoprim-sulfamethoxazole.
Bacteria identification were performed by matrix-assisted laser desorption–ionization time of flight mass spectrometry (MALDI-TOF MS) using the Vitek® MS system (bioMérieux, Marcy l’Etoile, France), according to the manufacturer's recommendations.
The antimicrobial susceptibility tests were performed by disc diffusion in Mueller-Hinton Agar (Oxoid), as recommended by the Clinical Laboratory Standards Institute (CLSI).5 Thirty-two different antibiotic discs (Oxoid) were tested: amikacin (30μg), aztreonam (30μg), cefaclor (30μg), cefepime (30μg), cefixime (5μg), cefotaxime (30μg), ceftriaxone (30μg), ceftazidime (30μg), cefuroxime (30μg), ciprofloxacin (5μg), chloramphenicol (30μg), doripenem (10μg), doxycycline (30μg), ertapenem (10μg), gentamicin (10μg), imipenem (10μg), levofloxacin (5μg), lomefloxacin (10μg), meropenem (10μg), minocycline (30μg), nalidixic acid (30μg), nitrofurantoin (300μg), norfloxacin (10μg), ofloxacin (5μg), piperacillin-tazobactam (100/10μg), streptomycin (10μg), sulphonamide (300μg), tetracycline (30μg), ticarcillin-clavulanate (75/10μg), trimethoprim (5μg), tobramycin (10μg) and trimethoprim-sulfamethoxazole (1.25/23.75μg). The strains E. coli ATCC® 25922 and ATCC® 35218, and Pseudomonas aeruginosa ATCC 27853 were used as quality control for these experiments. Although cefuroxime is considered as intrinsic resistance for E. cloaceae and E. aerogenes by the CLSI,5 this antibiotic was added in the study because it is recommended by Magiorakos et al.6 for these species. The isolates were classified as multi-drug resistant (MDR), extensively drug-resistant (XDR) and pandrug-resistant (PDR), according to the criteria established by Magiorakos et al.6 Isolates which did not fit the previous definitions were designated as not classified (NC).
Genomic DNA was extracted using the phenol/chloroform method as described by Covone et al.7 The concentration and purity were determined using a DS-11+Spectrophotometer (DeNovix, USA).
PCR reactions were performed for the detection of the following β-lactamase encoding genes: blaCTX-M-Gp1, blaCTX-M-Gp2, blaCTX-M-Gp8, blaCTX-M-Gp9, blaSHV, blaPER, blaVEB, blaGES, blaKPC, blaVIM, blaOXA-48-like, blaIMP, blaSPM, blaSIM, blaGIM, and blaNDM using primers and protocols previously described.8–11 All PCR were performed using positive and negative controls.
Detection of virulence genes were performed by PCR for the following genes: allS (allantoin metabolism), entB, ybtS and iutA (siderophores), kfu (iron transport and phosphotransferase function), mrkD (adhesin type 3 fimbriae), rmpA (regulator of mucoid phenotype A) according to Compain et al.,12 and ycfM (outer membrane lipoprotein) and fimH (adhesive subunit of type 1 fimbriae) according to El Fertas-Aissani et al.13 All PCR were performed using positive and negative controls.
Confirmation of the amplified genes’ identity was performed by sequencing. One of each amplicon was randomly selected and purified using the Illustra™ GFX PCR DNA and Gel Band Purification Kits (GE Healthcare, Buckinghamshire, UK). The sequencing was performed in an automated sequencer ABI 3500×L Genetic Analyzer platform (Applied Biosystems, USA). The sequences were analyzed using ChromasPro version 1.7.6 software (Technelysium Pty. Ltd) and compared with the sequences available in the BLAST algorithm (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Enterobacterial Repetitive Intergenic Consensus PCR (ERIC-PCR) analysis was performed to evaluate the genetic similarity among the bacterial isolates of this study. ERIC-PCR was performed as previously described by Versalovic et al.14 using primers ERIC1R (5′-ATGTAAGCTCCTGGGGATTCAC-3′) and ERIC2 (5′-AAGTAAGTGACTGGGGTGAGCG-3′). Band profile analysis was performed using the BioNumerics version 5.1 program (AppliedMaths, Keistraat, Belgium) for construction of the similarity dendrogram by the unweighted pair group mean method and Dice's similarity coefficient. Only bands representing amplicons between 500 and 3000bp were considered for this analysis. The ERIC-PCR assays were performed in triplicate to be sure of the reproducibility of the amplified bands.
Among the 14 isolates, eight (57.1%) were classified as MDR according to the criteria established by Magiorakos et al.,6 being five isolates of E. cloacae (EC02, EC03, EC04, EC05 and EC06) and three E. aerogenes (EA01, EA05 and EA06) (Fig. 1). All isolates showed non-susceptibility (i.e. either intermediate or resistance) to sulphonamide; 12 (85.7%) isolates were non-susceptible to cefaclor and nitrofurantoin; 11 (78.6%) to doxycycline; 11 (78.6%) to cefixime, trimethoprim-sulfamethoxazole, trimethoprim and nalidixic acid; nine (64.3%) to streptomycin; eight (57.1%) to cefuroxime; six (42.9%) to lomefloxacin, ertapenem and minocycline; five (35.7%) to ceftazidime, cefepime, ceftriaxone and piperacillin-tazobactam; four (28.6%) to amikacin, gentamicin, tobramycin, ofloxacin, cefotaxime, aztreonam and ticarcillin-clavulanate; three (21.43%) to ciprofloxacin, levofloxacin, norfloxacin, doripenem and tetracycline; two (14.3%) to meropenem and just one isolate (7.14%) to chloramphenicol (Table 1).
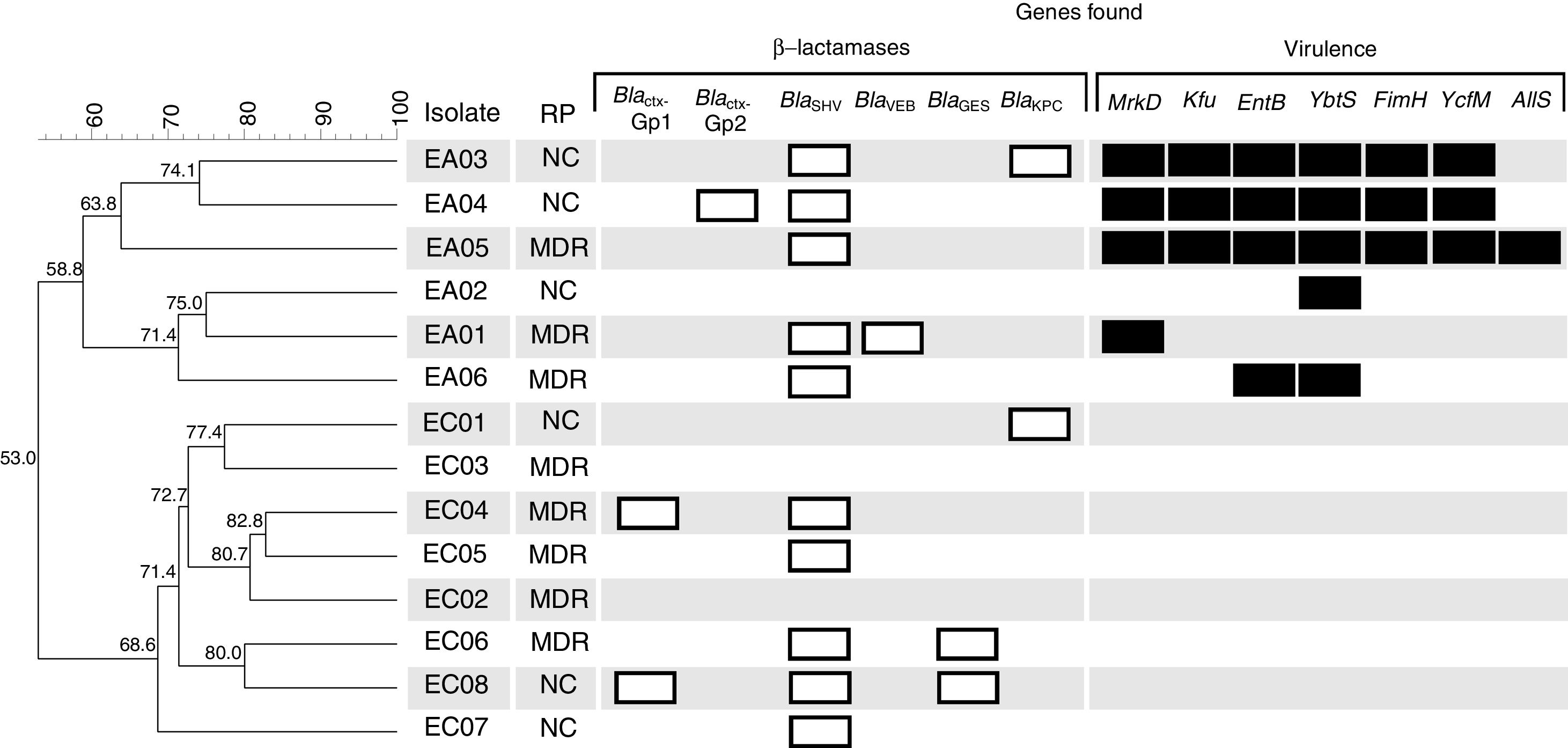
Dendrogram obtained by enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) analysis of six E. aerogenes and eight E. cloacae studied isolates from Brazil. Clusters were determined using the Unweighted Pair Group Mean (UPGMA) method and the Dice similarity coefficient. Similarity (%) among patterns is represented by the numbers beside the nodes. For each isolate their respective resistance profile classification (RP), β-lactamase (white bars) and virulence (black bars) encoding genes are represented. MDR, multi-drug resistant; NC, not classified.
The resistance profile found in the studied isolates corroborate with a study conducted by Cabral et al.,4 which analyzed a Brazilian collection of E. aerogenes and E. cloacae complex isolates, and found the lowest resistance rates in amikacin, gentamicin and tobramycin. However, they found low levels of resistance to trimethoprim–sulfamethoxazole while we found 71.4% resistance for this antibiotic. Fortunately, with the exception of isolate EA06, all isolates were sensitive to imipenem, since papers have described that this antibiotic remains one of the most effective in combating E. cloacae infections. However, there are several works reporting the adaptive response of clinical isolates of Enterobacter sp. to imipenem by regulation of porins.4,15
In this study 16 different β-lactamase encoding genes belonging to group 2 and 3, according to Bush and Jacoby16 were investigated, and a total of 18 amplicons from six different genes (blaCTX-M-Gp1, blaCTX-M-Gp2, blaSHV, blaVEB, blaGES, and blaKPC) were detected (Fig. 1). The most prevalent gene was blaSHV, detected in 10 isolates (EA01, EA03, EA04, EA05, EA06, EC04, EC05, EC06, EC07 and EC08). The following genes were found in two isolates each blaCTXM-Gp1 (EC04 and EC08), blaKPC (EC01 and EC05) and blaGES (EC06 and EC08). The blaCTX-M-Gp2 and blaVEB genes were found in just one isolate each (EA04 and EA01, respectively) (Fig. 1). The blaCTX-M-Gp8, blaCTX-M-Gp9, blaPER, blaVIM, blaOXA-48-like, blaIMP, blaSPM, blaSIM, blaGIM, and blaNDM were not detected. These results are in contrast to those described by Cabral et al.,4 which did not find blaSHV in Entorobacter sp. clinical isolates from Brazil, but they found a high prevalence of blaKPC. In addition, Pereira et al.17 studied 21 carbapenem-resistant E. aerogenes strains isolated from a Brazilian tertiary hospital in Juiz de Fora city located in southeast Brazil. They observed 52.4% of blaSHV and 28.6% of blaCTX-M groups in their collection.
Different mechanisms of resistance may be associated with non-susceptibility to β-lactam antibiotics in Gram-negative bacteria; however the enzymatic, for intrinsic and acquired β-lactamases, is the main mechanism.16,18 Among the isolates, EC01 and EA06 presented the blaKPC gene but did not present non-susceptibility to extended-spectrum cephalosporins or carbapenems, and the isolates EA05 and EA06 were non-susceptible to carbapenems and only the blaSHV gene was detected. Therefore, the diversity of mechanisms, such as porin loss, efflux pumps or lack of expression of acquired genes and the presence of other unsearched β-lactamases may be associated with these inconsistencies between genes found and expressed phenotype.3
Nine virulence genes were investigated and, surprisingly, all E. aerogenes isolates presented one or more genes, whereas no E. cloacae isolate presented virulence genes. The genetic similarity dendrogram constructed with the ERIC-PCR data showed the formation of two large clusters, denominated A and B, comprising the E. aerogenes and E. cloacae isolates, respectively (Fig. 1). The isolate EA05 presented seven virulence genes, followed by EA03 and EA04 with six, EA06 with two, and EA01 and EA02 with one gene (Fig. 1). The obtained virulence and β-lactamase encoding genes’ sequences were deposited in GenBank (www.ncbi.nlm.nih.gov/Genbank) with accession numbers MF622540 to MF622551.
The results found suggest a predominance of virulence genes in E. aerogenes when compared with E. cloacae. In addition to the few studies investigating virulence genes in Enterobacter sp.,19,20 to our knowledge there are no studies comparing the predominance of these genes in E. cloacae and E. aerogenes. However, as the number of isolates from this study does not allow establishing a statistical correlation on this, more detailed studies with a larger collection are required.
The role of the virulence genes found in the isolates is well described for Klebsiella pneumoniae strains. The genes fimH and mrkD encoding adhesins of type 1 and type 3 fimbriae and ycfM are often found in K. pneumoniae and play an important role in the process of bacterial adhesion in various human tissues and also in the formation of biofilms.13 The genes kfu, entB and ybtS are involved in the production of siderophores which are important molecules for the acquisition of iron for bacterial metabolism. These siderophores, especially Kfu, are often found in hypervirulent strains of K. pneumoniae.12 The allS gene associated with assimilation of allantoin is closely associated with K. pneumoniae isolates from liver abscesses.12 Hassan et al.19 studied thirty-two isolates of Enterobacter sp. obtained from clinical urine specimens and identified the fimH gene in 40% of the isolates. Hussain et al.21 studied some virulence factor encoding genes of 75 E. cloacae and nine E. sakazakiiand and observed that all isolates have the ability to produce siderophores. Despite the relevance of Enterobacter sp. as a nosocomial pathogen, the mechanisms of virulence of these species are still unclear due to the scarcity of studies in this area.3
In conclusion, a high prevalence of multi-drug resistance in the studied Brazilian Enterobacter species harboring several β-lactamase encoding genes was found. Moreover, several virulence genes were observed in E. aerogenes, contrasting with the E. cloacae isolates, which did not present any virulence genes. The combination of multi-drug resistance with β-lactamase encoding genes and association with virulence genes, especially in E. aerogenes, is worrying, since studies have shown an increasing incidence of these opportunistic pathogens causing nosocomial infections.
FundingThis work was supported by São Paulo Research Foundation – FAPESP [grant number, 2013/22581-5].
Ethical ApprovalEthical approval was received from the School of Pharmaceutical Sciences of Ribeirão Preto, University of São Paulo, Brazil [approval no.: CEP/FCFRP 362; CAEE 36031914.9.0000.5403].
We are thankful to Dr Johann Pitout and Dr Ana Cristina Gales for providing control strains. We also would like to thank John Carpenter (Ribeirão Preto, SP, Brazil) for the English revision.