MALDI-TOF mass spectrometry has become a reference method for the routine identification of bacterial isolates in clinical microbiology laboratories around the world. Its high specificity, user-friendliness and cost-efficiency, together with its ability to provide reliable results in less than 5min have favoured its implementation and further development. The amount of microbial species that can be identified by MALDI-TOF routinely has increased in the last few years and now it is possible to reliably identify non-tuberculous mycobacteria or closely-related species of Nocardia spp. Yeasts, both belonging to Candida and non-Candida genera can also be identified by MALDI-TOF, as well as filamentous fungi. In the latter case, both sample preparation methods and the available databases have been important factors in achieving accurate identifications at the species level.
The expertise acquired over time has allowed researchers to identify microorganisms directly from clinical samples, facilitating improved management of infected patients. This expertise has also been applied to the development of a MALDI-TOF-based methodology for the detection of different antimicrobial resistance mechanisms. Therefore, future applications such as bacterial strain typing, or the detection of virulence markers seems feasible to perform with this technology. Furthermore, other emerging mass spectrometry and spectroscopy technologies may assist MALDI-TOF in the near future to carry out important tasks that nowadays are performed by time-consuming and labour-intensive methods.
La espectrometría de masas MALDI-TOF se ha convertido en un método de referencia para la identificación de aislados bacterianos en la rutina de los laboratorios de microbiología clínica de todo el mundo. Su elevada especificidad, facilidad de aplicación y coste-eficacia, unido a su capacidad de facilitar resultados fiables en menos de 5min, han favorecido su implantación y desarrollo. La cantidad de especies microbianas que se pueden identificar mediante MALDI-TOF de forma rutinaria se ha ido incrementando en los últimos años, y actualmente es posible identificar de forma fiable micobacterias no tuberculosas o especies estrechamente relacionadas del género Nocardia. También se pueden identificar especies de levaduras —del género Candida y de otros géneros— y de hongos filamentosos. En este último caso, tanto los métodos de preparación de muestras como las bases de datos disponibles han sido factores importantes para conseguir identificaciones precisas hasta el nivel de especie.
La experiencia adquirida durante todo este tiempo ha permitido a los investigadores identificar microorganismos directamente desde muestras clínicas, lo que facilita el manejo optimizado de pacientes infectados. El conocimiento adquirido se ha aplicado también a desarrollar una metodología basada en MALDI-TOF para detectar diferentes mecanismos de resistencia a antimicrobianos. Este hecho permite creer que en el futuro se podría aplicar esta tecnología para realizar tipado bacteriano o detección de marcadores de virulencia. Además, otras tecnologías emergentes basadas en la espectrometría de masas y espectroscopía podrían, en un futuro cercano, asistir a MALDI-TOF para llevar a cabo tareas importantes que en la actualidad requieren métodos lentos y trabajosos.
MALDI-TOF MS (matrix-assisted laser desorption/ionization time of flight mass spectrometry) emerged almost two decades ago as a technique for the rapid identification of microorganisms and it has been established as a fundamental instrument in the clinical microbiology laboratory (CML).1,2 It is based on the unique protein pattern displayed by each microbial species and its detection through the application of a soft desorption/ionization – using a UV laser – to bacterial/fungal biomass embedded in an organic matrix – usually α-cyano-4-hydroxycinnamic acid, HCCA.3,4 The energy of the laser transforms the mixture into gas phase but does not fragment the molecules present. Thus, proteins ranging from 2 to 20kDa – and even bigger ones when specific matrixes are applied – can be detected. Ribosomal proteins can be found within this range and it is upon them that MALDI-TOF identification relies.
MALDI-TOF allows rapid, cost-effective and accurate identification of microorganisms, even those closely related.5–7 This goal was reached mainly due to the improvement of the sample processing methods applied prior to MALDI-TOF analysis.8 Optimized sample preparation has allowed the inclusion of mycobacteria and filamentous fungi in the list of microorganisms that can be routinely identified by MALDI-TOF.9,10
Comprehensive and updated databases containing common and rare species with clinical importance are now available. This is another key factor for the achievement of accurate identifications. Speed is also of paramount importance for clinicians to initiate directed antibiotic treatment when needed or to avoid the use of unnecessary antibiotics. The turnaround time for the identification of bacterial isolates from colonies ranges between 5 and 45min, depending on whether a protein extraction step is required, what happens less often every time due to the increasing reliability of the methodology.11 Besides, when MALDI-TOF is used for antibiotic resistance detection, this information is available to clinicians at least 24h earlier than when identification and/or resistance detection is performed using conventional methods.12 Thus, the implementation of MALDI-TOF has been shown to have a clinical impact, useful in epidemiologic studies and a support for the development of antimicrobial stewardship programs (ASP).13
Applications of MALDI-TOF for the identification of microorganismsBacterial identificationThe use of MALDI-TOF for the routine identification of bacterial isolates is the most widely spread and optimized so far. MALDI-TOF has become, over the last ten years, the reference method because of its great speed for identification, either from colonies grown on agar plates or directly from clinical samples (see below). The accuracy of MALDI-TOF has increased over time and the identification of most bacterial species commonly found in CMLs can be reliably identified nowadays using MALDI-TOF.14 Species belonging to the order Enterobacterales have been demonstrated to be accurately identified by MALDI-TOF. A collaborative study carried out in 201815 proved that all Salmonella spp. and Cronobacter spp. isolates included in the study were correctly identified at the genus level. Besides, closed-related genera (Aeromonas, Citrobacter, Escherichia, Plesiomonas, Providencia, Serratia and Yersinia) were tested in the same study so, the authors could rule out their misidentification as Salmonella spp. or Cronobacter spp. The most commonly encountered species within the Enterobacterales order (Citrobacter spp., Enterobacter spp., Escherichia spp., Klebsiella spp., Morganella spp., Proteus spp., Salmonella spp. and Serratia spp.) have been widely demonstrated to be accurately identified in the routine of CMLs using MALDI-TOF.16,17 However, MALDI-TOF has been reportedly shown to misidentify Shigella spp. as Escherichiacoli and to provide limited discrimination for the species belonging to the Citrobacter freundii, Enterobacter cloacae, Klebsiella pneumoniae, Klebsiella oxytoca complexes and to Salmonella enterica serovars.16,18
The identification of non-fermenting gramnegative rods by MALDI-TOF is currently implemented in the routine of most CMLs. The most commonly species encountered in the clinical environment (Pseudomonas aeruginosa, Stenotrophomonas maltophilia, Moraxella catharralis) have been shown to be identified accurately by different MALDI-TOF systems in 93.6–96.6% of the analyzed cases.19 However, other genera of this group have been only identified at the complex level (i.e. Burkholderia cepacia complex and Acinetobacter baumannii complex) or at the genus level (Achromobacter spp., Chryseobacterium spp. or Ralstonia spp.) due to the high homology displayed by species within the same complex and genus, respectively. These species represent a challenge also for the DNA-based methods but, in most cases, the identification at the species level is not relevant from the clinical perspective. When identification at the species level is required in order to establish the antibiotic susceptibility of a certain isolate, improvements in the database are requested using reference strains correctly identified by molecular methods at the species level. However, a higher level of accuracy may be achieved more easily in the future, thanks to the improvement in the databases and to spectra analysis.
Other gramnegative bacteria such as the HACEK group has also been successfully identified by MALDI-TOF with rates of 100% species level accuracy when enriched databases are used.20 In one study, a commercial database provided 93.0% correct genus level assignment but only 66.0% accurate species level identification.20 This is also true for the genus Neisseria since different species belonging to the normal oral microbiota have been misidentified as Neisseria meningitidis.21 Only the use of updated and enriched libraries can prevent these misidentifications and warrant an accurate identification. The level of accuracy achieved for the identification of gramnegative bacteria using MALDI-TOF is so high currently that can be coupled with the detection of resistance mechanisms – also performed by MALDI-TOF – and both goals can be achieved within 1h,22 as shown in other section of this review.
Grampositive bacteria have always posed a challenge for MALDI-TOF. Both their complex cell wall composition and the close-relatedness of the species within the main genera makes difficult to differentiate some of them. The use of formic acid for a rapid, on-plate protein extraction has greatly increased the successful identifications of these microorganisms at the species level.11 However, some species can only be reliably resolved to the complex level so far. This is the case of Streptococcus pneumoniae and the species belonging to the Streptococcus mitis complex. Despite the improvements in the available libraries, the discrimination among these Streptococcus species is not always easy. Efforts have been applied to the detection of specific peaks that allow their accurate differentiation.6 Besides, and algorithm applied to the top ten identifications provided by MALDI-TOF has been developed for the same purpose.23
Regarding anaerobic bacteria, MALDI-TOF has shown to provide species-level accuracy similar to molecular techniques, but in a rapid and cost-effective way.24 Great improvements have been achieved for the reliable identification of anaerobes thanks to a multicentric study called ENRIA, which allowed the enrichment of the Biotyper V6 library (6903 main spectrum profiles, MSPs) with well-characterized anaerobic isolates from more than 60 different genera.25 The impact of the updated library was especially significant for the identification of grampositive anaerobic isolates at the species-level, whose identification was known to be hindered by the lack of reference spectra from this group of bacteria in previous versions of the commercial libraries.
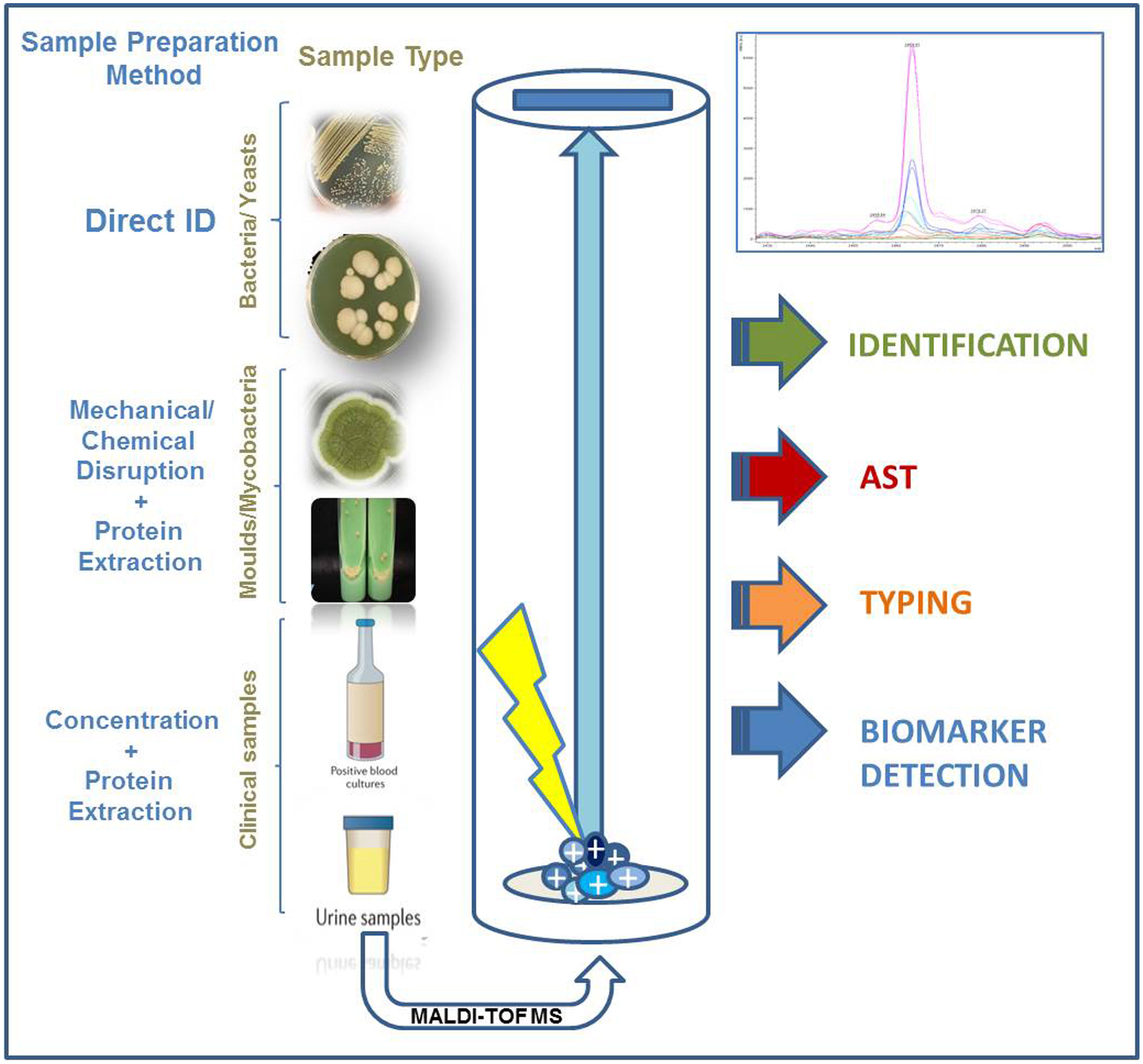
Identification of isolates belonging to the order ActinomycetalesAlthough the discrimination of the species belonging to the Mycobacterium tuberculosis complex remains one of the main challenges that MALDI-TOF should overcome in the near future, discrimination of the growing number of non-tuberculous Mycobacterium species using MALDI-TOF has been achieved in the last years thanks to the availability of updated and comprehensive databases and to the development of standardized sample processing methods.9 An “in vitro diagnostic” (IVD) procedure developed by Bruker Daltonics (Bremen, Germany) based on mechanical disruption of the bacterial biomass using crystal beads followed by protein extraction with formic acid and acetonitrile has been shown to provide high-confidence species level assignment (Fig. 1). Besides, the application of a sonication or high-performance beating step to the heat-inactivated mycobacteria cells have also shown to provide highly reliable results at the species-level. Differentiation of species belonging to complexes such as Mycobacterium avium complex is being successfully achieved thanks to the detection of species-specific marker peaks.26 Altogether, MALDI-TOF provides highly reliable identification of non-tuberculous Mycobacterium species. The accuracy of these identifications is comparable to those provided by molecular methods but the turnaround time of MALDI-TOF is around 1h, due to the need to heat-inactivate the samples during 30min. Despite the clear advantages of MALDI-TOF for the identification of these microorganisms, many users declare that the preprocessing of the samples discourages them from applying MALDI-TOF routinely for the identification of non-tuberculous mycobacteria. However, in the last years, the increasing number of publications on this subject demonstrates that this application of MALDI-TOF is becoming widely accepted.
Regarding the order Actinomycetales, clinical interest is focused on two genera: Nocardia and Streptomyces. Similar to mycobacteria, these microorganisms require disruption of their cell wall in order to be accurately identified by MALDI-TOF. However, their complicated taxonomy makes it difficult to discriminate closely related species. This is important for the correct administration of the optimal antibiotic treatment in the case of Nocardia due to species specific susceptibility patterns. The implementation of mechanical disrupture and protein extraction is advisable for routine identification of Actinomycetales, although improvements in the most updated commercial and in-house databases allow species-level identification from isolated colonies using on-plate formic acid extraction.27
Identification of yeasts and filamentous fungiThe routine identification of yeasts using MALDI-TOF is totally implemented in CMLs nowadays.2 Although the use of protein extraction was described in old studies, the identification of yeasts, especially these belonging to commonly found genera as Candida or Cryptococcus, is performed directly from colonies grown on agar media and from clinical samples (Fig. 1). Literature concerning the identification of non-Candida genera is scarce, but most authors recommend the use of in-house libraries for this purpose.
In the case of filamentous fungi, similar events have taken place to those explained above for non-tuberculous mycobacteria: the identification of these microorganisms required previous disruption in order to break their cell wall, the complexity of these eukaryotic species is higher than in the case of bacteria, and the pattern of expressed proteins changes with time. In addition to these difficulties, the lack of comprehensive commercial databases was a fact until very recently. Thus, most authors eventually constructed their own in-house libraries using specific sample preparation methods.10 As a result, no standardized method for the identification of filamentous fungi is available so far, but MALDI-TOF users have applied similar combinations of mechanical and chemical disruption with successful results.28 The use of the enriched databases has proved useful, although in most cases the reference spectra included by only one research centre was very limited. This approach also presented the handicap of being time consuming and requiring research personnel with high mass spectrometry expertise. A more global attempt has been developed recently by Normand et al.29 A database containing over 11,000 reference spectra from 938 fungal species of clinical interest have been constructed and it can be accessed on-line in order to compare spectra from unidentified fungal isolates. The database has been shown to provide correct identification of clinically important species from the genus Aspergillus, Trichophyton or Microsporum and allows CMLs to implement the routine identification of filamentous fungi in a rapid and reliable manner.
The implementation of MALDI-TOF for the identification of other groups of microorganisms such as viruses or parasites has been developed recently. Further information about this topic can be found in the proceedings published recently by the Spanish Society of Clinical Microbiology and Infectious Diseases (https://seimc.org/contenidos/documentoscientificos/procedimientosmicrobiologia/seimc-procedimientomicrobiologia65.pdf).
Direct identification of microorganisms from clinical samplesOne of the most clinically impacting applications of MALDI-TOF is the direct identification of microorganisms from clinical samples. The correct identification of the microorganisms present in blood cultures has been widely evaluated. Its implementation has allowed the change of the empirical therapy for an optimized treatment in 35.1% if the cases.30 The clinical impact of MALDI-TOF is even higher when the detection of resistance mechanisms is associated with microbial identification since the information is obtained 24–48h earlier than when conventional methods are applied.12
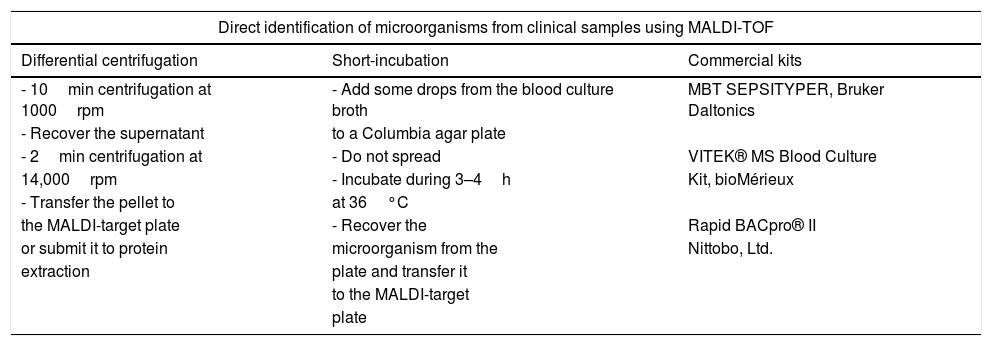
The identification of the microorganisms present in blood cultures requires a preprocessing of the cultured blood sample aiming at concentrating the pathogens and eliminating blood cells and resins present in the blood culture bottles (Fig. 1). These goals were achieved in early studies by differential centrifugation which consisted on a first centrifugation step at low speed (1000rpm) to remove substances present in the blood culture that may interfere with the identification of the pathogens. Then, a second centrifugation step at high speed (14,000rpm) is applied in order to concentrate the microorganisms present. Later, the pellet can be directly applied onto the MALDI target plate or submitted to protein extraction before their analysis by MALDI-TOF (Table 1). Other authors have avoided this sample processing by culturing some drops of the blood culture on Columbia agar medium and incubating the plates for 3–4h. The identification was carried out successfully by scrapping the microorganisms grown on the agar Plates.31 However, later studies have shown that the use of ad-hoc commercial kits provide standardization of the method and increased species and genus level identification.32
Different methods applied for the direct identification of microorganisms from blood cultures and other clinical samples.
| Direct identification of microorganisms from clinical samples using MALDI-TOF | ||
|---|---|---|
| Differential centrifugation | Short-incubation | Commercial kits |
| - 10min centrifugation at 1000rpm | - Add some drops from the blood culture broth | MBT SEPSITYPER, Bruker Daltonics |
| - Recover the supernatant | to a Columbia agar plate | |
| - 2min centrifugation at | - Do not spread | VITEK® MS Blood Culture |
| 14,000rpm | - Incubate during 3–4h | Kit, bioMérieux |
| - Transfer the pellet to | at 36°C | |
| the MALDI-target plate | - Recover the | Rapid BACpro® II |
| or submit it to protein | microorganism from the | Nittobo, Ltd. |
| extraction | plate and transfer it | |
| to the MALDI-target | ||
| plate | ||
Regardless of the method used, the rate of correct identifications at the species level using different MALDI-TOF platforms is above 80% of the analyzed samples, fulfilling the requirements for the implementation of this method in the routine of CMLs.1 Although some microbial species, such as yeasts, have represented a challenge for MALDI-TOF,13 the use of commercial kits have started to overcome this problem. The limitation of this method so far is the reliable identification in the case of polymicrobial bacteremia.1,13
Other clinical samples such as urine and cerebrospinal fluid (CSF) have been tested with MALDI-TOF, although literature concerning this subject is scarce.13 The main limitations in these cases are the low concentration of pathogens in this type of samples (<1.5×105–106), insufficient volume of sample, the presence of other substances that may interfere with the identification and polymicrobial samples. Despite the difficulties, successfully identification of bacteria has been performed from urine and CSF samples. The differential centrifugation method has been applied in both cases, and the implementation of commercial kits for sample preparation did not only enhanced the rate of reliable species-level identifications from urine samples, but allowed the detection or antimicrobial resistance mechanisms.33
Clinical and economic impact of MALDI-TOF on rapid diagnosis and integration into antimicrobial stewardship programs (ASPs)Beyond the contribution made by MALDI-TOF to the reliability and accuracy of microbiological identification, which after 10 years of experience is beyond doubt, there are still fields to be explored. Currently, the balance between costs and savings is the cornerstone for introducing new technologies in clinical practice, as there is a reduction in the budget of health care systems and there is need of a reduction of costs without sacrificing quality.34,35 There have been several studies evaluating the cost savings of MALDI-TOF in clinical practice. Most studies focus on measuring the benefits of microbial identification in bacteremia cases, due to its clinical impact on the patients. Although the methodology for the direct identification microorganisms grown in blood cultures is not yet fully standardized and great variability is shown depending on the volume of blood culture used, the method of extraction of microorganisms (centrifugation, commercial methods, tubes with separator gel, etc.), the results suggest that the rapid analysis of positive blood cultures using MALDI-TOF, by any of these methodologies, is a valuable contribution. The most important clinical benefits observed in the literature are an early administration of adequate antimicrobial therapy, reduced morbidity and mortality, a reduction in hospital stay and a reduction in costs per hospitalized patient.
One of the first studies to evaluate the clinical impact of MALDI-TOF on patient outcome was conducted by Huang et al.36 These researchers proposed integrating MALDI-TOF into the workflow of an ASP. The study included 501 patients with bacteremia or candidemia for evaluation, and 245 patients in the intervention group. In patients included in the MALDI-TOF group, the time to organism identification and time to effective antibiotic therapy were reduced, optimal antibiotic therapy was provided, mortality was lower and the length of intensive care unit stay and risk of recurrent bacteremia were reduced.
As mentioned in the previous epigraph, the study of Clerc et al. demonstrated that in 35% of the episodes (n=202), the information generated by MALDI-TOF had a clinical impact by modifying the empirical therapy. The most remarkable consequence was the correct broadening of the antibiotic spectrum in 43.7% of cases. In those cases, in which MALDI-TOF did not have a documented impact, modification of the therapy would have been possible in 24% of cases.30 Factors involved in not taking the MALDI-TOF results into account included younger age of the patient, admission of patient in intensive care units, male sex and lack of confidence shown by medical staff in the use of an unfamiliar technology. Verroken et al. evaluated the impact on antimicrobial prescription of a workflow in which the identification was provided by MALDI-TOF.12 Partial susceptibility results were performed regarding the previously obtained identification results and were provided on the same day, as identification from positive blood cultures of Enterobacterales, P. aeruginosa and Staphylococcus aureus. The mean time to identification was reduced by around 65%, mean time to complete susceptibility results was reduced by 27%, and the mean time to optimal antimicrobial treatment was decreased to 16h. Beganovic et al. evaluated the impact of MALDI-TOF in combination with antimicrobial stewardship intervention on patient outcome.37 The primary outcome was the time to optimal therapy, which was significantly reduced by the use of MALDI-TOF. The mean time to microbiological clearance, the mean length of hospital stay, the mean stay in a critical care unit, the number of patients with recurrence of the same bacteremia, and the mean length of antimicrobial therapy were also reduced.
A very recent study comparing the cost/effectiveness of different rapid methods for the diagnosis of bacteremia (especially molecular methods) combined or not with an ASP38 shows that the diagnosis of bacteremia by the classical methodology, combined with an ASP, does not significantly improve survival either globally or adjusted for quality of life (quality-adjusted life years, QALY) compared to the classical diagnosis not combined with an ASP, although it reduces costs by 25%. On the other hand, the combination of rapid diagnosis using MALDI-TOF associated with an ASP not only reduces costs by an additional 25% compared to the classical ASP methodology, but also, compared to the simple conventional methodology, improved overall survival by 7% and improved QALY by 0.94 years. The authors conclude that, even in comparison with the combination ASP using molecular techniques, coupling MALDI-TOF and ASP is the most cost-effective option, and the one associated with greater savings per QALY gained.
Further cost-effective multicentre studies are required to evaluate the impact of identification by MALDI-TOF MS compared to the previously reported methodologies and others yet to be developed, but we also expect interesting research in new fields as resistance detection and detection of high-risk clones which are increasingly introduced in the routine of microbiology laboratories.
Other applications of MALDI-TOFThe versatility of MALDI-TOF favours the emergence of more and more applications based on this technology. The most promising ones that are ready or almost ready to be used in clinical practice are the detection of antimicrobial resistance, the typing studies and the detection of virulence markers.
Detection of antimicrobial resistance mechanismsOne of the most widespread application related to the detection of antimicrobial resistance by MALDI-TOF MS is the detection of β-lactamases and carbapenemases by measuring the hydrolysis of different antibiotics in the presence of resistant bacteria.39 If the antibiotic is hydrolyzed, the peak pattern of the antibiotic shifts with respect to the native antibiotic, so we have two different spectra for each form of the antibiotic. This way we can compare the results of our sample spectra to a positive and negative control. If the spectra have the same peak pattern as the positive control, the antibiotic is hydrolyzed and the bacteria analyzed carries a β-lactamase and/or carbapenemase. If the spectra have the same peak pattern as the negative control, the antibiotic is not hydrolyzed, and the bacteria does not carry a β-lactamase and/or carbapenemase.
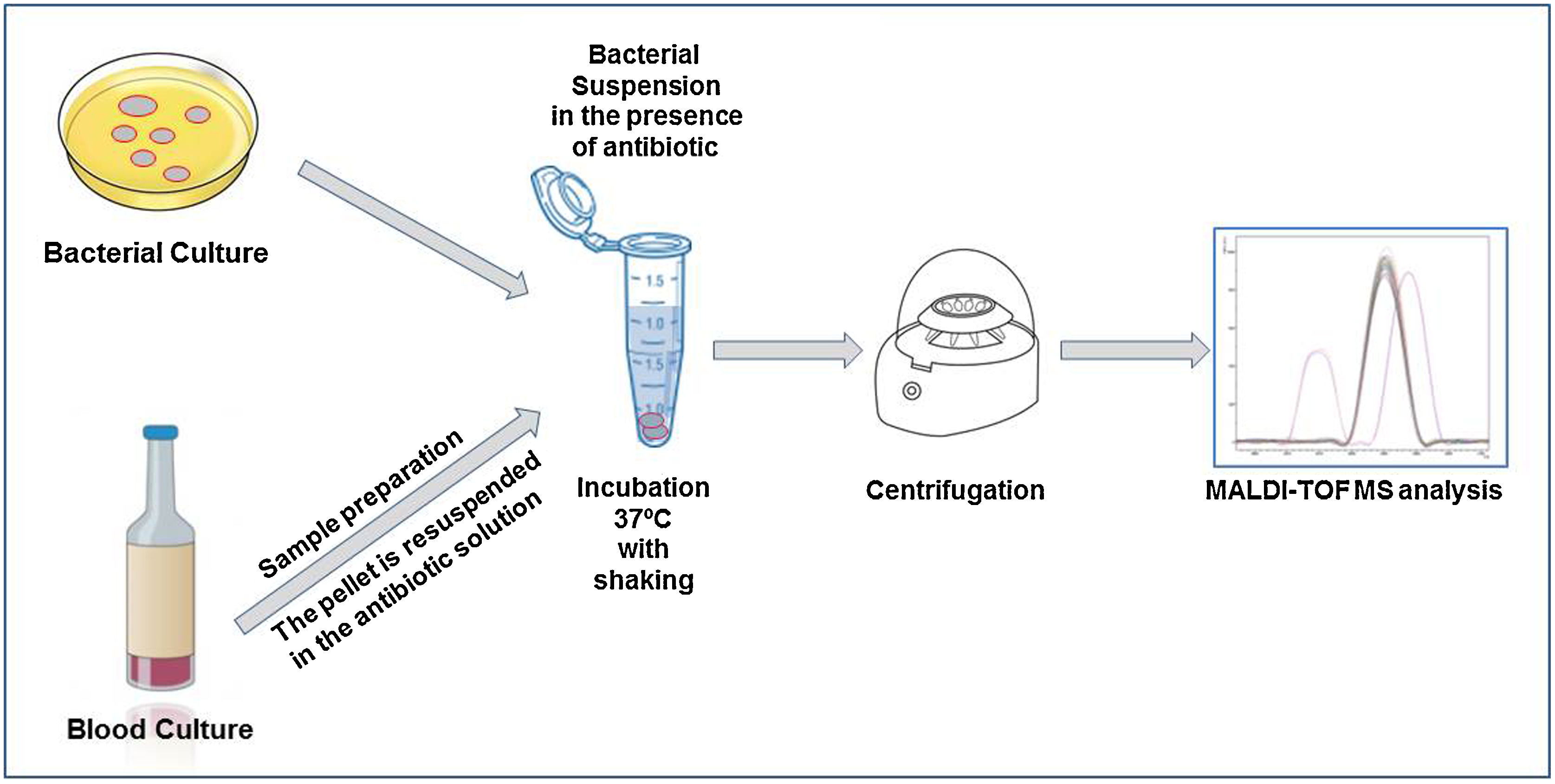
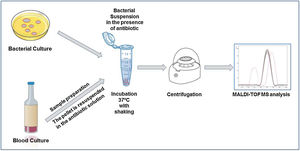
The direct detection of the β-lactamase activity is carried out in a very similar way in all the published works. A fresh bacterial culture is resuspended in an antibiotic buffer and incubated at 37°C under agitation. At the end of the incubation time, the mixture is centrifuged and the supernatant is deposited on the MALDI-TOF target plate. Once dried, the matrix can be added and the sample analyzed by MALDI-TOF.
Within cephalosporins, the mass spectrum of cefotaxime, ceftazidime, ceftriaxone, cefpodoxime and cefepime and their hydrolysis products have been described.22 Cefepime has been discarded for the detection of β-lactamases by MALDI-TOF due to its low specificity, as it presents false positives in the hydrolysis. Ceftriaxone is the antibiotic that has shown the highest sensitivity and specificity, as well as the shortest detection time. With 30min of incubation, we would observe a positive hydrolysis both in the case of bacteria that produces extended spectrum β-lactactamases (ESBL) or AmpC type β-lactactamases (Fig. 2). In the case of cefotaxime and cefpodoxime, the results are similar, although there is more experience when using cefotaxime. Both have sensitivity levels above 90%, but in both cases the recommended incubation time is 1h. If using ceftazidime, hydrolysis and subsequent detection by MALDI-TOF is much less effective, so it is not recommended as a first line antibiotic for the analysis of β-lactactamases. The sensitivity is less than 90% and the recommended incubation time is 3h.
In the case of carbapenem antibiotics, studies analyzing the hydrolysis of ertapenem, meropenem and imipenem have been described by using mass spectrometry. For ertapenem and meropenem, the sensitivity is about 90–95% but with a recommended incubation time of about 2h. In the case of ertapenem the mass peaks of the hydrolysis metabolite are perfectly characterized and appear in the mass spectrum. This is not the case for meropenem and imipenem. In order to solve this problem, two alternatives have been proposed. In the case of meropenem, the use of dihydroxybenzoic acid (DHB) as a matrix has been proposed.40 This matrix allows the observation of the mass peaks corresponding to meropenem and its hydrolyzed product. The disadvantage of this methodology is that DHB provides a less homogeneous crystallization than HCCA, being necessary to perform manual readings in many cases, which makes it difficult to automate the process and to implement its use in routine. In the case of imipenem, the fact of not observing hydrolysis peaks generated certain uncertainties. This problem has been solved using an internal standard in the matrix, so that we can semi-quantify the decrease in the mass peaks of imipenem with respect to the internal standard.32
In the MALDI Biotyper IVD (Bruker Daltonik) there is a resistance module, the MBT STAR-BL IVD software that together with the MBT STAR-Carba IVD Kit and the MBT STAR-Cepha IVD Kit allows, in an automated way, the interpretation of spectra and provides a combined report with the identification of the microorganism and its susceptibility to imipenem. This allows the automation of the interpretation of hydrolysis facilitating the use of this technique for users who are not experts in mass spectrometry.
In a different approach, susceptible and resistant microorganisms from the same species can be differentiated by peak analysis. There are mass peaks in the spectrum associated with a certain pattern, due to the expression of a peptide or protein directly or indirectly related to a resistance phenotype. The presence of these mass peaks helps the user to differentiate between susceptible and resistant microorganisms.
Research in this field have identified the presence of KPC-producing K. pneumoniae, by searching the peak mass at 11.109Da related to the expression of protein p019.41 It can also be used to detect methicillin-resistant S. aureus by detecting the mass peak at 2.415Da corresponding to the soluble phenyl modulin (PSM).42 Resistance to colistin -either mediated by the mcr-1 gene or not- has been detected by MALDI-TOF by observation of the lipid A or the modified lipid A by addition of a phosphoethanolamine group in the bacterial spectrum.43 Associated with a specific peak pattern and not only a mass peak is the presence of Bacteroides fragilis with the cfiA resistance gene44 and also the presence of vanA-producing Enterococcus faecium.45
This procedure has not only been applied in bacteria, but also in mycobacteria and fungi with good results.46,47 Although the sensitivity and specificity of the method is high, the amount of biomass needed and the laboriousness of the technique for these microorganisms hamper its future clinical application.
Finally, a new approach has been developed based on the measure of the relative bacterial growth in the presence of an antimicrobial drug. Susceptible isolates will not be able to grow, so a recognizable spectrum will not be observed, whereas resistant microorganisms will show certain growth proved by the typical spectrum obtained by MALDI-TOF. The procedure consists of 4-h incubation of the suspected microorganism together with an antimicrobial solution. This test has been performed both on bacteria from plate cultures and directly from positive blood cultures. Different antibiotics have already been tested such as piperacillin, oxacillin, cefotaxime, meropenem, gentamicin, ciprofloxacin and vancomycin with good results. This procedure has also been automated so that the hands-on-time is reduced and the technique is less hands-dependent.48 However, it has the disadvantage of requiring a minimum bacterial growth, so the incubation time is higher than the usual time required for rapid techniques.
Regarding the detection of antifungal resistance mechanisms, advance studies have been performed by de Carolis et al. in which the expression of resistance mechanisms of Candida glabrata isolates from clinical origin to caspofungin could be detected by MALDI-TOF.49 The isolates were incubated in growing concentrations of caspofungin and the resulting protein spectra were compared to each other using a composite correlation index (CCI). The minimal profile changing concentration, i.e., the concentration of antifungal that makes the yeasts not viable, could be detected by this statistical method and correlated with the minimal inhibitory concentration against each strain. Further studies have been carried out in order to shorten the incubation times and adapt the methodology for routine use, but available results refer only to a limited number of Candida and Aspergillus species so far.
Typing studiesPeak analysis of the spectra obtained by MALDI-TOF and its correlation of a specific peak pattern within certain microbial populations opens a new field of research for the detection of important characteristics of an isolate in a few minutes and in a cost-efficient way. The most limiting factor for MALDI-TOF when applied to typing of microorganisms is reproducibility, which can be obtained by maintaining the same working conditions such as incubation time or culture medium. Following a standardized analysis procedure with MALDI-TOF it has been demonstrated that results are reproducible and also very simple to develop.50 MALDI-TOF typing does not require highly qualified personnel to be carried out.
Specific software for spectra analysis, such as BioNumerics (Applied Maths, bioMérieux), Mass-Up,51 Clover MS Data Analysis (http://www.clovermsdataanalysis.com/) or MALDIquant (http://strimmerlab.org/software/maldiquant/) is now available. Besides, MALDI Biotyper Subtyping software module (Bruker Daltonik) allows the differentiation between Mycobacterium chimaera and the Mycobacterium intracellulare complex and the differentiation of Listeria monocytogenes from related species with high sensitivity and specificity as part of the identification process.52
So far, the application of MALDI-TOF for the typing of bacteria has made it possible to accurately and quickly identify five high-risk clones of Pseudomonas aeruginosa, like the sequence type 111 (ST111), ST175, ST235, ST253, and ST395.53 In addition, O4 serotyping and MALDI-TOF biomarker peak analysis, proved to be sensitive and specific methods that could be easily incorporated in the routine workflow for the early detection of P. aeruginosa ST175 high risk clone.54Clostridioides difficile typing has been made possible by a MALDI-TOF method based on the high molecular weight (HMW) protein profiling (mass range of 30–50kDa) from whole cells, followed by analysis of peaks in the 2–20kDa range from protein extractions, in comparison with the results obtained with the PCR ribotyping as the reference method.55 An E. coli outbreak has also been typed correctly using MALDI-TOF in comparison with PFGE with a simple full protein extraction.56
From an epidemiological point of view, the use of MALDI-TOF as a typing tool would mean a very important advance when applied to the infection control programmes of healthcare facilities, as isolates involved in outbreaks would be quickly and efficiently detected since the information is acquired almost in real time.
Detection of virulence markersSince many bacterial pathogenicity factors are proteins, MALDI-TOF can definitely be considered as a tool for their detection. However, this option has been barely exploited so far. Admittedly, there are some intrinsic difficulties, such as the amount of protein needed (MALDI-TOF MS is not a particularly sensitive technique) or the size of the proteins, which may lie out of the default detection window and require constant adjustment and calibration of the instrument.
Nevertheless, several attempts have been performed in this regard, such as the detection of Clostridioides difficile toxins.57 Differentiation between 2 genotypes, cdtA(+) and cdtB(+) was 100% accurate by a 6-peak cluster analysis of spectra generated. In addition, agr-positive methicillin resistant S. aureus58 or Clostridium botulinum neurotoxin have been detected by MALDI-TOF.59 This technology has also been applied for detection of the anthrax lethal factor (LF), a virulence factor produced by Bacillus anthracis, the causative agent of anthrax. The sensitivity of the test was 100%.60
ConclusionsMALDI-TOF mass spectrometry has become a reference method for the identification of a wide range of microorganisms. Its application for the detection of antimicrobial resistance mechanisms has also been widely established, reducing the turnaround time and simplifying the workflow in CMLs.
Although for the moment the information obtained by MALDI-TOF regarding bacterial typing or the detection of virulence markers requires confirmation through much slower molecular techniques, the potential of MALDI-TOF to become a first line technology in the near future is high. In combination with other emerging technologies such as Fourier-Transform Infrared Spectroscopy, MALDI-TOF can provide clinically impacting information rapidly and cost-effectively.
Conflict of interestThe authors report no conflicts of interest. The authors alone are responsible for the content and the writing of the paper.
The authors are supported by the Juan Rodés Contract JR18/00006 (MO) and the Miguel Servet Contract CPII19/00002 (BR-S) from the Health Research Fund (FIS) of the Carlos III Health Institute (ISCIII), Madrid, Spain, partially financed by the European Regional Development Fund (FEDER) ‘A way of making Europe.’ The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.