Pseudomonas aeruginosa causes severe infections, particularly in healthcare settings and immunocompromised patients in whom MDR and XDR isolates are more prevalent. The aim of this study is to validate a method based on MALDI-TOF spectra analysis for early detection of the ST175 high-risk clone (HRC).
MethodsThe MALDI-TOF spectra of the first 10 P. aeruginosa clinical isolates from each of the 51 participating Spanish hospitals were analyzed (n=506). Resistance profiles were determined by broth microdilution, and clonal epidemiology was assessed by PFGE analysis and multilocus sequence typing (MLST) in a previous study.
ResultsAmong all the isolates, 14.2% were XDR and 26.9% were non-susceptible to meropenem, while rates of resistance to ceftolozane/tazobactam (3.6%) and colistin (5.7%) were low. Up to 41.7% of all XDR isolates belonged to the ST175 clone, and most of them were only susceptible to ceftolozane/tazobactam and colistin. However, most of the resistance to ceftolozane/tazobactam among isolates belonging to this HRC was observed in carbapenemase-producing isolates. A model based on the presence of two MALDI-TOF biomarker peaks at m/z 6911 and 7359 yielded a negative predictive value (NPV) and a positive predictive value (PPV) of 99.8% and 91.9%, respectively, and sensitivity and specificity values of 97.1% and 99.4%, respectively.
ConclusionsMALDI-TOF spectra analysis using a model based on the presence of two biomarker peaks proved to maintain high sensitivity and specificity for early detection of the ST175 HRC in a large collection of isolates from all Spanish regions. These data support the use of this model in a clinical setting; however, the consequences of detection of the ST175 HRC, such as choice of empirical antibiotic therapy, must be consistent with local epidemiology and the prevalence of certain resistance patterns of this HRC, such as carbapenemase production, in a given geographical area.
P. aeruginosa causa infecciones graves, particularmente asociadas a cuidados sanitarios y en pacientes inmunodeprimidos, donde los aislamientos MDR o XDR son más frecuentes. El objetivo de este estudio es validar el método basado en el análisis de espectros MALDI-TOF para la detección precoz del clon de alto riesgo ST175.
MétodosSe analizaron los espectros de MALDI-TOF de los primeros 10 aislados clínicos de P. aeruginosa pertenecientes a cada uno de los 51 hospitales españoles participantes (n=506). En un trabajo previo se determinaron los perfiles de resistencia mediante microdilución en caldo y se estableció su relación clonal mediante electroforesis en campo pulsante (PFGE) y multilocus sequence typing (MLST).
ResultadosDel total de los aislamientos el 14,2% fueron XDR y el 26,9% resultaron ser no sensibles a meropenem, mientras que la resistencia al ceftolozano-tazobactam (3,6%) y la colistina (5,7%) fue baja. Hasta el 41,7% de todos los aislamientos XDR pertenecieron al clon ST175 y la mayoría de ellos solo resultaron ser sensibles a ceftolozano-tazobactam y a colistina. No obstante, la mayor parte de la resistencia a ceftolozano-tazobactam observada entre los aislados pertenecientes a este clon de alto riesgo se debió a la producción de carbapenemasas. El modelo basado en la presencia de dos picos de biomarcadores MALDI-TOF en m/z 6911 y 7359 obtuvo un valor predictivo negativo y positivo (VPN/VPP) del 99,8/91,9% y valores de sensibilidad y especificidad del 97,1/99,4%, respectivamente.
ConclusionesEl análisis de los espectros de MALDI-TOF utilizando el modelo basado en la presencia de dos picos de biomarcadores ha demostrado poseer una alta sensibilidad y especificidad para la detección precoz del clon de alto riesgo ST175 en una gran colección de aislados clínicos representando todo el territorio español. Estos datos, por tanto, respaldan el uso de este modelo en el entorno clínico; no obstante, las consecuencias derivadas de la detección del ST175, como la elección de un tratamiento antibiótico empírico, deben ser acordes a la epidemiología local y a la prevalencia de ciertos patrones de resistencia de este clon de alto riesgo en una determinada área geográfica, como puede ser la producción de carbapenemasas.
Pseudomonas aeruginosa causes severe infections, particularly in health care settings and in immunocompromised patients from whom multidrug-resistant (MDR) or extensively drug-resistant (XDR) isolates are more prevalent. In this scenario, these infections are associated with high morbidity and mortality given the limited therapeutic options.1,2P. aeruginosa has an epidemic population structure composed by a limited number of the so-called “high-risk” clones (HRC) disseminated in hospitals worldwide, among which the ST175 is one of the most widespread.3,4 A recent single-center pilot study described applicable methods in the routine workflow of a clinical microbiology laboratory for the rapid detection of ST175 MDR/XDR clinical isolates based on the MALDI TOF spectra analysis and/or O4 serotyping.5 The objective of this work is to validate the model in a previously characterized collection of clinical isolates belonging to a nationwide multi-center study involving 51 Spanish hospitals, where the ST175 HRC is the most prevalent.
MethodsBacterial strains: The first ten P. aeruginosa clinical isolates collected during October 2017 from each of the 51 Spanish hospitals (covering all 17 Spanish regions) participating in the GEMARA-SEIMC/REIPI project6 were blindly studied. One of the 51 hospitals could only contribute with 6 isolates which is reflected in the total number of isolates studied (n=506).
MALDI-TOF analysis: Strains were cultured for 18h at 37°C in Columbia agar supplemented with 5% of sheep blood. Protein extracts were obtained from the colonies after on-plate extraction with formic acid followed by the addition of the matrix solution, following Bruker™ recommendations. Two spectra for each strain were manually analyzed using Bruker™ provided software (FlexAnalysis™), as described previously.5
Susceptibility testing and clonal epidemiology: The MICs of piperacillin–tazobactam, ceftazidime, ceftolozane–tazobactam, cefepime, imipenem, meropenem, ciprofloxacin, tobramycin, amikacin and colistin had been determined by broth microdilution in the previous work.6 Clinical susceptibility categories were interpreted following v8.1 2018 EUCAST guidelines (www.eucast.org). MDR and XDR profiles were defined according to previously described criteria.7 AmpC hyperproduction, OprD deficiency, and the presence of horizontally acquired β-lactamases had been assessed in the previous work. The clonal relatedness of XDR isolates was previously evaluated by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST)6; MDR isolates were analyzed using the same procedure in the present work.
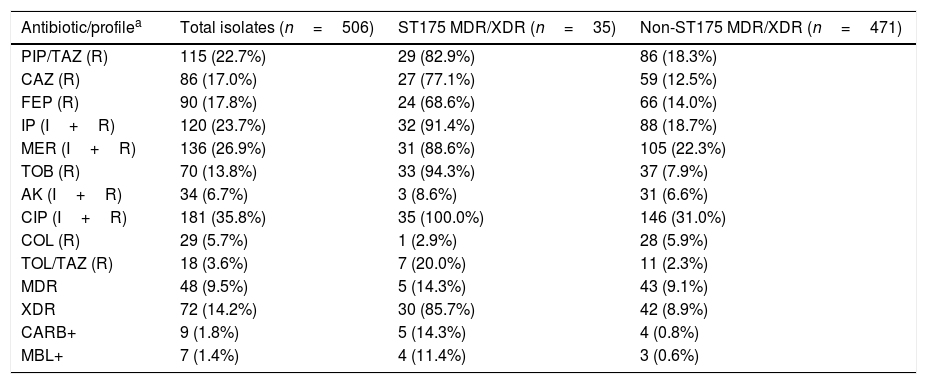
ResultsTable 1 shows the susceptibility data for the 506 clinical isolates tested, which represents a subset of the previously studied collection.6 Up to 9.5% of the isolates were MDR whereas 14.2% of them were categorized as XDR. Non-susceptibility to meropenem and amikacin was observed in the 26.9% and 6.7% of the isolates, respectively, whereas non-susceptibility to ceftolozane–tazobactam and colistin was documented in 3.6% and 5.7% of the total isolates, respectively. Phenotypic tests along with specific PCR and sequencing revealed that nine isolates (1.8%) were carbapenemase-producers, of which seven (1.4%) were metallo-beta-lactamases (three blaVIM-2, two blaVIM-20, one blaVIM-1 and one blaIMP-1), and two (0.4%) were serine-beta-lactamases (blaGES-5). Molecular typing of MDR/XDR isolates revealed that 35 (6.9%) belonged to the ST175 clone. All of them were XDR (5.9%), except 5 that were MDR (1.0%). The ST175 clone accounted for 41.7% and 29.2% of all XDR and XDR+MDR clinical isolates from this collection, respectively. Up to 88.6% of all ST175 MDR/XDR isolates were non-susceptible to meropenem and 8.6% were amikacin non-susceptible. Only one ST175 isolate was resistant to colistin and ceftolozane–tazobactam resistance was documented in 7 isolates (20.0%), of which 5 was due to carbapenemase production (Table 1).
Resistance profiles and characteristics of the collection of clinical isolates tested.
| Antibiotic/profilea | Total isolates (n=506) | ST175 MDR/XDR (n=35) | Non-ST175 MDR/XDR (n=471) |
|---|---|---|---|
| PIP/TAZ (R) | 115 (22.7%) | 29 (82.9%) | 86 (18.3%) |
| CAZ (R) | 86 (17.0%) | 27 (77.1%) | 59 (12.5%) |
| FEP (R) | 90 (17.8%) | 24 (68.6%) | 66 (14.0%) |
| IP (I+R) | 120 (23.7%) | 32 (91.4%) | 88 (18.7%) |
| MER (I+R) | 136 (26.9%) | 31 (88.6%) | 105 (22.3%) |
| TOB (R) | 70 (13.8%) | 33 (94.3%) | 37 (7.9%) |
| AK (I+R) | 34 (6.7%) | 3 (8.6%) | 31 (6.6%) |
| CIP (I+R) | 181 (35.8%) | 35 (100.0%) | 146 (31.0%) |
| COL (R) | 29 (5.7%) | 1 (2.9%) | 28 (5.9%) |
| TOL/TAZ (R) | 18 (3.6%) | 7 (20.0%) | 11 (2.3%) |
| MDR | 48 (9.5%) | 5 (14.3%) | 43 (9.1%) |
| XDR | 72 (14.2%) | 30 (85.7%) | 42 (8.9%) |
| CARB+ | 9 (1.8%) | 5 (14.3%) | 4 (0.8%) |
| MBL+ | 7 (1.4%) | 4 (11.4%) | 3 (0.6%) |
PIP/TAZ: piperacillin–tazobactam; CAZ: ceftazidime; FEP: cefepime; IP: imipenem; MER: meropenem; TOB: tobramycin; AK: amikacin; CIP: ciprofloxacin; COL: colistin; TOL/TAZ: ceftolozane–tazobactam; MDR: multi-drug resistant; XDR: extensively-drug resistant; CARB+: carbapenemase producers; MBL+: metallo-β-lactamase producers.
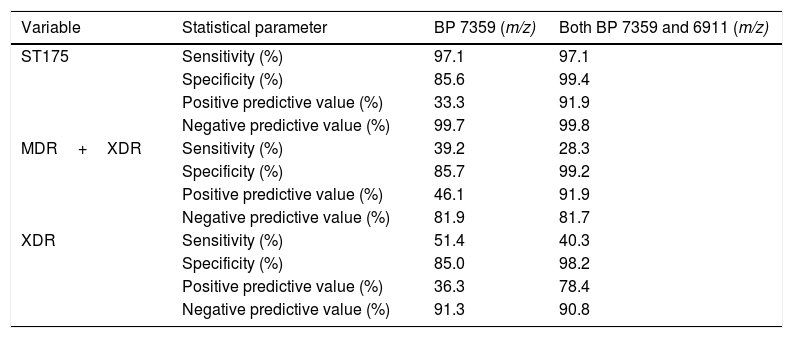
As shown in Table 2, all but one confirmed MDR/XDR ST175 isolates showed the biomarker peak at m/z 7359 described by Cabrolier et al.8 (97.1% sensitivity), however it was also present in 68 isolates not belonging to the MDR/XDR group, yielding a specificity of 85.6% and a PPV of 33.3%. Using the updated model based in the presence of two biomarker peaks,5 false positives decreased from 68 to 3 isolates, increasing the specificity to 99.4% and the PPV to 91.9%. These 3 false positive isolates were further studied and were not related to the ST175 cluster and showed a non-MDR/XDR resistance profile. The m/z 6911 biomarker peak was present in all confirmed ST175 MDR/XDR but given that the peak at m/z 7359 was absent in one of them, correct identification could only be achieved in 34 out of 35 isolates using this model (sensitivity 97.1%, NPV 99.8%).
Performances of the presence of each biomarker peak (BP) alone and in combination, as recognizing methods of the ST175 high-risk clone.
| Variable | Statistical parameter | BP 7359 (m/z) | Both BP 7359 and 6911 (m/z) |
|---|---|---|---|
| ST175 | Sensitivity (%) | 97.1 | 97.1 |
| Specificity (%) | 85.6 | 99.4 | |
| Positive predictive value (%) | 33.3 | 91.9 | |
| Negative predictive value (%) | 99.7 | 99.8 | |
| MDR+XDR | Sensitivity (%) | 39.2 | 28.3 |
| Specificity (%) | 85.7 | 99.2 | |
| Positive predictive value (%) | 46.1 | 91.9 | |
| Negative predictive value (%) | 81.9 | 81.7 | |
| XDR | Sensitivity (%) | 51.4 | 40.3 |
| Specificity (%) | 85.0 | 98.2 | |
| Positive predictive value (%) | 36.3 | 78.4 | |
| Negative predictive value (%) | 91.3 | 90.8 |
Given that the ST175 of P. aeruginosa is very prevalent and it is strongly linked to a MDR/XDR profile,4,6 the ability to predict an XDR+MDR or an XDR profile through the presence of one or both biomarker peaks was also evaluated in this collection. As shown in Table 2, sensitivity and specificity using the model based in two biomarker peaks for detecting XDR+MDR isolates were 28.3% and 99.2%, and for detecting XDR isolates were 40.3% and 98.2% respectively. On the other hand, PPV and NPV for detecting XDR+MDR isolates were 91.9% and 81.7% and 78.4% and 90.8% for the XDR isolates alone, respectively.
DiscussionIn the recent years MALDI-TOF analysis has become a useful, inexpensive, first-line epidemiological tool for typing bacteria, however only few studies establish the bases aimed at the standardization of a working methodology.9,10 The first model aimed at detecting the ST175 was described by Cabrolier et al.8 and it considered the presence of the peak at m/z 7359 and the absence of some others. In agreement with our findings this peak at m/z 7359 alone indeed yielded high sensitivity and NPV but analogously it was also present in many non-ST175 isolates. The addition of a second biomarker peak at m/z 6911 described by Mulet et al.5 proved here to improve the previous model increasing the specificity from 85.6% to 99.4% and the PPV from 33.3% to 91.9% without modifying the high sensitivity and NPV.
Our results showed that increasing the studied area up to 51 hospitals spread throughout the Spanish geography did not impact sensitivity (96.4% Vs 97.1%) and specificity (97.8% Vs 99.4%) values significantly, nor the PPV (87.1% Vs 91.9%) and NPV (99.4% Vs 99.8%) when compared to the results of a single-center pilot study.5 This finding suggests that the ST175 is highly conserved, at least among the Spanish population.
In our study, ST175 clone accounted for 41.7% of all XDR isolates belonging to 51 Spanish hospitals and the vast majority was susceptible to colistin, ceftolozane/tazobactam and amikacin and non-susceptible to other antipseudomonal agents including third-generation cephalosporins, carbapenems and fluoroquinolones. These data are in agreement with other multicenter studies in Spain which showed even higher prevalence of this HRC among their XDR isolates.4,11 Altogether, this prompted us to explore the capability to predict an XDR or XDR/MDR phenotype through the presence of both biomarker peaks. The results suggest that the prediction of the ST175 HRC could be used as a specific surrogate marker of multidrug resistance; however the sensitivity and NPV would be linked to the prevalence of this HRC in a geographical area.
Thus, ST175 early detection methods like MALDI TOF spectra analysis represent a useful tool not only for guiding antibiotic therapy 24h prior to obtaining susceptibility data but also for adopting early epidemiological containment measures in the hospital, especially in geographic areas where this high-risk clone is prevalent, such as Spain and France.3,6,12
Additionally, recent works demonstrate that O4 serotype is strongly linked to the MDR/XDR profile of the widespread ST175.13 Thus, O4 serotyping might also represent a useful tool in its early detection in a clinical lab. However, this work also shows that while O4 serotype is the most prevalent among the XDR isolates (40.9%), 30% of all O4-positive isolates were not MDR or XDR, and 18.4% of the XDR ST175 isolates in this work where non-typeable. These data suggest that O4 serotyping is not sensitive and specific enough but could represent an inexpensive useful tool when used in combination with MALDI TOF spectra analysis in the clinical microbiology lab.
In summary, the results from this national multicenter evaluation of a MALDI-TOF approach for the presumptive identification of the ST175 high-risk clone of P. aeruginosa supports the use of this model in clinical settings. However, the consequences derived from the detection of the ST175 such as guiding empirical antibiotic therapy need to be consistent with the local epidemiology and the prevalence of certain resistance patterns of this HRC in a given geographical area, such as the production of carbapenemases.
FundingThis work was supported by Merck Sharp and Dohme; Plan Nacional de I+D+i 2013–2016 Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía, Industria y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD16/0016); and grants PI15/00088 and PI18/00076 co-financed by European Development Regional Fund ERDF ‘A way to achieve Europe’, Operative program Intelligent Growth 2014–2020.
Conflict of interestsThe authors declare that they have no conflicts of interest.
Members of the GEMARA-SEIMC/REIPI Pseudomonas study group:
Germán Bou, Luis Martínez-Martínez, Fátima Galán, Irene Gracia, Manuel Antonio Rodríguez, Lina Martín, Juan Manuel Sánchez, Laura Viñuela, Ma Victoria García, José Antonio Lepe, Javier Aznar, Inma López-Hernández, Cristina Seral, Francisco Javier Castillo-García, Ana Isabel López-Calleja, Carmen Aspiroz, Pedro de la Iglesia, Susana Ramón, Elena Riera, María Cruz Pérez, Carmen Gallegos, Jorge Calvo, María Dolores Quesada, Francesc Marco, Yannick Hoyos, Juan Pablo Horcajada, Nieves Larrosa, Juan José González, Fe Tubau, Silvia Capilla, Mar Olga Pérez-Moreno, Ma José Centelles, Emma Padilla, Alba Rivera, Ferrán Navarro, Raquel Elisa Rodríguez-Tarazona, Noelia Arenal-Andrés, María del Pilar Ortega, Gregoria Megías, Inmaculada García, Cristina Colmenarejo, José Carlos González, Nora Mariela Martínez, Bárbara Gomila, Salvador Giner, Nuria Tormo, Eugenio Garduño, José Andrés Agulla, Alejandro Seoane, Julia Pita, Isabel Paz Vidal, David Mauricio Guzmán, Marta García, María Luisa Pérez del Molino, Gema Barbeito, Fernando Artiles, José Manuel Azcona-Gutiérrez, Yolanda Sáenz, José Antonio Oteo, Ana González, Jennifer Villa, Fernando Chaves, Emilia Cercenado, Teresa Alarcón, Nelly Daniela Zurita, Irene Merino, María Isabel Morosini, Rafael Cantón, María Isabel Sánchez, Laura Moreno, Genoveva Yagüe, José Leiva, José Luis Barrios, Andrés Canut, Jesús Oteo.