HLA-B27 is strongly associated with ankylosing spondylitis (AS) and its presence helps to confirm AS diagnosis. Due to the high HLA polymorphism and the differentiated contribution of alleles and molecules encoded by them, HLA-B*27 allele identification is relevant in the clinical follow-up, diagnosis, and treatment of this spondyloarthropathy. Inexpensive genotyping techniques with high specificity and sensitivity are of great interest in histocompatibility laboratories. This work aimed to optimize HLA-B*27 genotyping by Polymerase Chain Reaction Sequence-specific Primer (PCR-SSP), which is an accessible and inexpensive technique.
METHODS:The PCR-SSP was standardized using 26 HLA-B*27 positive and 3 HLA-B*27 negative samples previously defined by Polymerase Chain Reaction Sequence-specific Oligonucleotide Probes (PCR-SSOP) (medium resolution, One Lambda®) and primers described by Duangchanchot et al. (2009). For validating the technique, 397 samples were genotyped using PCR-SSP as well as PCR-SSOP.
RESULTS:The PCR-SSP technique was standardized for identifying the alleles HLA-B*27:02, HLA-B*27:CAFRW (05/13/16/17/28/37/38/39/42), HLA-B*27:CAFRZ (08/26/40), HLA-B*27:09 and HLA-B*27:12, which were found in 90 positive samples (22.67%). There was 100% agreement between the two techniques for heterozygous samples; however, two homozygous samples could not be detected by PCR-SSP.
CONCLUSION:The HLA-B*27 genotyping using PCR-SSP, an easy-to-use, specific, and affordable technique, was optimized for heterozygous samples. This technique may contribute to AS diagnosis.
Leukocyte antigen system (HLA), a major histocompatibility complex (MHC), is located on the short arm of chromosome 6 in humans (1). This system has an extensive polymorphism for class I and II genes, characterized by a high number of alleles. As of October 2019, 25,756 HLA alleles have been described, of which 18,691 are class I alleles and 7,065 are class II alleles. HLA-B is the most polymorphic locus of the HLA system, with 7,053 alleles identified (2). Class I human leukocyte antigens are expressed on most nucleated cell surfaces. They carry endogenous peptides to the cell surface for recognition by T-cell receptors and their functions are involved in immune responses. Thus, many diseases are associated with HLA (3).
Ankylosing spondylitis (AS), a chronic inflammatory disease of the spondyloarthropathy group, is strongly associated with the HLA-B27 antigen. This association was first described in 1973 and is the largest genetic factor contributing to the disease (4). Early genetic association studies in Caucasians showed that HLA-B*27 was present in approximately 90% of the individuals with AS. Further studies confirmed this association in other population, but with different strengths of association; thus, 50-90% of the individuals with this disease have the HLA-B*27 gene (5–7). The HLA-B*27 allelic group has more than 290 known alleles (8), although most alleles are not associated with AS. Studies have shown that HLA-B*27:05, HLA-B*27:04, and HLA-B*27:02 are associated with AS in the Caucasian, Chinese, and Mediterranean populations, respectively (4). The HLA-B*27:06 and HLA-B*27:09 alleles were not found to be associated with AS (4,9,10).
Inexpensive genotyping techniques with high specificity and sensitivity are of great interest in histocompatibility-testing laboratories. Duangchanchot et al. (2009) described specific primers for genotyping 42 HLA-B*27 alleles (B*27:01-B*27:21 and B*27:23-B*27:43) using high-resolution polymerase chain reaction with sequence-specific primer (PCR-SSP) (11). PCR-SSP has been reported to be a simple, fast, inexpensive, specific, and highly sensitive method (12).
The identification of the HLA-B*27 alleles is relevant in the clinical follow-up, diagnosis, and treatment of AS (9). This work aimed to optimize HLA-B*27 genotyping using PCR-SSP, an easy-to-use and affordable technique.
MATERIALS AND METHODSSample selectionTwenty-six samples previously known as HLA-B*27 positive and three HLA-B*27 negative samples using PCR-SSOP® (medium resolution; One Lambda; Canoga Park, CA, USA) were collected from the UEM Immunogenetic Laboratory database (https://www.onelambda.com/en/products-services/products/molecular-typing/labtype.html) and used to standardize the method. For validation, samples were collected from the individuals with AS (N=160) and psoriatic arthritis (PsA; N=57) due to the high frequency of HLA-B*27 in this population; samples from individuals without the disease (N=180) were added to increase the randomness of the process. The patients with AS and PsA were classified through clinical, laboratory and radiological criteria according to the ASAS 2009/2011 criteria (13,14) and CASPAR (15), respectively, by rheumatologists from the Regional Maringá University Hospital. All participants were from the northwestern region of Paraná, southern Brazil (22°29′30–26°42′59- S and 48°2′24–54°37′38- W) and classified as ‘mixed ethnicity with predominantly European origin,‘ based on the ethnic constitution of Paraná, as previously described (16) and confirmed for this region (17). The samples were collected sequentially from May 2014 to December 2016. This study was approved by the Research Ethics Committee of the State University of Maringá (UEM), number CAAE 27723114 and all participants signed the consent form.
DNA extractionDNA was extracted from whole blood or buffy coat collected in 5-mL tubes containing EDTA using the salting out method and/or DNA extraction kit BIOPUR® (Mobius; Curitiba, Paraná, Brazil). DNA purity and concentrations were determined by NanoDrop® 2000 UV-Vis spectrophotometer (Thermo Fisher; Wilmington, DE, USA). The concentration was adjusted to 50-100 ng/μL.
Standardization of HLA-B*27 genotypingThe primers used for standardizing the PCR-SSP reaction were constructed according to the sequences described by Duangchanchot et al. (2009) and are shown in Table 1.
Primers for genotyping HLA-B*27 (Mix SC1 and SC2) and specific alleles (Mix 2, 3, 4, 5, 7, 8, 9, 10, 12).
| Mix | Name | P | Sequence (5′ - 3′) | Location | Position | HLA-B*27 alleles amplified |
|---|---|---|---|---|---|---|
| SC1 | F167T | 1 | GCT ACG TGG ACG ACA CGC T | Exon 2 | 149-167 | 01-11, 13-15, 17, 19-21, |
| R272G | 2 | GTC TGT GCC TTG GCC TTG C | Exon 2 | 272-290 | 24-28, 30-43 | |
| SC2 | F204A | 3 | GAC GCC GCG AGT CCG AGA | Exon 2 | 187-204 | 01-06, 08-10, 12-13, 15-18, |
| R362A | 4 | CAC GTC GCA GCC ATA CAT AT | Exon 3 | 362-381 | 20, 23, 25-29, 31, 35-42 | |
| 2 | F311T | 6 | ACC GAG AGA ACC TGC GGA T | Exon 2 | 293-311 | 02 |
| R362A | 4 | CAC GTC GCA GCC ATA CAT AT | Exon 3 | 362-381 | ||
| 3 | F167T | 7 | GCT ACG TGG ACG ACA CGC T | Exon 2 | 149-167 | 03 |
| R247C | 8 | GTG TCT CCC GGT CCC AAT G | Exon 2 | 247-265 | ||
| 4 | F362A | 9 | GGT CTC ACA CCC TCC AGA A | Exon 3 | 344-362 | 04, 06, 10, 15, 18, 20, 25 |
| R527A | 10 | CTC TCA GCT GCT CCG CCT | Exon 3 | 527-544 | ||
| 5 | F272G | 11 | ACC GGG AGA CAC AGA TCT G | Exon 2 | 254-272 | 01-05, 08, 10, 12-17, 19, 25-26 |
| R418G | 12 | CTT GCC GTC GTA GGC GTC | Exon 3 | 418-434 | 28, 30-32, 36-40, 42 | |
| 7 | F301G | 14 | GCA CAG ACT GAC CGA GAG G | Exon 2 | 283-301 | 07, 32, 34, 43, B*0727, |
| R363C | 15 | CAC GTC GCA GCC GTA CAT G | Exon 3 | 363-381 | B*3707, B*3709 | |
| 8 | F311A | 16 | CCG AGA GAG CCT GCG GAA | Exon 2 | 294-311 | 08, 12, 18, 26, 40, 42, B*1802 |
| R362A | 4 | CAC GTC GCA GCC ATA CAT AT | Exon 3 | 362-381 | ||
| 9 | F272G | 11 | ACC GGG AGA CAC AGA TCT G | Exon 2 | 254-272 | 09 |
| R418C | 17 | CTT GCC GTC GTA GGC GTG | Exon 3 | 418-434 | ||
| 10 | F301G | 14 | GCA CAG ACT GAC CGA GAG G | Exon 2 | 283-301 | 03, 05, 09-10, 13, 16-17, 27-29 |
| R362A | 4 | CAC GTC GCA GCC ATA CAT AT | Exon 3 | 362-381 | 35, 37-39, 41-42, | |
| B*3702, B*4701, B*4705 | ||||||
| 12 | F277A | 19 | GGA GAC ACA GAT CTG CAA GA | Exon 2 | 258-277 | 12, 16, 18, 29, |
| R362A | 4 | CAC GTC GCA GCC ATA CAT AT | Exon 3 | 362-381 | B*3702, B*4704-05 |
Source: Adapted from Duangchanchot et al. (2009). P: primer identification.
Two primer mixes (SC1 and SC2) were used to assess the presence of HLA-B*27 allelic group and nine mixes were used to identify the alleles. Primers amplifying a 782 bp fragment in the third intron of HLA-DRB1 (C5: 5′-TGCCAAGTGGAGCACCCA-3′; C3: 5′- GCATCTTGCTCTGTGCAGAT-3′) were used as the internal control (18).
Same PCR conditions were used for all mixes (Table 2). The PCR mixture contained 1× standard Taq reaction buffer, 200 µM deoxyribonucleotide phosphates (dNTP), 2 ng/μL each specific primer, 1 ng/μL each internal control primer, and 40-200 ng template DNA in a 10-μL volume. The final concentrations of MgCl2 and Taq DNA polymerase were different in the mixes and are described in Table 3. PCRs were performed with a final DNA concentration of 40-200 ng, and the sensitivity for this DNA concentration range was same.
Thermocycling conditions for the HLA-B*27 genotyping using polymerase chain reaction with sequence-specific primer (PCR-SSP).
| Cycle | Denaturation | Annealing | Extention |
|---|---|---|---|
| 1 Cycle | 96°C; 2 min | — | — |
| 5 Cycles | 96°C-30 s | 68°C-60 s | 72°C-40 s |
| 21 Cycles | 96°C-30 s | 65°C-60 s | 72°C-40 s |
| 4 Cycles | 96°C-30 s | 55°C-75 s | 72°C-120 s |
| 1 Cycle | — | — | 72°C; 10 min |
Min: minutes. s: seconds.
Concentration of reagents for different primer mixes used to define the HLA-B*27 allelic variants and the expected amplified fragment size.
| MIX | Primer Identification | MgCl2 (mM) | Taq DNA Polymerase (U) | Band size |
|---|---|---|---|---|
| SC1 | 1 and 2 | 1.5 | 0.5 | 142 |
| SC2 | 3 and 4 | 2.0 | 0.5 | 436 |
| 2 | 6 and 4 | 3.0 | 1.0 | 330 |
| 3 | 7 and 8 | 1.5 | 0.5 | 117 |
| 4 | 9 and 10 | 1.5 | 0.8 | 201 |
| 5 | 11 and 12 | 1.5 | 0.8 | 423 |
| 7 | 14 and 15 | 1.5 | 0.8 | 333 |
| 8 | 16 and 4 | 1.5 | 0.8 | 329 |
| 9 | 11 and 17 | 2.0 | 0.5 | 383 |
| 10 | 14 and 4 | 1.5 | 0.8 | 340 |
| 12 | 19 and 4 | 2.0 | 0.8 | 365 |
Primer identifications are described in Table 1.
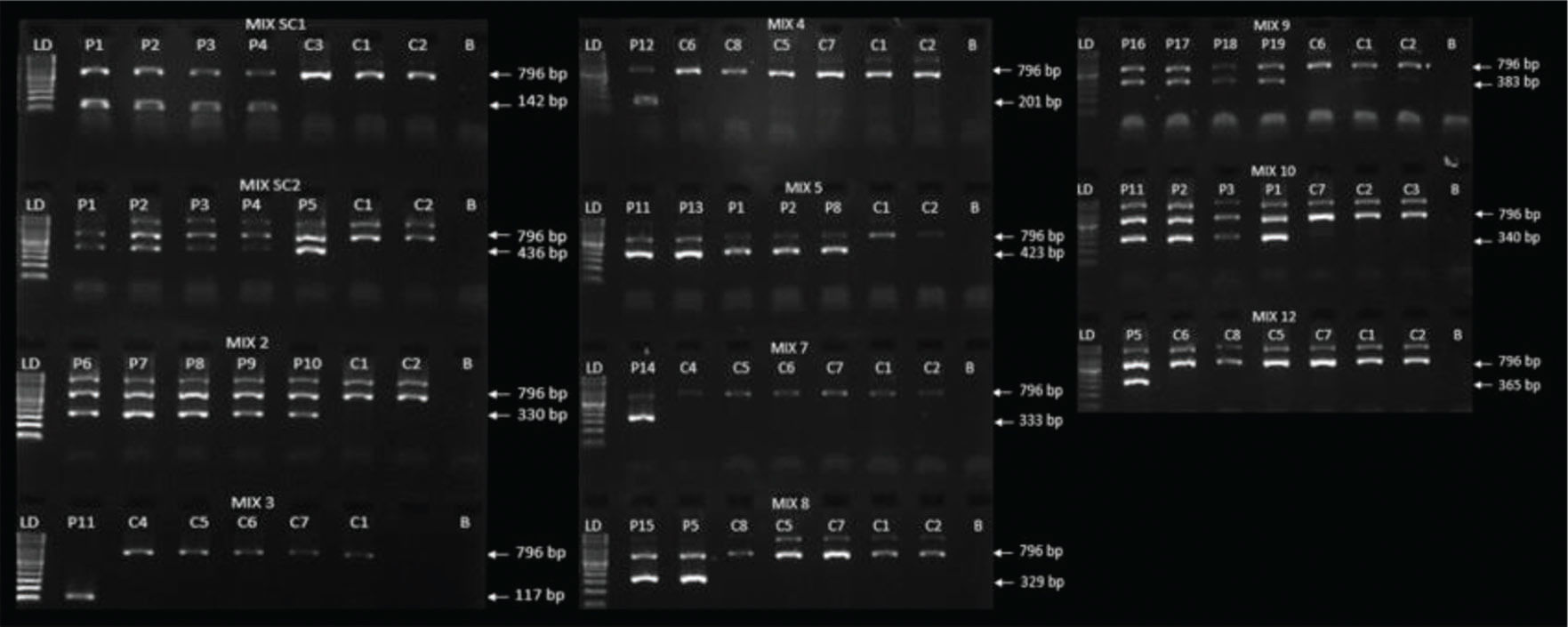
The PCR products were analyzed using 2% agarose gel electrophoresis stained with SYBR™ Safe DNA Gel Stain dye (Invitrogen; Carlsbad, CA, USA), after running at 100 V, 300 mA, 150 W for 20 minutes. Visualization and photo documentation were performed on the Quantum ST4 transilluminator (Vilber Lourmat; Collegien, France). Molecular weight markers with 100 base pairs (bp) (DNA Ladder, Thermo Fisher; Vilnius, Lithuania) was included to ensure the band sizes.
ValidationFor validating the technique, 397 samples were genotyped using the standardized PCR-SSP technique and confirmed using PCR-SSOP®, which is a routine method used in our laboratory (LIG-UEM). To avoid biased results, PCR-SSOP® was performed after genotyping all samples using standardized PCR-SSP.
RESULTSThe standardized PCR-SSP was performed using the same thermocycling conditions for all primers and a low final volume of reagents. Only the final concentrations of MgCl2 and Taq DNA polymerase were different for each primer mix used.
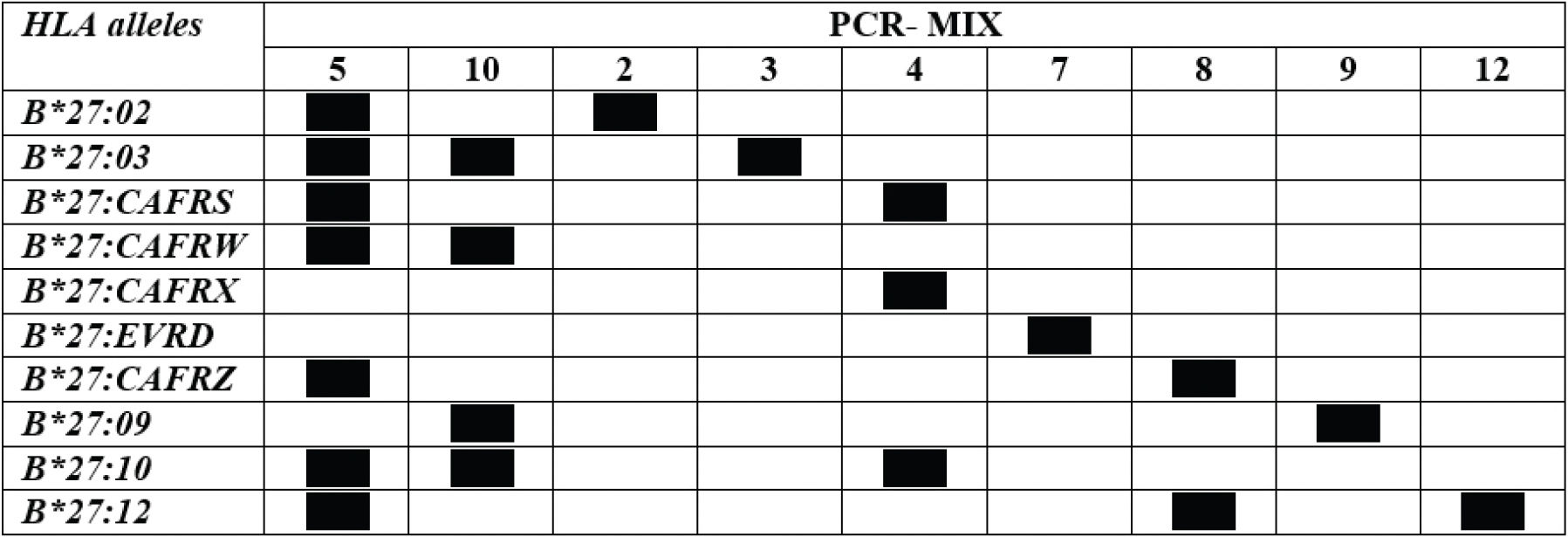
To validate the technique, 397 samples were genotyped using PCR-SSP and PCR-SSOP®. Using PCR-SSP, 90 samples (22.7%) were positive for mix SC1 and/or SC2, therefore being positive for HLA-B*27. The frequency of HLA-B*27 was 44.4%, 15.8%, and 5.5% in individuals with AS, PsA, and no spondyloarthropathy, respectively; the distribution of frequency was as expected for that observed in Brazilians (19–21) allowing to validate the technique. To define the HLA-B*27 alleles, these samples were genotyped with primer mixes 2, 3, 4, 5, 7, 8, 9, and 10. Mix 12 was used only when samples were positive after genotyping with mix 8. The genotyping results are shown in Table 4. HLA-B*27:02, HLA-B*27:02/HLA-B*27:CAFRW, HLA-B*27:CAFRW, HLA-B*27:CAFRZ, HLA-B*27:09, and HLA-B*27:12 alleles were identified in 2, 1, 84, 1, 1, and 1 individuals, respectively, using PCR-SSP. HLA-B*27:CAFRW allele frequency was significantly different between the patients with AS and PsA (21.6% and 7.0%, respectively) and individuals without spondyloarthropathies (2.8%).
Number and definition of HLA-B*27 alleles identified using PCR-SSP and PCR-SSOP®.
| PCR-SSP N=397 | PCR-SSOP® N=397 | |
|---|---|---|
| HLA-B*27 negative | 307 | 307 |
| HLA-B*27 positive | 90 | 90 |
| HLA-B*27:02 | 2 | 2 |
| HLA-B*27:CAFRW | 84 | 82 |
| HLA-B*27:CAFRZ | 1 | 1 |
| HLA-B*27:09 | 1 | 1 |
| HLA-B*27:12 | 1 | 1 |
| HLA-B*27:02/B*27:CAFRW | 1 | 1 |
| HLA-B*27:CAFRWB*27:CAFRW | 0 | 2 |
N: number of individuals. CAFRW: 05/13/16/17/28/37/38/39/42. CAFRZ: 08/26/40.
After performing PCR-SSP, these same samples were genotyped using PCR-SSOP®. There was 100% agreement between the two techniques for the heterozygous samples. However, two homozygous HLA-B*27:CAFRW samples were not identified by standardized PCR-SSP (Table 4).
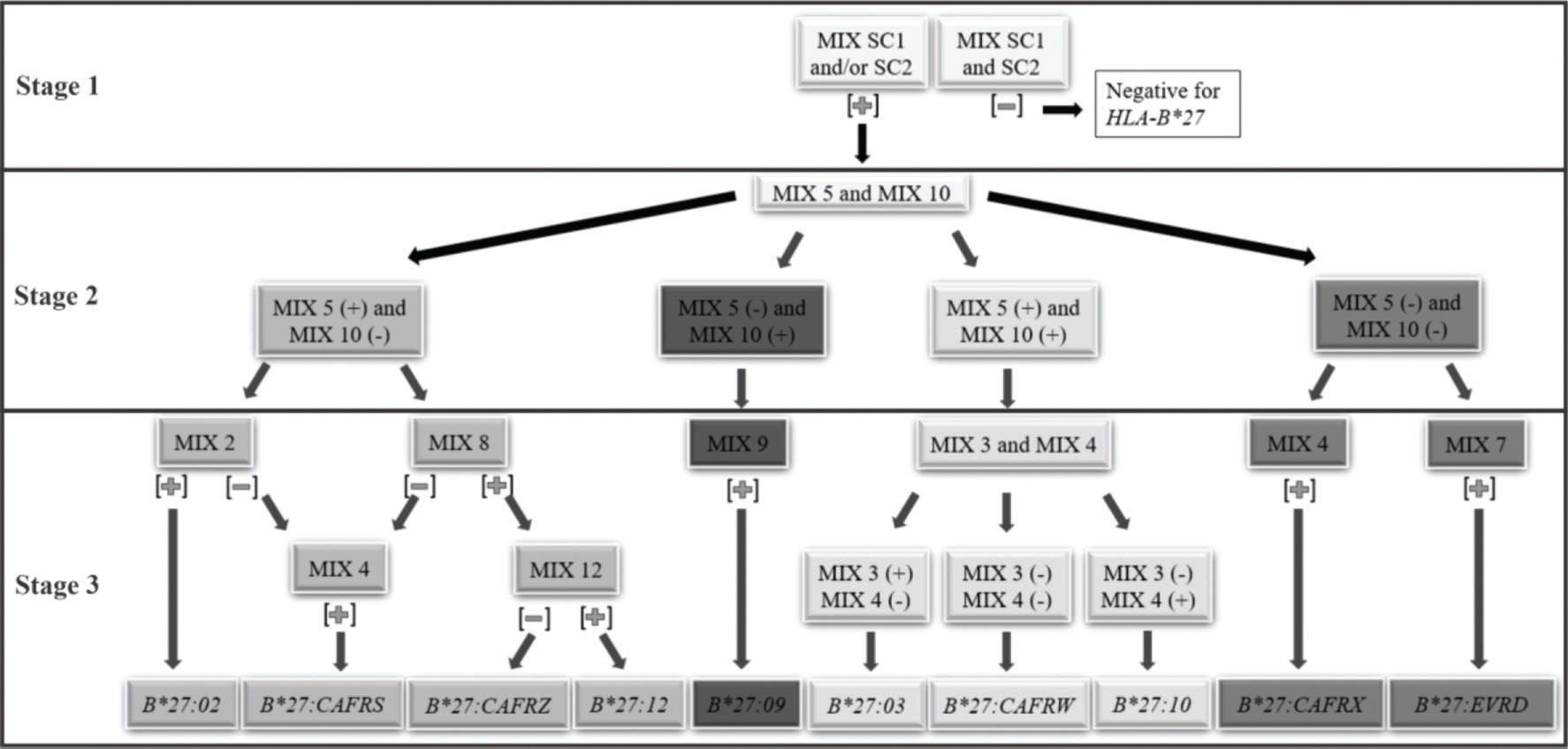
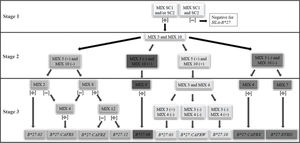
A facilitator was proposed to define the reaction sequence for HLA-B*27 genotyping using the in-house PCR-SSP (Figure 1).
Thus, we proposed a short path for the definition of HLA-B*27 alleles and allelic variants. The reaction can be performed in three stages. 1st stage: PCR-SSP with primer mixes SC1 and SC2. Positive samples for at least one of them define the positive HLA-B*27 genotype, which will be used for the next stage reactions. 2nd stage: PCR-SSP with primer mixes 5 and 10. According to the results obtained, the different paths described in the next stage shall be used. 3rd stage:i. mix 5 positive and mix 10 negative: PCR-SSP with primer mixes 2, 4, 8, and 12; ii. mix 5 negative and mix 10 positive: PCR-SSP with primer mixes 9; iii. both mix 5 and 10 positive: PCR-SSP with primer mixes 3 and 4; iv. both mix 5 and 10 negative: PCR-SSP with primer mixes 4 and 7. The third-stage reactions can be performed concurrently to facilitate the process. The results must be interpreted according to step 3 of the flowchart (Figure 1).
The worksheet for HLA-B*27 PCR-SSP and band patterns of positive and negative samples using standardized PCR-SSP are shown in Appendix figures S1 and S2, respectively.
DISCUSSIONHLA-B*27 allelic genotyping has become important in clinical practice for the treatment and management of spondyloarthropathies. In AS, HLA-B*27 is the most important genetic marker and some alleles such as HLA-B*27:05, B*27:02, B*27:04, and B*27:07, have been associated with the disease (4,9). Through the standardized PCR-SSP technique, it was possible to genotype the HLA-B*27 alleles frequent in the Brazilian population. This technique can be used as an auxiliary method for diagnostic purposes, as well as in genetic association studies to estimate the HLA-B*27 frequency in a specific population.
To define the HLA-B*27 allele with minimal PCRs, a flow chart has been proposed (Figure 1). The first stage defines the positivity for the HLA-B*27 allelic group and the specific allele is identified in samples heterozygous for the HLA-B locus in two further steps. We consider the methodology of high resolution, although some alleles, such as HLA-B*27:05 more frequent in our population, could not be identified without some alleles with low frequencies.
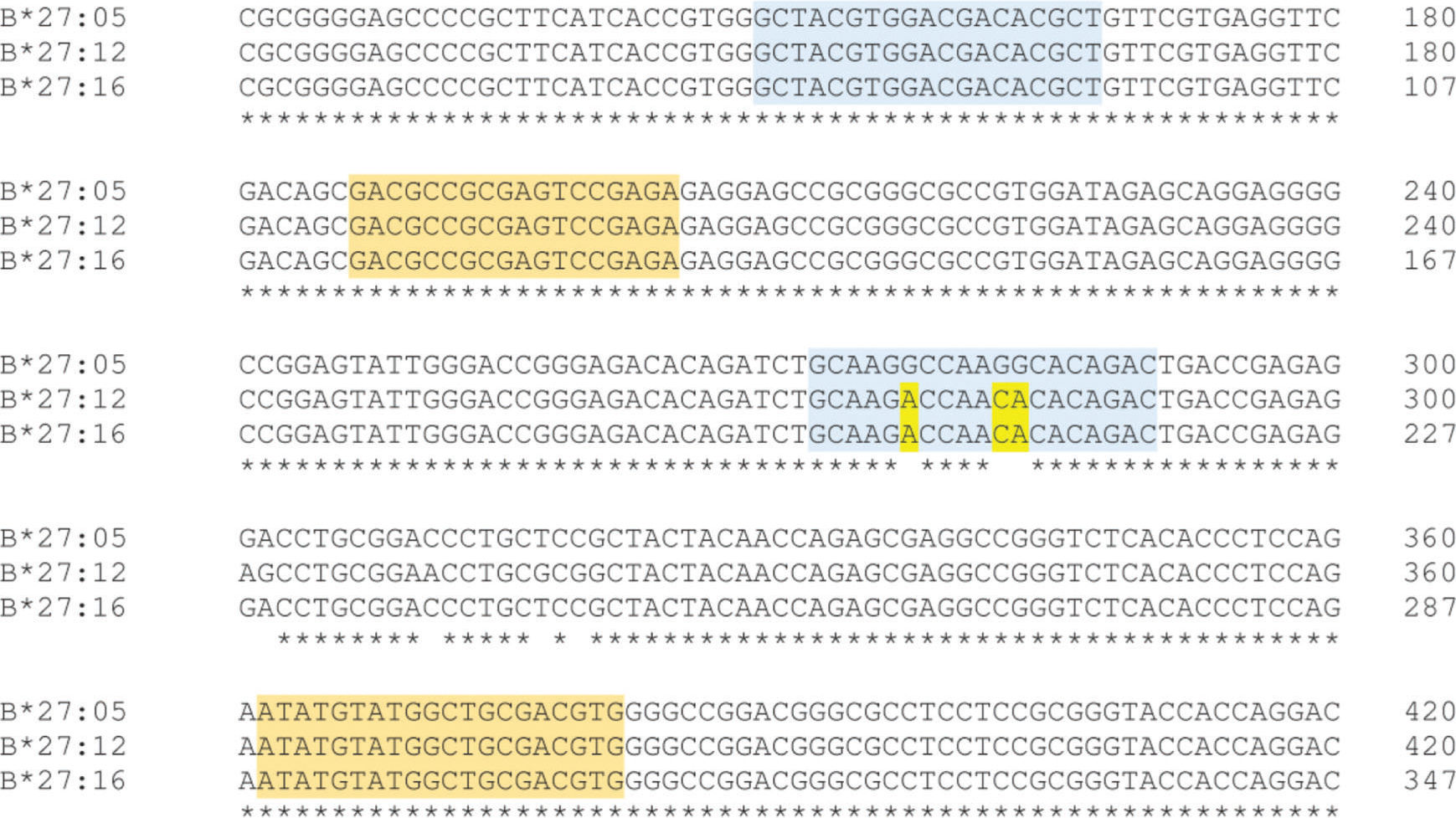
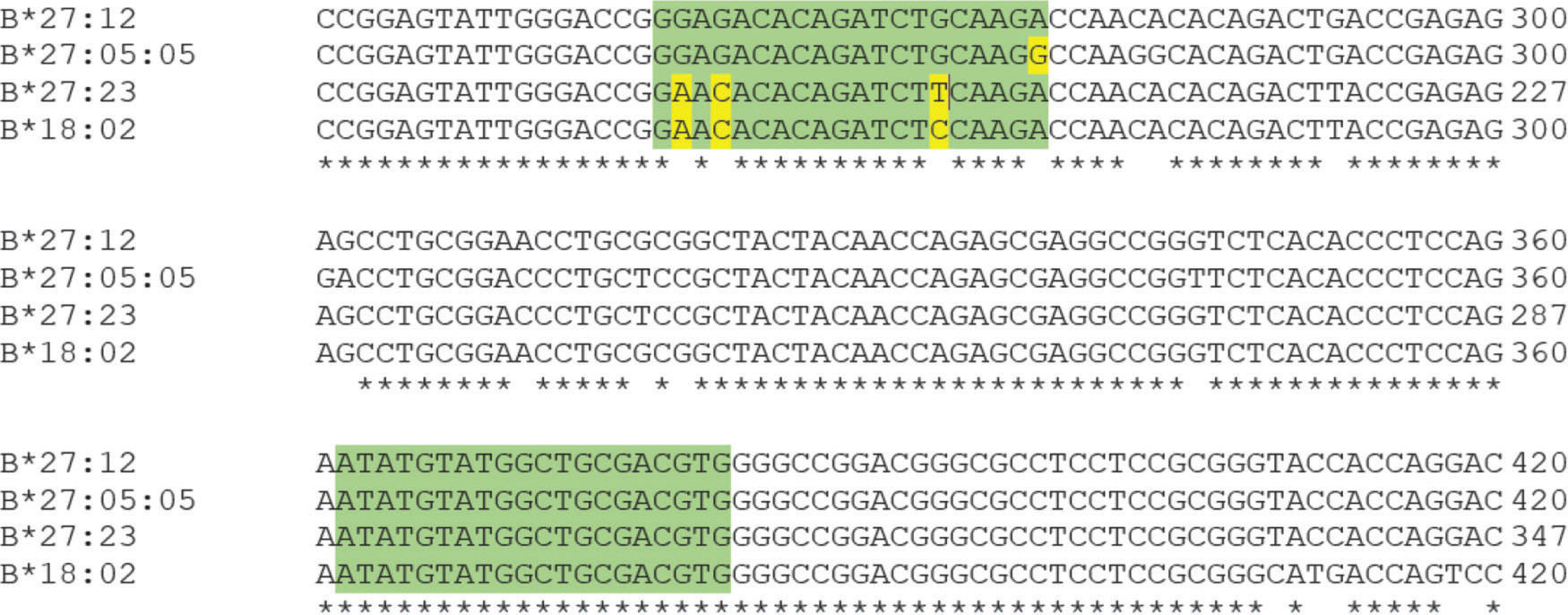
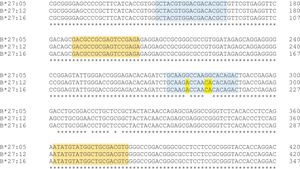
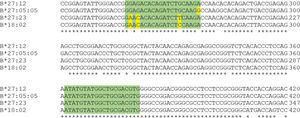
According to Duangchanchot et al. (2009), SC1 and SC2 mixes can be used to detect whether the samples are HLA-B*27-positive or -negative (11). Our results were in agreement with this, but there was a discordance for SC1 mix, which did not amplify HLA-B*27:12 and HLA-B*27:16 alleles. As shown in Appendix Figure S3, there are three nucleotides that diverge between the sequence of SC1 reverse primer and the sequence of HLA-B*27:12 and B*27:16 alleles. Since the PCR is performed at high annealing temperatures (68°C and 65°C) for almost the entire amplification process, the SC1 reverse primer is unable to bind to the template due to these mismatches, which can lead to inefficient or no amplification of those alleles. Appendix Figure S4 shows a similar situation for PCR-SSP with primer mix 12. In this situation, the alleles HLA-B*27:05:05, B*27:23, and B*18:02 have a low amplification efficiency and Duangchanchot et al. (2009) describes a possible amplification for these alleles.
PCR-SSP has been described as being more economical, relatively simple, fast, and highly sensitive and specific than other methods, (12,22). In our study, the standardized PCR-SSP technique showed 100% sensitivity and specificity for HLA-B heterozygous samples, as measured by the concordance of the PCR-SSP and PCR-SSOP results. The main advantage of this technique is that it can be performed in small laboratories with simple equipment, such as a thermal cycler, PCR workstation, and electrophoresis apparatus.
Rare alleles were not detected using the standardized PCR-SSP method and this limitation was due to the database used for standardizing the technique. Another limitation of the technique is that it cannot detect homozygous HLA-B*27 samples. The frequency of individuals homozygous for HLA-B*27 varies in different populations. Zou et al. (2015) identified one (0.4%) homozygous patient (HLA-B*27:05/27*05) among 247 Chinese patients with AS (23). Yi et al. (2013) studied 336 positive HLA-B*27 Korean patients with AS and identified 9 (2.67%) to be homozygous (24). In this study, we identified two (2.2%) homozygous individuals (HLA-B*27:CAFRW/*27:CAFRW) among 90 HLA-B*27 positive individuals. Studies have reported that individuals who were homozygous for HLA-B*27 are more susceptible to develop AS (5,25,26), but it was not linked to severe clinical manifestations of the disease (26–28). Other genotyping techniques, such as sequencing or high-resolution PCR-SSOP, should be performed to identify the homozygosis. In addition, to define heterozygosis in homozygous samples for the HLA-B*27 allelic variants, all PCR-SSPs (Figure 1) must be performed.
The standardized technique may be used as an auxiliary method in the diagnosis of AS and other diseases as well as in future genetic association studies.
CONCLUSIONThis study standardized and optimized a PCR-SSP method for HLA-B*27 genotyping in heterozygous individuals, which was considered of high definition, with good sensitivity and affordability.
AUTHOR CONTRIBUTIONSLara-Armi FF carried out the writing of the manuscript and performed the standardization and the validation of the PCR-SSP; Visentainer JE, Colli CM and Moliterno RA were responsible for the study design; Alves HV carried out the PCR-SSOP for validation; Rocha-Loures MA and Neves JS were responsible for sample collection; Lima Neto QA was responsible for in silico analyses. Sell AM was responsible for the study design, writing and revision of the manuscript.
We thank everyone for participating in the study: patients and controls, the Laboratory of Immunogenetics of the State University of Maringá, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and Fundação Araucária do Estado do Paraná, Brazil. This work was supported by the Laboratory of Immunogenetics of the State University of Maringá (LIG-UEM: Proc. n°. 1589/2017-CSD-UEM).
No potential conflict of interest was reported.