The development of sonication protocols over the last few years has improved the sensitivity of conventional cultures for the diagnosis of prosthetic-joint infection (PJI). However, the development of a new, specifically designed kit for the molecular diagnosis of PJI could provide a major improvement in this field.
MethodsProstheses retrieved from patients who underwent implant removal from May 2014 to May 2015 were sent for culture, and processed according to a previously defined protocol that included sonication. Furthermore, 180 microlitres of sonication fluid were used to carry out the multiplex PCR test (Unyvero i60 system®). A comparison of the sensitivity, specificity, positive (PPV) and negative (NPV) predictive value, was performed. The study was approved by the Clinical Research Ethics Committee.
ResultsThe analysis included 88 prostheses from 68 patients (1.29 prostheses/patient). The type of prostheses studied were knee (n=55), total hip (n=26), partial hip (n=5), and shoulder (n=2). Twenty-nine patients were diagnosed with a PJI (15 delayed, 12 acute, and 2 haematogenous infections). In 24 cases, the result of the PCR was positive, all but 1 corresponding to patients with clinical criteria of PJI. Nine resistance mechanisms were detected from 5 samples. The Unyvero i60 system® showed slightly better results than traditional culture in terms of specificity and PPV.
ConclusionsThe Unyvero i60 system® may play a role in rapid diagnosis of PJI, due to its high specificity and PPV. However, despite these results, cultures have to be performed to detect organisms not detected by the system.
El desarrollo de la sonicación durante los pasados años ha incrementado la sensibilidad de los cultivos convencionales para el diagnóstico de Infecciones de Prótesis Articulares (IPA). Sin embargo, el desarrollo de un nuevo kit, diseñado específicamente para el diagnóstico de las IPA podría suponer un avance significativo en este campo.
MétodosTodas las prótesis retiradas de pacientes entre mayo 2014 y mayo 2015 fueron enviadas para cultivo mediante un protocolo de procesamiento que incluye la sonicación del implante. Además, se emplearon 180 microlitros del líquido de sonicado en la realización de una PCR múltiple (Unyvero i60®). Se realizó una comparación de la sensibilidad, especificidad, valor predictivo positivo (VPP) y negativo (VPN). El estudio fue aprobado por el Comité de Ética en Investigación Clínica.
ResultadosSe analizaron 88 prótesis de 68 pacientes (1,29 prótesis/paciente). Las prótesis estudiadas fueron rodillas (n=55), total de cadera (n=26), parcial de cadera (n=5), y hombro (n=2). Veintinueve pacientes fueron diagnosticados de IPA (15 crónicas, 12 agudas y 2 hematógenas). En 24 casos, el resultado de la PCR fue positivo, siendo todas menos 1 de estas de pacientes con criterios de IPA. Se detectaron además 9 mecanismos de resistencia en 5 muestras. El sistema Unyvero i60® mostró resultados ligeramente superiores al cultivo tanto en especificidad como en VPP.
ConclusionesEl sistema Unyvero i60® puede tener un papel en el diagnóstico rápido de IPA debido a su elevada especificidad y VPP. Sin embargo, a pesar de estos resultados, debe realizarse cultivo para detectar organismos no detectados por el sistema.
Diagnosis of prosthetic-joint infections (PJI) is still a challenge for microbiology laboratories in spite of the advances achieved over the last few years. Although implant sonication has improved the sensitivity of conventional cultures, there are still some infected patients who remain undiagnosed using this technique.1,2 In an attempt to obtain better results, molecular biology has been used with different samples, most with good results in terms of specificity and sensitivity.2–7 However, most of these studies are based on locally-developed PCR methods which can have unsatisfactory standardization,4 which is why the development of a commercial system that features higher reproducibility would improve PJI diagnosis in most laboratories. Aiming to achieve this, several studies have used commercial PCR techniques8–13 with good results, despite the fact that most of them were not designed for the diagnosis of PJI, but rather to identify organisms isolated from blood cultures.8–12 However, some frequently isolated species from patients with PJI cannot be detected by these kits (notably Propionibacterium acnes14).
Material and methodsDesign of the studyBetween May 2014 and May 2015, a prospective study was carried out to compare two microbiological methods for the diagnosis of PJI basing on parameters of sensitivity, specificity, and predictive values. Traditional diagnosis, which includes culture of sonicated prostheses and culture of synovial fluid and peri-prosthetic tissue, was compared with a new commercial molecular biology technique (Unyvero i60 system (Curetis AG, Holzgerlingen, Germany). This method is specifically designed for the diagnosis of bone and joint infections and it is able to detect not only the main microorganisms responsible for PJI, but also some mechanisms of resistance.
Patients and definitionsPatients included were those diagnosed with PJI or aseptic loosening. They were diagnosed as having implant-related infection when at least one of the following commonly accepted criteria15 was met: (1) a draining sinus; (2) presence of acute inflammation identified by histopathological examination; (3) presence of macroscopic purulence around the implant; (4) presence of two or more positive cultures from high-quality samples (synovial fluid, peri-prosthetic tissue, blood cultures); or (5) presence of acute or chronic pain in the absence of a mechanical problem AND at least one altered blood parameter (including erythrocyte sedimentation rate (ESR), CRP, or synovial cell count).
Definitions of the different types of PJI (based on the time to infection after the last surgery) were those recommended by Tande et al.2 Infections are classified as early (<3 months), delayed (3–12 or 24 months), or haematogenous one (it is a late-onset PJI, occurring >12–24 months after surgery).
From every patient, prostheses were removed (culture and PCR were performed from sonicated fluid) and synovial fluid and peri-prosthetic tissue analyzed by culture.
Microbiological proceduresProstheses—in those cases where different parts of the implant were submitted separately, they were processed separately—were sonicated and processed according to a previously described and published protocol.16,17 A quantitative inoculation was made in different culture media (Tryptic soy agar with 5% sheep blood, chocolate agar, MacConkey agar, Schaedler agar with 5% sheep blood, and Sabouraud-chloramphenicol agar, all from bioMérieux (Marcy L’Etoile, France)) and incubated in different gas mixtures and temperatures during 1 week, with the exception of Sabouraud-chloramphenicol agar, which was maintained at 30°C during 1 month.16
Together with the implants, samples of synovial fluid and 3–6 samples of peri-prosthetic tissue samples were also submitted. Periprosthetic tissue samples were processed by grinding, and the samples were then inoculated in the same media as the sonicated samples. Synovial fluid was inoculated in Bact/Alert® blood culture bottles (bioMérieux, Marcy L’Etoile, France).
All organisms isolated from these samples were identified using the MALDI-ToF methodology (Vitek MS, bioMérieux, Marcy L’Etoile, France), and susceptibility testing was performed using the Vitek 2 system (bioMérieux, Marcy L’Etoile, France) or E-test strips (bioMérieux, Marcy L’Etoile, France), depending on the organism isolated.
The concentrated sonicate fluids were frozen at −20°C until multiplex PCR was performed.
Molecular techniqueIt was carried out, usually 24–48h after the arrival of the sample to the microbiology laboratory, using the Unyvero i60 system® (Curetis AG, Holzgerlingen, Germany) and according to the manufacturer's instructions. This system is designed to detect 27 species or groups of organisms (Table 118). The system allows simultaneous detection of 19 resistance genes (Table 118). Briefly, 180μl of the sonicated fluid were pipetted into the sample tube and placed in the lysis incubator for 30min. Subsequently, the sample was placed in the Unyvero i60 cartridge with the Master mix vial. Then, the loaded cartridge was inserted in the system hardware for amplification and hybridization. After a period of 4.5hours of analysis, the results were displayed on the screen of the system. This technique includes an internal control to determinate whether the result is valid or not.
Microorganisms and resistance genes detected by the Unyvero i60 System.
| Detected pathogens |
|---|
| Staphylococcus aureus |
| Staphylococcus epidermidis |
| Coagulase-negative Staphylococcus1 |
| Streptococcus mitis group2 |
| Streptococcus anginosus group3 |
| Streptococcus salivarius group4 |
| Streptococcus agalactiae |
| Streptococcus pyogenes5 |
| Enterococcus faecalis |
| Enterococcus sp.6 |
| Granulicatella adjacens |
| Abiotrophia defective |
| Corynebacterium sp.7 |
| Escherichia coli |
| Enterobacter cloacae complex |
| Enterobacter aerogenes |
| Proteus sp.8 |
| Klebsiella pneumoniae9 |
| Pseudomonas aeruginosa |
| Acinetobacter baumannii |
| Propionibacterium acnes |
| Propionibacterium avidum/granulosum |
| Finegoldia magna |
| Bacteroides fragilis group10 |
| Candida sp.11 |
| Candida parapsilosis |
| Candida albicans |
| Antibiotic resistance markers |
|---|
| mec A |
| mec C (LGA251) |
| aac(6′)/aph(2″) |
| ermA |
| ermC |
| vanA |
| vanB |
| rpoB |
| ctx-M |
| vim |
| imp |
| kpc |
| ndm |
| aacA4 |
| gyrA |
| oxa-23 |
| oxa-24 |
| oxa-48 |
| oxa-58 |
1 Includes Staphylococcus epidermidis, Staphylococcus haemolyticus, Staphylococcus lugdunensis, Staphylococcus saprophyticus, Staphylococcus capitis, and Staphylococcus hominis.
2 Includes Streptococcus mitis, Streptococcus australis, Streptococcus cristatus, Streptococcus gordonii, Streptococcus infantis, Streptococcus oligofermentans, Streptococcus oralis, Streptococcus parasanguinis, Streptococcus paroris, Streptococcus pseudopneumoniae, Streptococcus sanguinis, and Streptococcus sinensis.
3 Includes Streptococcus anginosus, Streptococcus constellatus, and Streptococcus intermedius.
4 Includes Streptococcus salivarius, Streptococcus vestibularis, and Streptococcus termophilus.
5 Includes Streptococcus pyogenes and Streptococcus dysgalactiae subsp. equisimilis.
6 Includes Enterococcus casseliflavus, Enterococcus faecium, Enterococcus hirae, Enterococcus faecalis, Enterococcus sulfurens, Enterococcus saccharolyticus, Enterococcus mundii, Enterococcus gallinarum, Enterococcus flavescens, Enterococcus durans, Enterococcus dispar, Enterococcus columbae, Enterococcus cecorum, and Enterococcus asini.
7 Includes Corynebacterium genitalium, Corynebacterium casei, Corynebacterium striatum, Corynebacterium jeikeium, Corynebacterium aurimucosum, Corynebacterium singularis, Corynebacterium simulans, Corynebacterium accolens, Corynebacterium amycolatum, Corynebacterium minutissimum, Corynebacterium macginleyi, and Corynebacterium auriscanis.
8 Includes Proteus mirabilis, Proteus vulgaris, and Proteus penneri.
9 Includes Klebsiella pneumoniae subsp. Ozenae.
10 Includes Bacteroides fragilis, Bacteroides ovatus, and Bacteroides thetaiotaomicron.
11 Includes Candida albicans, Candida krusei, Candida dublinensis, Candida tropicalis, and Candida glabrata.
The results of both studies (culture and molecular biology) were blindly obtained by 2 different authors, each of whom had no knowledge of the results obtained by the other until all results were available.
Data analysisMicrobiological and clinical dataAfter molecular and conventional studies were carried out, a retrospective analysis of clinical charts was performed using a predefined protocol. All aspects of the study were approved by the Ethics in Research Committee of the institution (ref. 45/2014_FJD).
StatisticsA descriptive analysis of clinical and microbiological data was made. Furthermore, a comparison of the sensitivity (Sn), specificity (Sp), positive (PPV) and negative (NPV) predictive value of both methods was performed. These parameters were calculated using patients without clinical criteria of infection as control group. Furthermore, these data were compared with those of culture of periprosthetic tissue and synovial fluid, since neither molecular biology nor even sonication techniques, are not always available in sanitary centres for the diagnosis of PJI.
ResultsDuring the study period, 88 samples from prostheses of 68 patients were analyzed. In some cases, 2 or more samples were taken from the same patient, in particular, 1 patient with 4 samples, 5 with 3, 9 with 2 and 51 with only one sample studied (1.29 samples/patient). In 10 cases, results of PCR technique were not valid since this set in particular was defective. The PCR was repeated with other set and results were then valid.
PatientsThere were 39 patients without clinical criteria of infection and 29 with it. Among the last group, the mean age was 72.75 years, 19 of them were women and 10 men. The most frequent type of infection was delayed (n=15), followed by acute, (n=12) and haematogenous (n=2). Only 1 patient received a combination of vancomycin and ceftazidime, undergoing this treatment for the 2 days prior to the operation.
SamplesThe type of prostheses studied included knee (n=55), total hip (n=26), partial hip (n=5), and shoulder (n=2).
Of the 88 samples, 38 corresponded to patients with clinical criteria of PJI, while 50 did with patients without criteria of PJI.
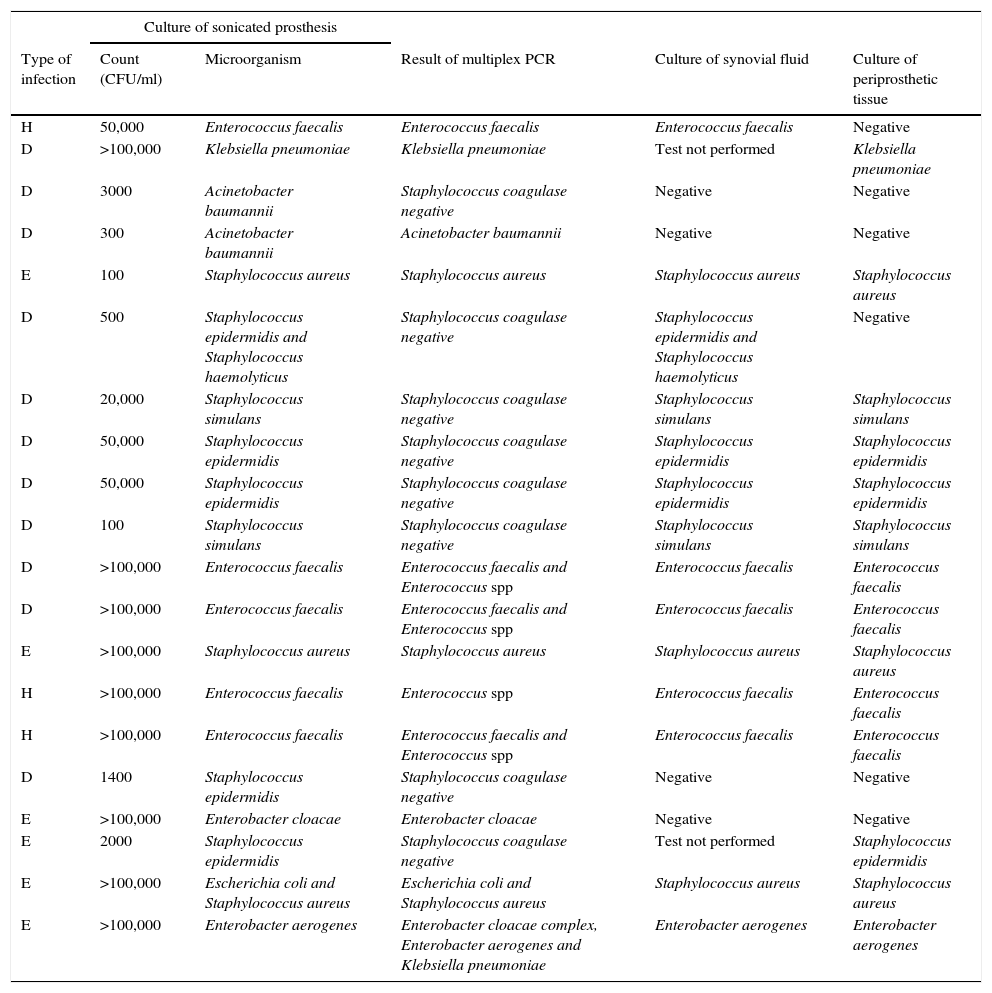
Data among samples related to patients with clinical criteria of PJI (n=38)Twenty samples had both culture of sonicated implant and multiplex PCR positive. Data are shown in Table 2 together with results of culture of periprosthetic tissue and synovial fluid.
Comparison of results of all microbiological methods for samples from patients with clinical criteria of PJI. Culture of sonicated implant and PCR positive (n=20).
| Culture of sonicated prosthesis | |||||
|---|---|---|---|---|---|
| Type of infection | Count (CFU/ml) | Microorganism | Result of multiplex PCR | Culture of synovial fluid | Culture of periprosthetic tissue |
| H | 50,000 | Enterococcus faecalis | Enterococcus faecalis | Enterococcus faecalis | Negative |
| D | >100,000 | Klebsiella pneumoniae | Klebsiella pneumoniae | Test not performed | Klebsiella pneumoniae |
| D | 3000 | Acinetobacter baumannii | Staphylococcus coagulase negative | Negative | Negative |
| D | 300 | Acinetobacter baumannii | Acinetobacter baumannii | Negative | Negative |
| E | 100 | Staphylococcus aureus | Staphylococcus aureus | Staphylococcus aureus | Staphylococcus aureus |
| D | 500 | Staphylococcus epidermidis and Staphylococcus haemolyticus | Staphylococcus coagulase negative | Staphylococcus epidermidis and Staphylococcus haemolyticus | Negative |
| D | 20,000 | Staphylococcus simulans | Staphylococcus coagulase negative | Staphylococcus simulans | Staphylococcus simulans |
| D | 50,000 | Staphylococcus epidermidis | Staphylococcus coagulase negative | Staphylococcus epidermidis | Staphylococcus epidermidis |
| D | 50,000 | Staphylococcus epidermidis | Staphylococcus coagulase negative | Staphylococcus epidermidis | Staphylococcus epidermidis |
| D | 100 | Staphylococcus simulans | Staphylococcus coagulase negative | Staphylococcus simulans | Staphylococcus simulans |
| D | >100,000 | Enterococcus faecalis | Enterococcus faecalis and Enterococcus spp | Enterococcus faecalis | Enterococcus faecalis |
| D | >100,000 | Enterococcus faecalis | Enterococcus faecalis and Enterococcus spp | Enterococcus faecalis | Enterococcus faecalis |
| E | >100,000 | Staphylococcus aureus | Staphylococcus aureus | Staphylococcus aureus | Staphylococcus aureus |
| H | >100,000 | Enterococcus faecalis | Enterococcus spp | Enterococcus faecalis | Enterococcus faecalis |
| H | >100,000 | Enterococcus faecalis | Enterococcus faecalis and Enterococcus spp | Enterococcus faecalis | Enterococcus faecalis |
| D | 1400 | Staphylococcus epidermidis | Staphylococcus coagulase negative | Negative | Negative |
| E | >100,000 | Enterobacter cloacae | Enterobacter cloacae | Negative | Negative |
| E | 2000 | Staphylococcus epidermidis | Staphylococcus coagulase negative | Test not performed | Staphylococcus epidermidis |
| E | >100,000 | Escherichia coli and Staphylococcus aureus | Escherichia coli and Staphylococcus aureus | Staphylococcus aureus | Staphylococcus aureus |
| E | >100,000 | Enterobacter aerogenes | Enterobacter cloacae complex, Enterobacter aerogenes and Klebsiella pneumoniae | Enterobacter aerogenes | Enterobacter aerogenes |
Type of infection: H (haematogenous), D (delayed), and E (early).
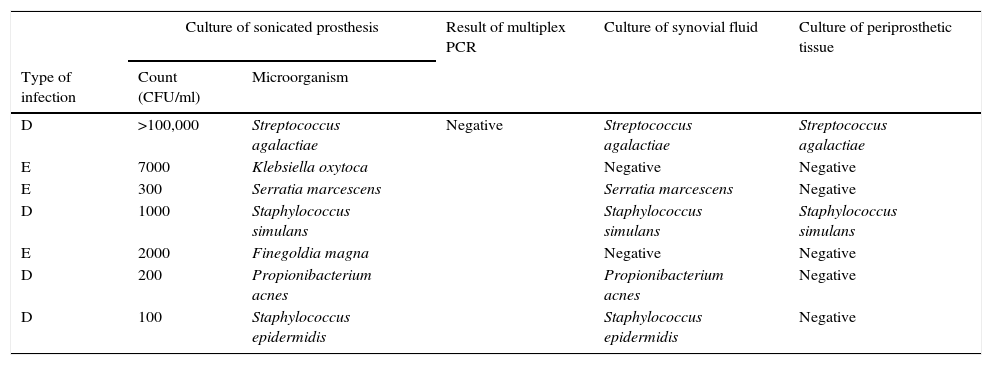
Seven samples showed culture of sonication positive but PCR negative (Table 3). These discrepancies are explained as follows: (a) 1 patient with 105CFU/ml of S. agalactiae; (b) 1 patient with 300CFU/ml of S. marcescens (not included among the organisms detected by PCR); (c) 1 patient with 100CFU/ml of S. epidermidis (regarded as a contaminant), with infection due to Mycobacterium tuberculosis which grew 8 weeks later in mycobacterial cultures; (d) 1000CFU/ml of S. simulans; (e) 7000CFU/ml of K. oxytoca; (f) 2000CFU/ml of Finegoldia magna; and (g) 200CFU/ml of Propionibacterium acnes.
Comparison of results of all microbiological methods for samples from patients with clinical criteria of PJI. Culture of sonicated implant positive but PCR negative (n=7).
| Culture of sonicated prosthesis | Result of multiplex PCR | Culture of synovial fluid | Culture of periprosthetic tissue | ||
|---|---|---|---|---|---|
| Type of infection | Count (CFU/ml) | Microorganism | |||
| D | >100,000 | Streptococcus agalactiae | Negative | Streptococcus agalactiae | Streptococcus agalactiae |
| E | 7000 | Klebsiella oxytoca | Negative | Negative | |
| E | 300 | Serratia marcescens | Serratia marcescens | Negative | |
| D | 1000 | Staphylococcus simulans | Staphylococcus simulans | Staphylococcus simulans | |
| E | 2000 | Finegoldia magna | Negative | Negative | |
| D | 200 | Propionibacterium acnes | Propionibacterium acnes | Negative | |
| D | 100 | Staphylococcus epidermidis | Staphylococcus epidermidis | Negative | |
Type of infection: H (haematogenous), D (delayed), and E (early).
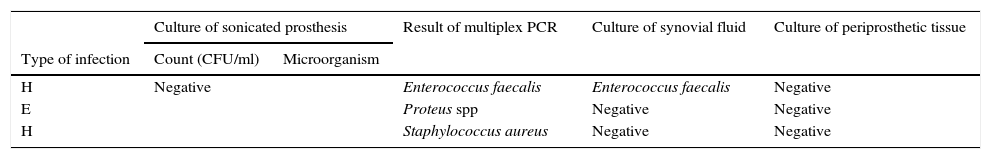
Three samples showed positive result by multiplex PCR but negative by culture of sonicated implant (Table 4). PCR were positive for E. faecalis (also isolated in cultures of periprosthetic tissues), S. aureus (not isolated in any other cultures) and P. mirabilis (this patient had a positive culture for P. mirabilis from a debridement surgery 1 month before the retrieval of the implant).
Comparison of results of all microbiological methods for samples from patients with clinical criteria of PJI. PCR positive but culture of sonicated implant negative (n=3).
| Culture of sonicated prosthesis | Result of multiplex PCR | Culture of synovial fluid | Culture of periprosthetic tissue | ||
|---|---|---|---|---|---|
| Type of infection | Count (CFU/ml) | Microorganism | |||
| H | Negative | Enterococcus faecalis | Enterococcus faecalis | Negative | |
| E | Proteus spp | Negative | Negative | ||
| H | Staphylococcus aureus | Negative | Negative | ||
Type of infection: H (haematogenous), and E (early).
In 8 cases, both techniques were negative. Only 1 sample from an infected patient was probably culture-negative due to previous antibiotic therapy.
Data among samples related to patients without clinical criteria of PJI (n=50)In one case, multiplex PCR gave a positive result while sonication of implant was negative (considered to be a probable false positive). The detected organism was a Coagulase-negative staphylococcus. As all cultures were negative, the patient was diagnosed with probable aseptic loosening. However, the patient received an antibiotic-loaded spacer (vancomycin+gentamicin) and second-stage surgery was performed 1 month after the first stage. Although the result may represent the detection of a true pathogen, because the patient was not treated with antibiotics and the final diagnosis was aseptic loosening, we decided to include it as a false positive of the technique.
Two positive cultures (100CFU/ml of S. aureus and 100CFU/ml of S. epidermidis) with PCR negative were considered as contaminants because of several factors: a very low colony count (<500CFU/ml); no other positive cultures from other samples; no clinical diagnosis of infection; and no presentation of complications despite the fact that no treatment was administered in these cases.
Mechanisms of resistanceConsidering antimicrobial susceptibility, 9 resistance mechanisms were detected in 5 samples using PCR: 3 mecA, 2 with fluoroquinolone resistance (1 gyrA83 and 1 gyrA87 mutations), 1 ctx-M extended-spectrum beta-lactamase, 1 aminoglycoside resistance (1 aac A4), 1 ermA, and 1 ermC gene. All these findings were confirmed by phenotypic antimicrobial susceptibility assay. Phenotypic susceptibility testing showed 1 isolate of S. simulans to be resistant to erythromycin and clindamycin, going undetected by the system.
Data about culture of synovial fluid and periprosthetic tissueAmong patients with clinical criteria of PJI (n=38), culture of periprosthetic tissue was always made, being positive in 16 cases and negative in 38. However, sample of synovial fluid was taken only from 35 patients and 20 had a positive result whereas 15 a negative one.
From the 50 patients without clinical criteria of PJI, culture of synovial fluid was negative in 48 cases and of periprosthetic tissue in every sample.
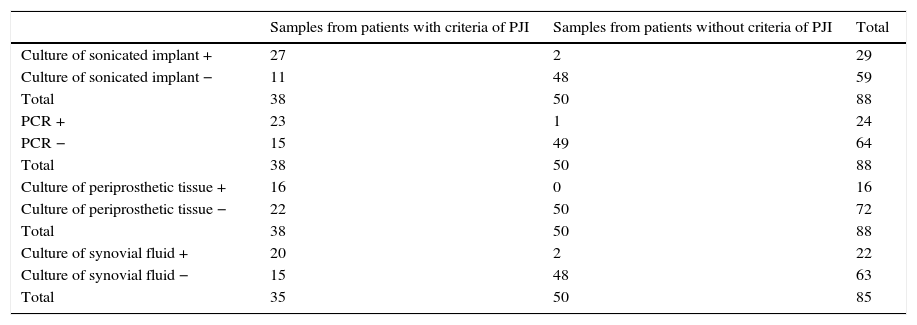
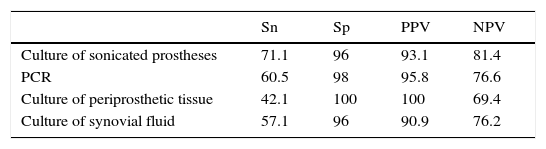
Statistical parametersData from which statistical parameters of Sn, Sp, PPV and NPV were calculated, are shown in Table 5 and final results in Table 6.
Data to calculate statistic parameters of culture of sonicated prostheses multiplex PCR, culture of periprosthetic tissue and culture of synovial fluid for the diagnosis of PJI.
| Samples from patients with criteria of PJI | Samples from patients without criteria of PJI | Total | |
|---|---|---|---|
| Culture of sonicated implant + | 27 | 2 | 29 |
| Culture of sonicated implant − | 11 | 48 | 59 |
| Total | 38 | 50 | 88 |
| PCR + | 23 | 1 | 24 |
| PCR − | 15 | 49 | 64 |
| Total | 38 | 50 | 88 |
| Culture of periprosthetic tissue + | 16 | 0 | 16 |
| Culture of periprosthetic tissue − | 22 | 50 | 72 |
| Total | 38 | 50 | 88 |
| Culture of synovial fluid + | 20 | 2 | 22 |
| Culture of synovial fluid − | 15 | 48 | 63 |
| Total | 35 | 50 | 85 |
Sensitivity of PCR method was not very high (60.5%) as well as NPV (76.6%). However, the same values for culture of sonicated implant, although slightly higher (Sn=71.1% and NPV=81.36%), were not very great.
The main advantage of Unyvero i60 system® is that it improves values of Sp and PPV in relation to sonicated implant culture (Sp=98% and PPV=95.8%, versus Sp=96% and PPV=93.1%, respectively).
When PCR technique was compared with culture of periprosthetic tissue, in general it had better values, since, although the last method had Sp and VPP of 100%, Sn and VPN were very low (42.1% and 69.4% respectively).
With regard to culture of synovial fluid inoculated in blood bottles, it had similar statistic parameters as PCR, but the last one improves all of them (see Table 6).
DiscussionKnowledge of the aetiology of PJI is an essential aspect of patient management, as antibiotic therapy is selected based on the isolated microorganism/s.2,19,20 The advent of implant sonication in the last decade has improved the conventional methodology, introducing a technique that allows microbiologists to detect bacteria (and fungi) attached to the implant.2,21,22 However, there are still some patients in whom no microorganisms can be detected despite the use of conventional cultures. For these and other cases, molecular detection seems to be the obvious step to achieve a microbiological diagnosis of PJI.2,3,5 Many different studies have shown good results using different approaches (e.g., 16S broad-range primers followed by sequencing specific primers, and multiplex PCR), though all these techniques lack reproducibility between centres and require molecular biology laboratories and specialists—resources often not available in microbiology laboratories. Several commercial kits have been tested to overcome these problems. Since there was no kit specifically designed for PJI, kits designed for blood cultures that include most organisms involved in PJI have been used.8–12
The SeptiFast kit (Roche) was initially tested using sonicate samples from 37 patients with PJI,8 revealing a sensitivity of 78% compared to a 65% sensitivity for sonication. Since no non-infected cases were included in the study, no specificity result could be obtained. This kit was also tested by another team, which included aseptic loosening patients as negative controls (62 cases) as well as PJI patients (24 cases). In this study, sensitivity was higher (96%), with a specificity of 100% (no positive results among aseptic loosening cases). Another kit (GenoType BC, Hain Lifescience, Germany) was studied in our laboratory, using the kit to analyze sonicate samples from 79 patients without infection and 47 infected cases.10 In this research, the main problem was the high number of theoretically non-infected cases producing a positive PCR result and negative cultures, which may be due to the low specificity of the kit. Finally, the latest commercial multiplex PCR designed for blood cultures is the FilmArray kit (bioMérieux, Marcy L’Etoile, France),12 which has been shown to have a low sensitivity (53%), especially for coagulase-negative staphylococci. Sensitivity increased to 58% when only potentially detected cases were considered. The main advantage of this methodology was the rapid turnaround time (less than 1hour) and easy management in the laboratory due to the cartridge technology. This circumstance was also used in a previous study with the Xpert MRSA/SA kit for detection of Methicillin-resistant Staphylococcus aureus, yielding good results in spite of the obvious limitation of the kit (only some staphylococci detected).9 For all the above-mentioned studies8,10–12 except the last one, some important etiological agents of PJI do not appear in the list of each kit, especially Propionibacterium acnes and Corynebacterium sp.
Due to this absence, a specific kit for the diagnosis of PJI is desirable. A study performed with a kit with these requisites (Prove-it Bone and Joint StripArray assay, Mobidiag Ltd., Helsinki, Finland) was tested in periprosthetic tissue samples from 60 patients with a diagnosis of suspected PJI. Among these, 38 fulfilled the criteria of PJI, and in 31 the PCR test was positive (82% sensitivity). However, in 6 of the cases without PJI, the results were positive (74% specificity), a problem described in another of the aforementioned studies.10 More recently, a new test was designed specifically for the diagnosis of bone and joint infections, especially PJI. This test was previously tested with tissue samples23 from 54 patients (10 of them infected) and compared against a homemade broad-range 16S rRNA PCR. The commercial test was performed in only 28 patients, showing a high specificity (95.2%) but a low sensitivity (42.9%). In the present study, we used this kit and compared it with the cultures of the sonicated implants in a higher number of patients, all with a diagnosis and follow-up performed in accordance with international recommendations.15 This allowed us to compare the results not only against conventional cultures, but also against clinical and analytical diagnosis. Moreover, we have tried to test the kit in routine conditions, aiming to obtain a more realistic view of its actual usefulness. The results obtained showed a relatively low sensitivity (similar to that obtained in some of the previously mentioned studies8,12), but a high specificity, with only 1 false positive. In this case, coagulase-negative Staphylococcus was detected in a patient who underwent a 2-step exchange surgery, with only 4 weeks between surgeries and no antibiotic therapy, but with the use of an antibiotic-loaded spacer. For the false-negative results of the kit, most of them appeared in patients with a low colony count or in patients with a pathogen not included in the kit. There was an infection due to S. agalactiae (105CFU/ml) also isolated in other samples (periprosthetic tissues and synovial fluid), which was persistently negative with this kit. Unfortunately, we do not have an explanation for this result. However, we have detected 2 polymicrobial infections in which some of the organisms were not detected by culture (E. cloacae+E. aerogenes+E. coli infection) or were detected only after reincubation and careful isolation of a few colonies of an E. coli strain (S. aureus+E. coli).
We have also detected some resistance genes or mutations. Though most of them correlated well with the conventional antimicrobial susceptibility testing performed, we detected 1 potential discrepancy. Furthermore, some resistance phenotypes, as well as the bacterium S. marcescens, cannot be detected by this PCR because they are not included in this technique, thus marking a limitation of the Unyvero i60® system.
To sum up, the main advantages of this molecular biology technique against conventional microbiological diagnosis of PJI are, in first place, the high speed of the result, the possibility to detect mechanisms of resistance in the same step, and the easy management of the sample, which made a promising methodology for this purpose. Furthermore, it has better statistic parameters; in particular, it showed higher specificity and PPV versus culture of sonicated implant. In our opinion, it is of great importance, because a frequent criticism of molecular technology is the amount of false positive results. Secondly, when multiplex PCR was compared with culture of synovial fluid inoculated in blood culture bottles, PCR improved all statistic parameters. And although specificity of culture of peri-prosthetic tissue was higher than that obtained with PCR, the last one had better values in general.
With regard to other molecular biology techniques, Unyvero i60® system showed advantages against those designed for blood cultures (that include most organisms involved in PJI) and those specifically designed for the diagnosis of PJI. Among the first group (PCR for blood culture), the main limitations were: the impossibility to obtain specificity result due to the lack of control group (non-infected patients) in one case,8 the low specificity10 and sensitivity12 and the only detection of Methicillin-resistant Staphylococcus.9 Among the group of molecular techniques specifically designed for the diagnosis of PJI, Unyvero i60® system improved specificity of one of them (Prove-it Bone and Joint StripArray assay, Mobidiag Ltd., Helsinki, Finland), and the sensitivity and number of patients included of other one.23
However, this technique had limitations. The main was its relatively low sensitivity, which made a negative result inconclusive for the diagnosis of PJI using sonicated implants. Another one was the number of patients included, which, although higher than in other studies, were not high enough. In addition, there are some microorganisms as well as some resistance mechanisms not detected by this technique because they are not included; and one discrepancy in the mechanisms of resistance was found out. Further studies are necessary to evaluate its role with other samples (especially synovial fluid and periprosthetic tissue) for the diagnosis of PJI in a routine setting. Another important aspect is the price: studies on the economic impact and cost-effectiveness are also necessary, as this technology is costly, and rational use is essential in all cases. To achieve this, a close relationship between clinicians and microbiologists is essential in order to obtain the best possible management of the patient.
FundingThis work was funded by a grant from Laboratorios Leti SL and by a grant from the Spanish MINECO (Grant MAT2013-48224-C2-2-R).
Conflict of interestJE received travel grants from Laboratorios Leti SL, Pfizer, bioMérieux, and Alere, and research grants from Wyeth and Laboratorios Leti SL. RFR and IG received research grants from Wyeth and Laboratorios Leti SL. No other conflicts of interest are declared.
Ethical approval and Informed consentAll aspects of the study were approved by the Ethics in Research Committee of the institution (ref. 45/2014_FJD). No informed consent was required by the Committee.
We want to acknowledge Mr. Oliver Shaw for his help with the English edition of the article.
Parts of these results were presented at the 25th ECCMID (Copenhagen 2015) and 16th Effort (Prague, 2015) conferences.