Prompt detection of antibiotic resistance genes in healthcare institutions is of utmost importance in tackling the spread of multi-drug resistant micro-organisms. We evaluated the Antimicrobial Resistance (AMR) Direct Flow Chip Kit versus phenotypic screening assays for rectal and nasopharyngeal specimens upon ICU admission.
MethodsA total of 184 dual specimens (92 rectal and 92 nasopharyngeal swabs) from 92 patients were collected from 11/2017 to 8/2018. All swabs were subjected to both AMR and phenotypic tests according to their origin. The degree of agreement of the two methods was assessed by the kappa coefficient.
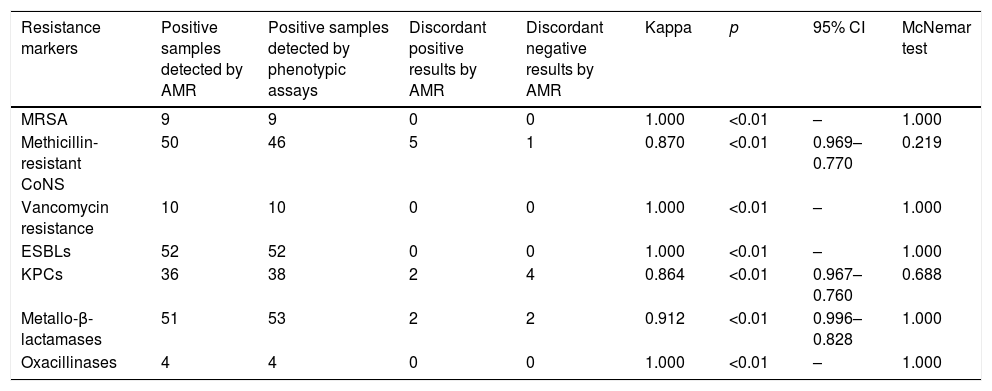
ResultsThe kappa coefficient showed perfect agreement for MRSA, ESBLs, oxacillinases and vancomycin resistance genes (1.000, p<0.01) and very good agreement for mecA-positive CoNS, KPC-carbapenemases and metallo-beta-lactamases (0.870, p<0.01; 0.864, p<0.01; and 0.912, p<0.01, respectively).
ConclusionThe AMR Direct Flow Chip Kit is a useful alternative to phenotypic testing for rapid detection of resistance markers.
La detección rápida de genes de resistencia a antibióticos en instituciones de salud es de importancia extrema para abordar la propagación de microorganismos multi-resistentes. Evaluamos el Antimicrobial Resistance Direct Flow Chip (AMR) versus los ensayos de detección fenotípica para muestras rectales y nasofaríngeas al ingreso en la UCI.
MétodosSe recogieron 184 muestras duales (92 rectales y 92 nasofaríngeos) de 92 pacientes durante 11/2017-8/2018. Todos los hisopos se sometieron a pruebas de AMR y fenotípicas según su fuente. El grado de acuerdo de los 2 métodos fue evaluado por el coeficiente kappa.
ResultadosEl coeficiente kappa mostró una concordancia perfecta para MRSA, ESBL, oxacilinasas y para genes de resistencia a vancomicina (1.000, p<0,01) y muy buena concordancia para CoNS mecA positivos, carbapenemasas KPC y metalo-beta-lactamasas (0,870, p<0,01; 0,864, p<0,01 y 0,912, p<0,01, respectivamente).
ConclusiónEl AMR es una alternativa útil a las pruebas fenotípicas para la detección rápida de marcadores de resistencia.
Antibiotic resistance, mainly among Gram negative nosocomial pathogens, Enterococci and Staphylococcus aureus is considered as a public health problem of major importance worldwide.1 Specific genes provide the bacteria with the ability to produce enzymes that inactivate antibiotics, express efflux pumps, reduce their outer membrane permeability, convert the drugs’ target sites or even alter the metabolic pathway of antimicrobial agents. In this regard, timely detection of antibiotic resistance determinants, especially for hospital associated infections, plays a crucial role for the treatment of critically ill patients and the containment of the spread of multi-drug resistant bacteria within health-care institutions.2
Although early detection is of great importance, in-house phenotypic tests are time-consuming whereas, PCR-based techniques are expensive and target only specific genes.3,4 The lately introduced DNA microarrays combined with multiplex PCR offer the advantage of simultaneous rapid detection of numerous predetermined resistance genes in a single test by clinical specimens or cultures.5 This advantage led to the development of various assays for the identification of resistance genes among Gram negatives and S. aureus.6–13
Antimicrobial Resistance Direct Flow Chip (AMR) (Máster Diagnóstica, Granada, Spain) is a novel microarray-based assay approved by the European Economic Area for in vitro resistance gene detection (CE IVD) by rectal and nasopharyngeal swabs. Recently, the performance of this assay using isolated colonies as a starting material has been also evaluated.14 In the present study, we evaluated the AMR using rectal and nasopharyngeal exudates from the same patient in a single assay and compared it with phenotypic screening tests.
MethodsAll rectal and nasopharyngeal swabs obtained upon admission from patients hospitalized in the intensive care unit (ICU) of AHEPA University Hospital over a nine month period (November 2019–August 2020) were included in the study. The samples were analyzed by the AMR assay and conventional phenotypic methods in order to identify patients colonized with carbapenemase/ESBL-producers and/or VRE in rectal exudates as well as patients with nasal MRSA colonization.
The AMR assay allows the rapid simultaneous detection of twenty antibiotic resistance genetic markers, closely associated with multi-drug resistant organisms such as extended-spectrum beta-lactamase (ESBL) producers, carbapenem-resistant Gram-negatives, methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococci (VRE). These markers belong to the gene families SHV, CTX-M, GES, SME, KPC, NMC/IMI, SIM, GIM, SPM, NDM, VIM, IMP, OXA23-like, OXA24-like, OXA48-like, OXA51-like, OXA58-like, MecA, VanA and VanB. Moreover, AMR allows the identification of S. aureus directly from clinical samples.
The test detects resistance-encoding genes directly from clinical samples (rectal and nasopharyngeal swabs) from colonized and infected patients or even subjects prone to colonization and therefore may be used for both clinical and surveillance purposes. The basic principle of the AMR Direct Flow Chip kit is the concurrent amplification of twenty resistance DNA targets by multiplex PCR, followed by manual or automatic hybridization on a membrane containing specific DNA probes. A major advantage of this test is that it provides results in a time frame lower than 5h.
In parallel with the AMR, all swabs were cultured using conventional phenotypic methods in accordance with the infection control policy of our hospital. More precisely, nasopharyngeal swabs were inoculated onto chromogenic media (Oxoid Brilliance MRSA 2 agar, Thermo Fisher Scientific) whereas, rectal swabs were placed onto chromID CARBA medium (bioMérieux, France), Brilliance ESBL agar (Oxoid, Basingstoke, United Kingdom), chromID VRE medium (Oxoid, Basingstoke, United Kingdom) and MacConkey agar with imipenem and ceftazidime discs. All agar plates were incubated at 37°C for 18–24h. Bacterial identification of the retrieved colonies from all media were identified by conventional tests (such as tube coagulase test) as well as by the automated VITEK2 system (bioMérieux, France). Antimicrobial susceptibility testing was performed to all suspected MRSA and VRE producing isolates in order to confirm oxacillin and vancomycin resistance respectively. Additionally, the cefoxitin screen test was used for MRSA detection. For carbapenemase and ESBL production, phenotypic disc synergy tests were applied.15 Screening for the presence of carbapenemases was performed with the Modified Hodge Test (MHT), while the type of either MBL or KPC was assessed by the combined-disc test (CDT) using meropenem discs with and without phenyl-boronic acid and/or EDTA. The likelihood of extended-spectrum-β-lactamases (ESBLs) co-production was assessed with the modified CLSI ESBL confirmatory test using cefotaxime and ceftazidime discs with and without clavulanate, on which both EDTA and PBA were dispensed.
In order to evaluate the degree of agreement between AMR and phenotypic methods, the kappa coefficient was calculated and the McNemar test was performed using IBM SPSS Statistics 21. Discrepant results were further evaluated by PCR using the respective specific primers.
ResultsDuring the study period, a total of 184 samples (92 rectal and 92 nasopharyngeal swabs) were subjected both to routine and AMR assay for the detection of antimicrobial resistance genes. With regard to Gram positives, perfect agreement of the two methods was shown for methicillin resistance among S. aureus isolates (in all cases, AMR was positive for both mecA gene and S. aureus) and VRE (vanA and/or vanB genes were identified among Enterococci presenting vancomycin resistance). Moreover, all but one nasal samples with a positive cefoxitin screen for Coagulase Negative Staphylococcus (CoNS), were mecA positive using the AMR (Table 1).
Degree of agreement of AMR and phenotypic methods.
| Resistance markers | Positive samples detected by AMR | Positive samples detected by phenotypic assays | Discordant positive results by AMR | Discordant negative results by AMR | Kappa | p | 95% CI | McNemar test |
|---|---|---|---|---|---|---|---|---|
| MRSA | 9 | 9 | 0 | 0 | 1.000 | <0.01 | – | 1.000 |
| Methicillin-resistant CoNS | 50 | 46 | 5 | 1 | 0.870 | <0.01 | 0.969–0.770 | 0.219 |
| Vancomycin resistance | 10 | 10 | 0 | 0 | 1.000 | <0.01 | – | 1.000 |
| ESBLs | 52 | 52 | 0 | 0 | 1.000 | <0.01 | – | 1.000 |
| KPCs | 36 | 38 | 2 | 4 | 0.864 | <0.01 | 0.967–0.760 | 0.688 |
| Metallo-β-lactamases | 51 | 53 | 2 | 2 | 0.912 | <0.01 | 0.996–0.828 | 1.000 |
| Oxacillinases | 4 | 4 | 0 | 0 | 1.000 | <0.01 | – | 1.000 |
Among Gram negatives, AMR detection of ESBLs (blaCTX-M and/or blaSHV) was in full concordance with the phenotypic assay results and as for carbapenemases, the agreement of the two assays was very good for both KPC- and MBL-producing isolates (0.864, p<0.01 and 0.912, p<0.01 respectively).
Overall, the degree of agreement of AMR and phenotypic methods for MRSA, mecA positive CoNS, ESBLs, oxacillinases, vancomycin resistance genes, KPC-carbapenemases and metallo-beta-lactamases as expressed by the kappa coefficient and the McNemar test is shown in Table 1. All ‘discordant AMR positives’ resulted as ‘true positives’ by PCR whereas, ‘discordant AMR negatives’ were confirmed as ‘true negatives’.
DiscussionWe evaluated the AMR in a large number of rectal and nasopharyngeal clinical specimens for the screening of patients admitted in the ICU, where rapid detection of carriers bearing resistant bacterial populations is of great importance to the implementation of infection control policies.
It has been already stated that the AMR performs comparably or even better than other commercially available DNA arrays presenting also the advantage of skipping the DNA extraction step.14 Moreover, this method allows the simultaneous detection of 20 resistance genes and the direct detection from clinical samples.
In our study, the degree of agreement of AMR and phenotypic methods was very good to excellent as shown by the kappa coefficient and the McNemar test. Our sample included a significant number of KPC- and MBL-producing isolates, showing that AMR is able to detect these major resistance determinants in most of the cases. Noteworthy also, the agreement regarding the detection of MRSA was excellent. Overall, it has been shown that AMR is a useful alternative to the resistance detection methods performed in our laboratory targeting simultaneously more resistance genes than simple PCR assays and being significantly faster than phenotypic tests. This faster detection of antibiotic resistance determinants could play a crucial role for the implementation of infection control measures such as patient isolation and cohorting, thus limiting the spread of multi-drug resistant bacteria in our hospital. Its cost is comparable to PCR methods but much higher than that of phenotypic assays.
The present study presents however some limitations that have to be mentioned. The AMR has been evaluated only against phenotypic assays and molecular confirmation was limited to discrepant results due to the large sample size. For the same reason, no clonality analyses were performed therefore, clonality bias may at some extent have influenced our results. Additionally, it should be mentioned that positive results in our study refer only to the genetic targets that are present in our geographic region and do not include the whole panel of the method.
In conclusion, our data demonstrate that the AMR Direct Flow Chip Kit is a useful tool for the rapid detection of antibiotic resistance determinants upon ICU and generally, hospital admission and may prove valuable for the prompt implementation of infection control strategies in hospital settings.
FundingFinancial support was partially provided by Máster Diagnóstica, Granada, Spain. The funding source had no involvement in the study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication. Part of the study was presented at the 29th ECCMID conference (Amsterdam, Netherlands).
Conflicts of interestNone.