The present study aimed to compare two MALDI-TOF identification methods [(a) direct sample identification after pre-incubation; or (b) use of bacteria isolated on pre-culture)] to standard, traditional bench microbiology. A total of 120 quarter milk samples from 40 Holstein lactating cows were screened based on culture-positive results obtained by microbiological culture (reference method) with the following numbers of quarters positive per cow: 4 cows with 1, 8 cows with 2, 12 cows with 3 and 16 cows with 4 infected quarters per cow. For direct identification method, quarter milk samples (n=120) were skimmed by centrifugation (10,000×g/10min) and pre-incubated at 37°C for 12h. After pre-incubation, quarter milk samples were submitted to total bacterial count by flow cytometry and for a preparation protocol for bacterial ribosomal protein extraction followed by MALDI-TOF MS analysis. The direct MALDI-TOF MS identification method compared to microbiological culture correctly identified isolates of coagulase-negative Staphylococci (27.2%), Streptococcus agalactiae (21.8%), Staphylococcus aureus (14.2%), and Streptococcus uberis (5.2%). The pre-incubation protocol of milk samples, associated to the direct identification method by MALDI-TOF MS, did not increase the identification at species level (score >2.0) of pathogens causing subclinical mastitis in comparison to the method without previous incubation.
Conventional microbiological culture of milk samples is based on biochemical tests for identification of microorganisms causing subclinical mastitis. Different bacterial characteristics are evaluated for identification of a single species, such as growth conditions, colony morphology, growth characteristics in selective medium, carbohydrate fermenting capacity, metabolic and antigenic characteristics, and antibiotic susceptibility.1 The procedures are time consuming, and it may take 2–7 days for the complete diagnosis of the causative pathogen of the intramammary infection (IMI).
The mass spectrometry (MS) technique using matrix assisted laser desorption/ionization source (MALDI) and a time-of-flight type mass analyzer (TOF) can be used for rapid identification of bacteria and yeast from colonies previously cultured on solid medium. Such methodology provides a rapid identification of mastitis-causing bacteria by means of the extraction of ribosomal proteins from bacterial colonies cultured on blood agar2 or using a direct transfer protocol.3 However, the MALDI-TOF MS method has been used for direct identification of bacteria from human blood samples as a rapid diagnostic tool in hospital laboratories, since it does not depend on previous bacterial isolation using microbiological culture.4,5 The direct classification of Gram-positive and Gram-negative bacteria at the genus level showed a 100% positive predictive value (PPV) when spectra consisting of joint ribosomal proteins (“fingerprints”) were acquired from blood samples.4,6
Urinary tract pathogens have been also successfully directly identified by MALDI-TOF MS using urine samples with bacterial counts >105cfu/mL. According to Ferreira et al.,7 the direct identification of microorganisms by MALDI-TOF MS in urine samples showed 91.8% agreement at the species level and 92.7% at the genus level, when compared to microbiological culture identification. These results suggested that MALDI-TOF MS allowed the identification of Gram-negative bacteria directly from urine samples in a short period of time when samples contained elevated bacterial counts.7 The MALDI-TOF MS method was also applied to clinical samples of cerebrospinal fluid and correctly classified the pathogens when bacterial counts in samples were between 104 and 106cfu/mL.8
The scores used for the direct (nonculture based) identification MALDI-TOF method (direct-MALDI-TOF) in blood samples were lower than those of isolates obtained from bacterial cultures obtained from hemoculture, indicating that these scores can be influenced by the bacterial count. Previous studies utilized serial dilutions and obtained excellent identification spectra when the bacterial count in the blood sample was ≥106cfu/mL, though the direct classification of microorganisms at the species level (75.8%) by MALDI-TOF MS was done considering scores ≥1.7.9
The total bacterial count is a critical factor for the direct identification of pathogens that cause mastitis by MALDI-TOF MS.3 According to Moussaoui et al.,10 the pre-incubation of blood samples enabled higher precision in the direct identification of bacteria using the MALDI-TOF MS method. Recently, our research group has evaluated the detection limit of MALDI-TOF MS for direct identification, without previous microbiological culture, of bovine mastitis-causing bacteria from milk samples.3 Therefore, we suggested that the non-culture-based protocol could be applied in diagnostic laboratories by subjecting all milk samples to direct MALDI-TOF, and those without a positive identification could be submitted to pre-incubation protocol, being identified by MALDI-TOF MS combined with standard bacteriology. However, the effect of pre-incubation of quarter milk samples from cows affected with subclinical mastitis has not been evaluated using direct identification of bacteria by the MALDI-TOF method. Thus, the objective of the present study was to compare two MALDI-TOF identification methods [(a) direct sample identification after pre-incubation protocol; or (b) use of bacteria isolated on pre-culture] to standard, traditional bench microbiology.
Material and methodsEthics approval was obtained through the Ethical Committee on the Use of Animals of the School of Veterinary Medicine and Animal Science (University of São Paulo, Brazil, protocol number 3002/2013) before the commencement of the study.
Sample collection and bacterial identificationComposite milk samples were collected from all cows on 2 commercial dairies located in the Midwest region of São Paulo State, Brazil. Based upon these results, milk samples were aseptically collected from all quarters of previously culture-positive cows. Microbiological culture (reference method) was performed using procedures consistent with National Mastitis Council guidelines. Briefly, 10μL of milk per sample (quarter) were inoculated on blood agar plates with 5% defibrinated bovine blood. Inverted plates were incubated aerobically at 37°C for 48h and observed every 24h for colony characteristics (shape, size, number, and color), hemolytic ability (presence and type). Gram stain, potassium hydroxide test (KOH) and catalase tests were performed to determine the morphology and differentiation between genera. Specific microbiology procedures such as coagulase, CAMP, esculin, bile esculin and pyr test were performed as described by Oliver et al.11 A total of 120 quarter milk samples from 40 cows were positive, with the following numbers of quarters positive per cow: 4 cows with 1, 8 cows with 2, 12 cows with 3 and 16 cows with 4 infected quarters per cow. All quarter milk samples were also submitted to the nonculture based identification MALDI-TOF method (direct sample identification-MALDI-TOF).
Direct sample identification of mastitis-causing pathogen by MALDI-TOF MSFor direct sample identification of mastitis causing pathogens, fat was removed (skimmed) from 1mL of milk samples by centrifugation (10,000×g/10min, followed by removal of the superior layer of fat. After skimming, milk samples were agitated in a vortex for 30s (Kasvi basic K45 2820, Paraná, Brazil) and submitted to pre-incubation at 37°C for 12h in a water bath with agitation (Solab, SL 155/10, São Paulo, Brazil). After the incubation period, one aliquot of each milk sample (40mL) was taken and submitted for total bacterial count (TBC) using a flow cytometry equipment BactoCount (Bentley Instruments, Chaska, MN, USA).
The remainder of each of the milk samples was submitted to a preparation protocol for bacterial ribosomal protein extraction using the MALDI Sepsityper® kit (Bruker Daltonik, Bremen, Germany). Initially, 200μL of the “Lysis Buffer” solution was added to 1000μL of milk sample, followed by a centrifugation step at 13,000×g during 2min. After centrifugation, the supernatant was discarded using a 1000μL pipette. Next, the pellet was re-suspended in 1000μL of distilled water and 200μL of the “Lysis Buffer” solution, followed by a second centrifugation step at 13,000×g. After discarding the supernatant using a 1000μL pipette, the pellet was diluted in 1000μL of “Washing Buffer” solution, followed by a third round of centrifugation at 13,000×g and disposal of the supernatant.
The bacterial pellet was diluted in 1200μL of a 75% ethanol/water solution (900μL/300μL) in order to inactivate the bacteria. The bacterial sediment was then centrifuged and the supernatant discarded by tube inversion. Next, a second centrifugation step was done to remove the remaining ethanol present in the sample and the supernatant discarded using a pipette. After pellet drying at room temperature, a 70% solution of formic acid was added in enough volume to cover the pellet (∼30–50μL) and lyse the bacterial cells, followed by addition of the same volume of 100% acetonitrile (∼30–50μL). During the final stage of preparation, the samples were centrifuged (13,000×g/2min) to separate the sediment of bacterial cells from the supernatant containing bacterial proteins, consisting mainly of ribosomal proteins (Biotyper 3.0 manual page 157; Bruker Daltonik, Bremen, Germany). Volumes of 1.0μL of the bacterial extract were applied onto steel target plates (mtp 384 Target Polished Steel; Bruker Daltonik, Bremen, Germany), followed by drying at room temperature. The dried supernatant was overlaid with 1.0μL of matrix solution, consisting of α-cyano-4-hydroxy-cinnamic acid diluted in 50% acetonitrile and 2.5% trifluoroacetic acid.
After the protocol for bacterial ribosomal protein extraction, the aliquots were submitted for MALDI-TOF MS analysis as described by Barreiro et al.2 The Bruker Bacterial Test Standard (BTS) was used for the mass calibration and instrument parameter optimization. Briefly, the mass spectra were obtained using an Autoflex III (Bruker Daltonik, Billerica, USA) mass spectrometer and were collected within a 2000–20,000m/z mass range. Spectral data processing was done using the Biotyper 3.0 (Bruker Daltonik, Bremen, Germany) computer software for microorganism identification (MBT version 6903 MPS library).
The result was given by means of an algorithm (score) obtained by the Biotyper 3.0 software, in which scores <1.7 were considered as non-reliable diagnoses; scores ≥1.7 but <2.0 were considered as reliable for genus identification; and scores ≥2.0 were reliable for identification of genus and bacterial species.
Standard protocol for mastitis pathogen identification by MALDI-TOF MSThe bacterial isolates (n=120) were also submitted for identification by MALDI-TOF MS using bacterial colonies grown on blood agar, and using the lysis protocol described by Barreiro et al.2 Briefly, the colonies were diluted in 1200μL of a 75% ethanol/water solution (900μL/300μL) for bacterial inactivation. The bacterial sediment was centrifuged and the supernatant discarded by tube inversion. A second round of centrifugation was done to remove remaining ethanol present in the sample, and afterwards, the supernatant was removed using a pipette. All centrifugation steps were performed at 13,000×g for 2min. The pellet was left drying at room temperature, and a 70% formic acid solution was added in enough volume to cover the pellet (∼30–50μL) and lyse the bacterial cells. After that, the same volume of 100% acetonitrile (∼30–50μL) was added. During the final stage of preparation, centrifugation was done in order to separate the sediments of the bacterial cells from the supernatant containing bacterial proteins, mainly ribosomal proteins (Biotyper 3.0 manual page 157; Bruker Daltonik, Bremen, Germany). After that, MALDI-TOF MS spectra were obtained and used for bacterial classification using the Biotyper 3.0 computer program for microorganism identification.
Statistical analysisStatistical tests were performed using McNemar test on the paired proportions12 and statistical significance was declared at P<0.05. Under the null hypothesis there was no difference between standard microbiological culture (used as the reference method) and non-culture direct-MALDI-TOF method of milk samples after pre-incubation protocol.
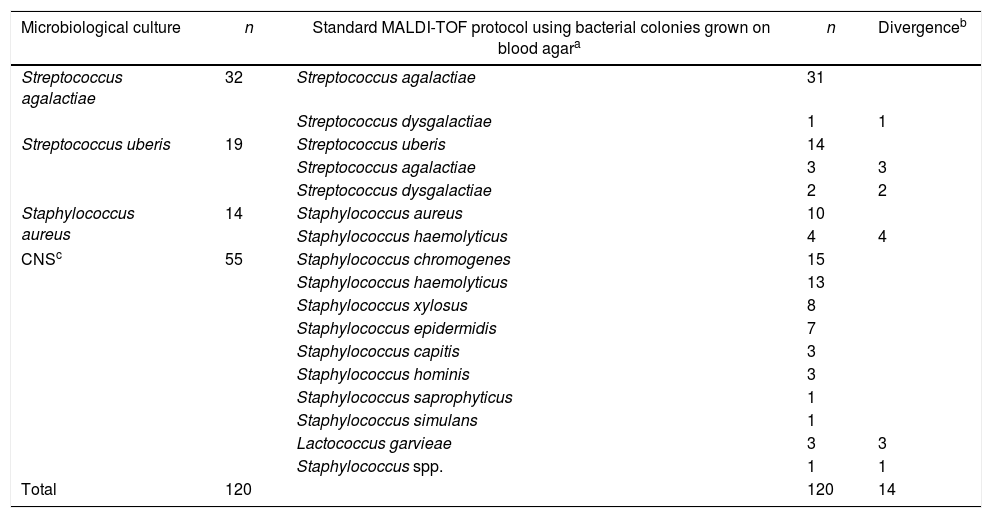
ResultsStandard MALDI-TOF MS protocol using bacterial colonies grown on blood agar vs. direct sample identification after pre-incubation protocolThe reference method, standard microbiological culture of the 120 quarter milk samples, identified Streptococcus agalactiae (n=32); Streptococcus uberis (n=19); Staphylococcus aureus (n=14) and coagulase-negative Staphylococci, CNS (n=55) (Table 1). Among Streptococcus agalactiae isolates identified by microbiological culture (n=32), the standard MALDI-TOF protocol using bacterial colonies grown on blood agar correctly identified 31 isolates as Streptococcus agalactiae and one isolate as Streptococcus dysgalactiae. For Streptococcus uberis isolates identified by microbiological culture (n=19), MALDI-TOF MS identified 14 isolates as Streptococcus uberis, and the remaining of isolates as Streptococcus agalactiae (n=3) and Streptococcus dysgalactiae (n=2). Considering Staphylococcus aureus isolates (n=14) identified by microbiological culture, 10 were identified by the standard MALDI-TOF protocol using bacterial colonies grown on blood agar as Staphylococcus aureus, and the remaining (n=4) as Staphylococcus haemolyticus. With respect to CNS identified by microbiological culture (n=55), the standard MALDI-TOF protocol using bacterial colonies grown on blood agar identified 51 CNS (various species, see Table 1), 3 isolates as Lactococcus garvieae and one as Staphylococcus spp., identified at the genus level (Table 1).
Results of the identification of mastitis causing pathogens (n=120) using the standard MALDI-TOF protocol using bacterial colonies grown on blood agar and microbiological culture.
| Microbiological culture | n | Standard MALDI-TOF protocol using bacterial colonies grown on blood agara | n | Divergenceb |
|---|---|---|---|---|
| Streptococcus agalactiae | 32 | Streptococcus agalactiae | 31 | |
| Streptococcus dysgalactiae | 1 | 1 | ||
| Streptococcus uberis | 19 | Streptococcus uberis | 14 | |
| Streptococcus agalactiae | 3 | 3 | ||
| Streptococcus dysgalactiae | 2 | 2 | ||
| Staphylococcus aureus | 14 | Staphylococcus aureus | 10 | |
| Staphylococcus haemolyticus | 4 | 4 | ||
| CNSc | 55 | Staphylococcus chromogenes | 15 | |
| Staphylococcus haemolyticus | 13 | |||
| Staphylococcus xylosus | 8 | |||
| Staphylococcus epidermidis | 7 | |||
| Staphylococcus capitis | 3 | |||
| Staphylococcus hominis | 3 | |||
| Staphylococcus saprophyticus | 1 | |||
| Staphylococcus simulans | 1 | |||
| Lactococcus garvieae | 3 | 3 | ||
| Staphylococcus spp. | 1 | 1 | ||
| Total | 120 | 120 | 14 |
Percentage agreement using standard MALDI-TOF protocol using bacterial colonies grown on blood agar for the identification of pathogens causing subclinical mastitis was of 96.8% for Streptococcus agalactiae; 73.6% for Streptococcus uberis; 71.4% for Staphylococcus aureus and 92.7% for CNS, when compared to microbiological culture results.
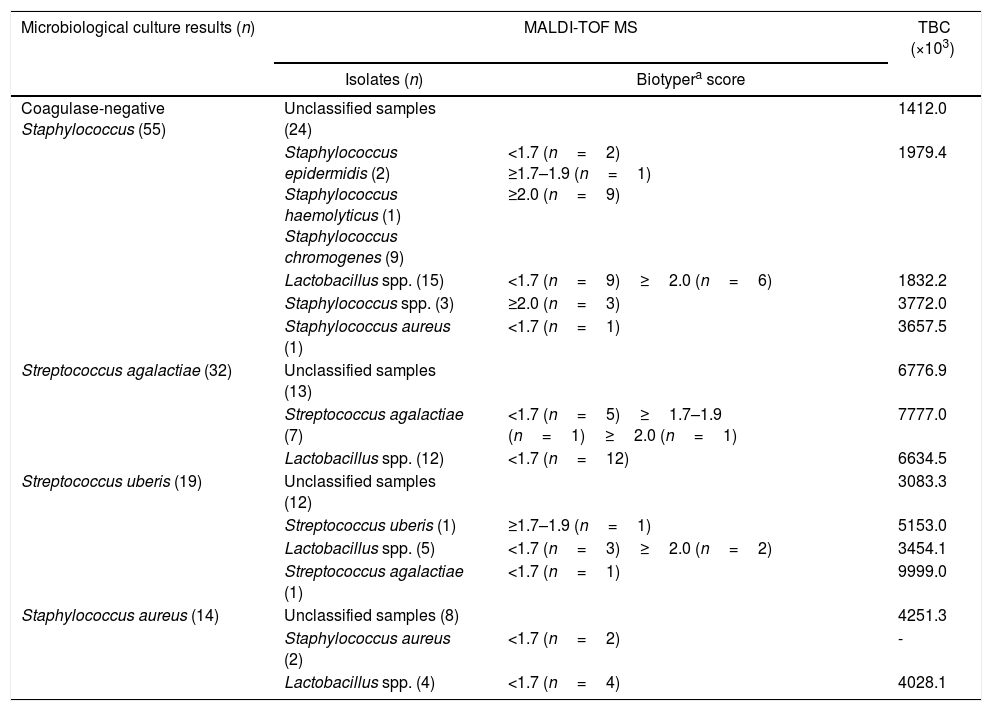
Mastitis pathogen identification using direct sample identification after pre-incubation protocolIn comparison to the reference microbiology method, the direct sample identification after pre-incubation protocol (direct MALDI-TOF MS method) only identified 15 (27.2%) of the 55 isolates as CNS (Table 2). This protocol identified CNS in 9/15 milk samples, at the genus and species level (score ≥2.0), 1/15 at the genus level (score ≥1.7–1.9), 2/15 milk samples were not reliably identified at the genus and species level (score <1.7), and 3/15 were identified as Staphylococcus spp. with scores ≥2.0 but without species suggestion by spectral data processing using the Biotyper 3.0. Among the remaining milk samples identified as CNS by microbiological culture, identification by direct-MALDI-TOF method was not possible in 24/55 (43.6%; not reliable identification by direct-MALDI-TOF), and for the other samples, there was disagreement between direct-MALDI-TOF and by microbiological culture. For example, 15/55 samples (27.2%) previously identified as CNS were identified using direct-MALDI-TOF as Lactobacillus spp., and 1/55 (1.8%) as Staphylococcus aureus with a score ≤1.7 (Table 2).
Efficiency of the direct sample identification of mastitis causing pathogens by MALDI-TOF MS in quarter milk samples (n=120) vs. microbiological culture.
| Microbiological culture results (n) | MALDI-TOF MS | TBC (×103) | |
|---|---|---|---|
| Isolates (n) | Biotypera score | ||
| Coagulase-negative Staphylococcus (55) | Unclassified samples (24) | 1412.0 | |
| Staphylococcus epidermidis (2) Staphylococcus haemolyticus (1) Staphylococcus chromogenes (9) | <1.7 (n=2) ≥1.7–1.9 (n=1) ≥2.0 (n=9) | 1979.4 | |
| Lactobacillus spp. (15) | <1.7 (n=9)≥2.0 (n=6) | 1832.2 | |
| Staphylococcus spp. (3) | ≥2.0 (n=3) | 3772.0 | |
| Staphylococcus aureus (1) | <1.7 (n=1) | 3657.5 | |
| Streptococcus agalactiae (32) | Unclassified samples (13) | 6776.9 | |
| Streptococcus agalactiae (7) | <1.7 (n=5)≥1.7–1.9 (n=1)≥2.0 (n=1) | 7777.0 | |
| Lactobacillus spp. (12) | <1.7 (n=12) | 6634.5 | |
| Streptococcus uberis (19) | Unclassified samples (12) | 3083.3 | |
| Streptococcus uberis (1) | ≥1.7–1.9 (n=1) | 5153.0 | |
| Lactobacillus spp. (5) | <1.7 (n=3)≥2.0 (n=2) | 3454.1 | |
| Streptococcus agalactiae (1) | <1.7 (n=1) | 9999.0 | |
| Staphylococcus aureus (14) | Unclassified samples (8) | 4251.3 | |
| Staphylococcus aureus (2) | <1.7 (n=2) | - | |
| Lactobacillus spp. (4) | <1.7 (n=4) | 4028.1 | |
Seven out of 32 (21.8%) milk samples were identified as Streptococcus agalactiae by direct-MALDI-TOF method, 1/32 (3.1%) isolate being identified at the genus and species level (score ≥2.0), 1/32 (3.1%) at the genus level (score ≥1.7–1.9), and 5/32 (15.6%) showed to be indicative of Streptococcus agalactiae but had an identification score of <1.7. For the remaining Streptococcus agalactiae identified by culture (n=25), 12/25 isolates (48%) were identified as Lactobacillus spp. by the direct-MALDI-TOF method, and for 13/25 (52%) isolates it was not possible to identify any pathogen using this method (Table 2).
The direct-MALDI-TOF method correctly identified 1/19 isolate (5.2%) of Streptococcus uberis, although it was only at the genus level (score ≥1.7–1.9). Among the remaining Streptococcus uberis identified by culture (n=18), for 12/18 milk samples there was no identification of any pathogen causing subclinical mastitis by the direct-MALDI-TOF (66.6%), 5/18 milk samples (27.7%) were identified as Lactobacillus spp. and 1/18 sample (5.5%) as Streptococcus agalactiae (Table 2).
The methodology of direct identification of pathogens causing subclinical mastitis by MALDI-TOF MS identified 2/14 isolates (14.3%) as Staphylococcus aureus, although with scores <1.7. Among the remaining Staphylococcus aureus identified by culture (n=12), for 8/12 isolates (66.6%) Staphylococcus aureus was not identifiable when submitted to direct-MALDI-TOF, and for 4/12 samples (33.3%), Lactobacillus spp. was identified by direct-MALDI-TOF, with scores ≤1.6 (Table 2).
The evaluated methods were significantly different, indicating disagreement of identification of subclinical mastitis-causing pathogens between both methods (P<0.05). The direct-MALDI-TOF method, when compared to microbiological culture, correctly identified isolates of coagulase-negative Staphylococci (27.2%), Streptococcus agalactiae (21.8%), Staphylococcus aureus (14.2%), and Streptococcus uberis (5.2%).
DiscussionThe method using a pre-incubation procedure for a non-culture direct sample-MALDI-TOF method showed low accuracy of identification of mastitis causing pathogens in our study when compared with the results obtained by microbiological culture. The identification of pathogens by direct sample-MALDI-TOF in clinical samples of human urine and cerebrospinal fluid was successfully described when the bacterial count was >104–106cfu/mL. These bacterial count values were obtained in patients with urinary tract infection.7 Similar to the urine samples, the cerebrospinal fluid was evaluated by direct-MALDI-TOF for identification of microorganisms that cause bacterial meningitis.8 In the present study, we observed TBC varied from 1.4 to 9.9×106cfu/mL after pre-incubation of milk samples. However, attaining a similar identification percentage of the pathogens causing subclinical mastitis by direct-MALDI-TOF was not possible in milk samples from mammary quarters, as described previously for urine and cerebrospinal fluid samples.8
The bacterial count in experimentally contaminated milk samples was a limiting factor that influenced the analytic sensitivity of the direct-MALDI-TOF method for identification of pathogens that cause mastitis.13 According to Moussaoui et al.10 the pre-incubation of blood samples allowed the identification of bacteria by the direct-MALDI-TOF method. Similarly, Ferreira et al.7 reported that the direct-MALDI-TOF method was efficient in bacterial identification in infected urine when the bacterial count was >105cfu/mL. Nonetheless, in the present study, using the cutoff of TBC published from Barreiro et al.3 combined with a 12-h pre-incubation protocol of the quarter milk samples did not result in higher identification of mastitis-causing pathogens, since the direct-MALDI-TOF method identified ≤20.8% (25/120 isolates) of specific pathogen identified by microbiological culture.
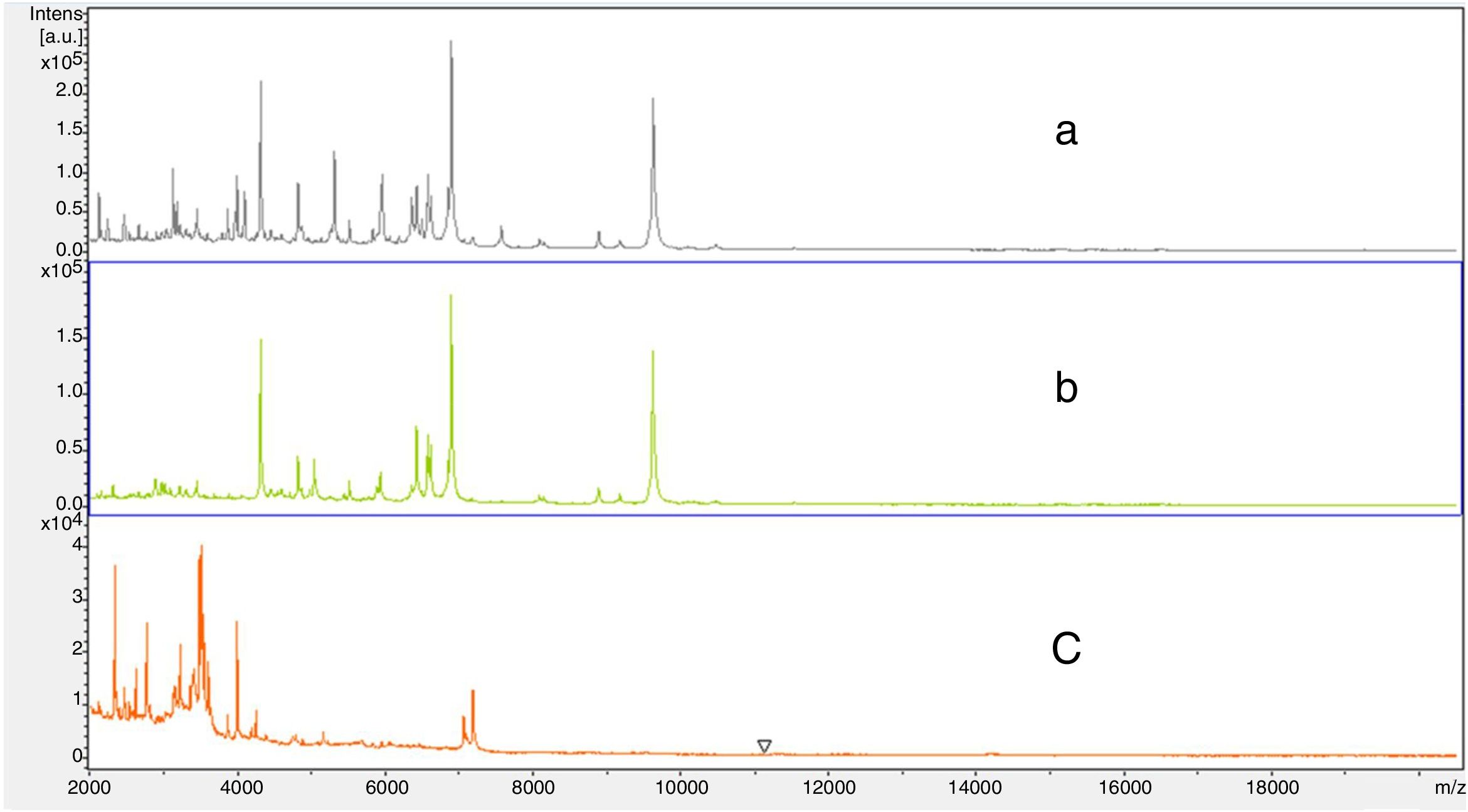
The TBC of the milk samples from naturally infected cows may have been an influencing factor in the reduced identification capacity of the pathogens by the direct-MALDI-TOF method. Also, the pre-incubation of the milk samples may have favored the growth of symbiotic or contaminant bacteria that could be present in milk samples, such as Lactobacillus spp., which would not allow sufficient bacterial count for the identification of pathogens causing subclinical mastitis by the direct-MALDI-TOF method. Another possible interfering factor was the presence of milk components (protein and fat), even after the washing protocol using Lysis Buffer, which may have interfered in spectra acquisition for the direct identification of the pathogens causing subclinical mastitis by direct-MALDI-TOF method. Milk proteins could affect classification results with the effective range between 3,000 and 20,000 m/z. While comparing mass spectra originating from the Standard MALDI-TOF protocol using bacterial colonies grown on blood agar vs. direct sample-MALDI-TOF identification protocol, interference peaks ranging between 2,000 and 7,000 m/z were observed for identification of Staphylococcus aureus (Fig. 1). One of the main limiting factors of the direct-MALDI-TOF method is related to the great heterogeneity and complexity of the set of proteins present in milk. Thus, the lower the concentration of the target proteins of identification, the higher the risk to be misidentified by direct-MALDI-TOF method, due to the detection limit and to the possible overlapping of milk proteins, which present similar molecular weight and isoelectric points.
Spectra ranging from 2000 to 20,000m/z: (a) Staphylococcus aureus identified by the direct MALDI-TOF method of an experimentally inoculated milk sample, (b) Staphylococcus aureus identified in the colony (ATCC 29213) by the standard MALDI-TOF protocol using bacterial colonies grown on blood agar, and (c) a quarter milk sample without microbiological growth.
The direct-MALDI-TOF method, when compared to microbiological culture, correctly identified 27.2%, 21.8%, 14.2% and 5.2% of the CNS, Streptococcus agalactiae, Staphylococcus aureus and Streptococcus uberis isolates, respectively. According to La Scola and Raoult,14 the agreement between MALDI-TOF MS results for the direct identification of bacteria in blood and in microbiological cultures was 59% of evaluated samples. However, the later study found that for samples in which Streptococcus spp. was isolated, there was low agreement with the reference method (4/25, 16%). After modifying the preparation protocol for blood samples, there was an increase of 67% in the efficiency of Gram-positive identification, mainly for Staphylococcus spp., though for Streptococcus spp., identification remained low (4/17, 23%).14 We believed that the decreased percentage of identification using the direct-MALDI-TOF method may be occurred because MALDI-TOF MS does not identify spectra from mixed cultures reliably. The correctness of classification is based on the false assumption that the “reference ID method” pathogen is the only organism present. Despite it remain a disadvantage, the direct MALDI-TOF MS method may be used as an alternative to conventional bacterial identification.
Regardless of the pathogen type studied, we observed the frequency of 36/120 Lactobacillus spp. (30%) identified by the direct-MALDI-TOF MS method combined with the pre-incubation protocol. The identification frequency of Lactobacillus spp. may have occurred due to the pre-incubation protocol of the milk samples, which allowed an increase in the bacterial count of these symbiotic microorganisms.
In the present study, regarding all isolates correctly identified as CNS (15 isolates) by the direct-MALDI-TOF MS method, 9/15 of them were identified at the genus and species level (score ≥2.0). With regard to the Streptococcus agalactiae isolates, only two were identified at the genus and species level (score >2.0). The level of identification reliability was even lower in Streptococcus uberis (score ≥1.7–1.9) and Staphylococcus aureus (score <1.7). Therefore, the direct-MALDI-TOF method for the identification of the pathogens causing subclinical mastitis showed low identification percentages when compared to microbiological culture.
In this study, the standard MALDI-TOF protocol using bacterial colonies grown on blood agar identified 88.3% (106/120) of the pathogens causing subclinical mastitis at the genus and species level (score ≥2.0), when compared to microbiological culture. The use of standard MALDI-TOF protocol using bacterial colonies grown on blood agar for the identification of bacteria in clinical bacteriology has become more common. Our results are similar to other studies on bacterial identification by MALDI-TOF using bacterial colonies, in which identification percentages at the genus level were 97–99%, and at the species level, 85–97%.15 On the other hand, the pre-incubation of the quarter milk samples for identification of pathogen causing subclinical mastitis by the direct-MALDI-TOF method did not display satisfactory results. Furthermore, the milk sample pre-incubation protocol may have favored symbiotic and other contaminant bacterial growth (e.g. Lactobacillus spp.), resulting in low frequency of identification by the direct sample-MALDI-TOF method.
In an previous study developed by our research group, we observed that a minimum of TBC varying of ≥106 to ≥108cfu/mL would be required for successful identification scores at the gender and species level, and it also varied according to the mastitis causing pathogen.3 However, the present study aimed to evaluate a rapid identification protocol (without culture) using the pre-incubation of milk samples followed by direct identification by MALDI-TOF MS. Therefore, milk samples were submitted to TBC in order to identify whether the microbial load before the pre-incubation protocol was within the range of TBC proposed in the previous study.3 Thus, it would be possible to use a rapid and direct sample protocol for identification of mastitis causing pathogens. However, we did not expect that the pre-incubation protocol would have favored symbiotic and bacterial growth (e.g., Lactobacillus spp.). These results suggest that we could have included the identification of milk samples after pre-incubation protocol by traditional bench microbiology and specific microbial counting methods (e.g. standard plate count – SPC) prior to the use of the milk sample incubation protocol, which we recognize as limitations of the present study. An alternative approach that may improve results would be to develop some method of inactivating symbiotic bacteria prior to pre-incubation.
ConclusionsThe nonculture based identification MALDI-TOF MS method, even after the 12h pre-incubation protocol of the quarter milk samples, did not accurately help to promote the rapid identification of mastitis pathogens. The pre-incubation protocol of milk samples, associated to the direct identification (nonculture-based) method by MALDI-TOF MS, did not increase the identification at species level (score >2.0) of pathogens causing subclinical mastitis in comparison to the method without previous incubation.
Conflict of interest statementThe authors declare no conflicts of interest.
We are grateful to the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Brazil, for a scholarship award (2011/14456-0) and research funding (2011/15815-4). The authors are grateful to Dr. Kevin L. Anderson (Department of Population Health and Pathobiology, College of Veterinary Medicine, North Carolina State University) who reviewed the English writing.