The main cause of cervical cancer is an infection of keratinocytes in the basal layer of the stratified epithelium of the cervix by human papillomavirus (HPV). Other than in cervical samples, HPV DNA has been found in serum and other fluids but its origin is unclear. Extracellular vesicles (EV) could be a conveyance of viral DNA given their emerging role in cellular communication. The content of EV derived from cervical cells has not been properly explored and it is not known whether or not they contain HPV DNA.
MethodsWe evaluated the DNA content of exosomes purified from cultures of HeLa cells by Next Generation Sequencing (NGS) and confirmed its presence by PCR. The presence of HPV DNA was also evaluated by PCR and NGS in EV from HPV-positive cervical samples without apparent lesion or with LSIL.
ResultsWe detected the integrated form of viral-DNA in exosomes from HeLa cells by NGS and confirmed its presence by PCR. The search for HPV sequences in EV obtained from cervical exudate samples without apparent lesion or with LSIL, where we expected to find the viral genome as an episome, indicated that HPV DNA, including the E6 and E7 oncogenes, is present in these EV.
ConclusionHPV DNA, including the viral oncogenes E6/E7, is found in exosomes regardless of the integration status of the virus in the infected cell.
La principal causa del cáncer de cérvix es la infección de los queratinocitos de la capa basal del epitelio estratificado del cuello uterino por el virus del papiloma humano (VPH). El ADN del VPH se ha encontrado en muestras cervicales, pero también en suero y otros fluidos, aunque su origen en estos últimos no está claro. Las vesículas extracelulares (VE) podrían ser el medio de transporte del ADN viral considerando su papel emergente en la comunicación celular. El contenido de las VE derivadas de células cervicales ha sido poco explorado y la presencia en ellas de ADN de VPH sigue siendo desconocida.
MétodosEvaluamos el ADN de exosomas purificados a partir de cultivos de células HeLa mediante secuenciación de nueva generación (NGS) y confirmamos su presencia a través de PCR. La presencia de ADN de VPH también se evaluó mediante PCR y NGS en VE de muestras cervicales positivas a VPH, sin lesión aparente o con LSIL.
ResultadosDetectamos la forma integrada del ADN viral en exosomas de células HeLa mediante NGS, y confirmamos su presencia a través de PCR. La búsqueda de secuencias de VPH en VE obtenidas a partir de muestras de exudado cervical sin lesión aparente o con LSIL, donde esperamos encontrar el genoma viral en forma episomal, indicó que el DNA de VPH incluyendo los oncogenes E6 y E7, está presente en estas VE.
ConclusiónEl ADN del VPH incluyendo el correspondiente con los oncogenes virales E6/E7 se encuentra en exosomas independientemente del estado de integración del virus en la célula infectada.
A persistent infection with high-risk human papillomavirus (HR-HPV) is the principal cause of cervical cancer and is associated with oropharyngeal squamous cell carcinoma.1 This infection is commonly cleared in 36 months but 27% of HR-HPV infections persist and cause lesions.2 The DNA of HPV has been found in cervical samples, but also in the serum of cervical cancer patients3 and other fluids such as nipple discharge4; however, the site of the infection in this last samples remains unclear. The detection of viral DNA in sites other than the infected cells is useful not only as a tumor marker but can also expand our knowledge about the mechanism of cancer progression. The infection and other factors in the microenvironment can influence the transformation and tumor progression. This microenvironment is composed of immune, endothelial, vascular, and all other cells in tissue but also cytokines and microvesicles.5,6
The extracellular vesicles (EV) are currently being analyzed due to their role in cell communication and their increase in different kind of cancers. The EV include microvesicles that bud from the cell membrane and exosomes, which are secreted from multivesicular bodies. The exosomes have been proposed as biomarkers along with some their content.6,7 The EV content includes RNA, DNA, and proteins; this content can be modified by HPV infection.7 The EV from cervicovaginal lavages of woman with HPV-positive cervical cancer contain elevated levels of miR-21 and miR-146,8 suggesting that miRNAs content in EV could be modified by oncoproteins E6 and E7.9,10 The viral oncoproteins could also induce changes in the content of lncRNAs 11 and proteins such as p53 and survivin in EV.12 However, to the best of our knowledge, there is no report about the content of genomic or viral DNA in the EV from cervical cells infected with HPV.
The transfer of genomic DNA13 or viral DNA in other kinds of cancer into exosomes has been demonstrated.14 According to this previous reports DNA viral of HPV should be contained into exosomes derived from keratinocytes infected. Next generation sequencing (NGS) has proved be useful to detect the presence of viral infections in EV from different samples including swabs. Thus, the main goal of this work was to evaluate the presence of HPV in EV derived from cell cultures where the viral genome is integrated as well as in EV derived from cervical samples without observed lesions but that are HPV-positive, where the viral genome is expected as episome.
Materials and methodsCell cultures and samplesHeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 100units/L penicillin, and 100μg/mL streptomycin. Cultures were maintained in a humidified atmosphere at 37°C and 5% CO2. We evaluated cervical samples taken from women attending the colposcopy service of the Medicine and Surgery Faculty, UABJO at different communities. The ages of the women were 18–65 years, and all were apparently healthy, not pregnant, no vaccination against HPV, and had colposcopy results showing no alterations associated with the infection or low-degree squamous intraepithelial lesion (LSIL). The samples of cervical scrapples were taken, placed in PBS and stored at 4°C until processing. The clinical and gynecological history were also obtained as well as informed consent. Samples with high-risk HPV infection or without HPV infection were included in this study. Several risk factors associated to the HPV infection were evaluated between the groups with and without infection but there was no significant difference. The risk factors considered were the lifetime number of sexual partners, age at first intercourse, age, cigarette smoking, oral contraceptive use and parity. All proceedings were performed in accordance with the Declaration of Helsinki and a protocol of the institutional ethics committee of the Medicine and Surgery Faculty, UABJO.
EV isolation from cervical samples or culture supernatantsHeLa cells were seeded at 1×106cells/plate in 100mm tissue culture plates and grown in Dulbecco's Modified Eagle's Medium (DMEM) (biowest) supplemented with 10% fetal bovine serum (biowest). After 48h, the culture media was replaced by serum-free medium and 0.5mM calcium chloride and maintained for 24h in humidified atmosphere with 5.0% CO2 at 37°C. The culture media was then collected and centrifuged at 2000×g for 20min twice. The pellets were discarded, and the supernatant was centrifuged at 95,000×g for 2h. The pellet was resuspended in PBS and centrifuged at 95,000×g for 2h. The supernatant was discarded, and the pellet containing the EV was used in the corresponding analysis. All centrifugation steps were at 4°C. Cervical samples were collected in tubes containing 5mL of PBS. They were centrifuged at 800×g for 5min. The pellets were employed for HPV typing, and the supernatant was centrifuged as described for culture media to obtain the EV.
Transmission electron microscopy (TEM)The EV were fixed with 2% glutaraldehyde (Electron Microscopy Science), and 10 microliters of fixed EV were placed onto carbon-coated copper grids. The EV were negatively stained with 2% uranyl acetate in water for 3min. Images were obtained with a JEM-1400 transmission electron microscope with an acceleration voltage of 80kV.
DNA isolation and HPV detectionThe EV isolated from HeLa cells or samples, as well as the pellets from cervical scrapples, were re-suspended in 200μL of lysis buffer (10mM Tris–HCl, 25mM EDTA, 100mM NaCl, 3.5% SDS) and Proteinase K (EO0491, Thermo). After 24h of incubation at 37°C, the samples were purified with the phenol–chloroform method. A fragment of the β-actin gene was amplified as control with oligonucleotides sense 5′ GCG TTA CAC CCT TTC TTG AC 3′ and antisense 5′ TTG TGA ACT TTG GGG GAT GC 3′. The typing was done with the universal oligonucleotides MY09/MY11 and GP5+/GP6+, and Sanger sequencing. The PCR reactions used Taq polymerase (EP402, Thermo), according to the instructions of the manufacturer. The positive samples were amplified to sequencing with Phusion Hot Start II DNA Polymerase (Thermo). The detection of HPV in EV used the oligonucleotides PU1M/2R15 or the PU1M/2R modified to HPV 18: Sense 5′ TGCCAGAAACCGTTGAATCC 3′ and Antisense 5′ GAGTCGCTTAATTGCTCGTG 3′; to detect a E6/E7 fragment of HPV. The identity of the amplified fragments was confirmed by Sanger sequencing.
DNA-Seq libraries preparation and sequencingA dual-indexed sequencing library was prepared using the Nextera XT DNA Sample Prep Kit (Illumina). For tagmentation, 1ng of DNA extracted from vesicles was mixed with 5μL of Tagment DNA Buffer and 2.5μL of Amplicon Tagment Mix, and incubated at 50°C for 10min. The reaction was stopped by addition of 2.5μL of Neutralize Tagment Buffer and incubation at room temperature for 5min. After tagmentation, the inserted DNA was amplified via a PCR reaction adding the i7/i5 indexes with an amplification program of 72°C for 3min; 95°C for 30s; 12 cycles of 95°C for 10s; 55°C for 30s; 72°C for 30s; and a final extension at 72°C for 5min. Libraries were purified with AMPure XP beads (Beckman Coulter) and quantified with Qubit dsDNA HS Assay Kit (Thermo). Additionally, the size distribution was evaluated using the 4200 Tape Station with D1000 ScreenTape (Agilent, Santa Clara, CA) and sequenced for 2×150 cycles (paired-end sequencing with dual-index of 8bp) on the NextSeq system (Illumina) as per the manufacturer's instructions.
Bioinformatics processingQuality control of raw sequences was conducted using FastQC and the Trimmomatic algorithm was used to remove adapter sequences and trim short and low-quality end-read sequences. By using Bowtie2 and SAMTools, we cleaned the sequence reads and then aligned them to the human reference genome (hg19) to remove any sequences derived from the human host.16,17 The remaining sequence reads were then subjected to taxonomic sequence classification by employing the Kraken algorithm.18 To confirm the Kraken results, we aligned raw sequence reads to the genome of HPV using the Bowtie2 aligner and SAMTools. The alignment was visualized using the Integrative Genomics Viewer19 and the resulting sequences were sorted out and BLASTed using the most updated NCBI nucleotide database.
Western blotExtracellular vesicles (EV) or HeLa cells (control) were incubated in RIPA buffer for 20min at 4°C. Samples were centrifugated at 12,000×g for 20min at 4°C, and the supernatant collected for quantification with the BCA Protein Assay Kit (Pierce), according to the manufacturer instructions. 40μg of protein was resolved by SDS-polyacrylamide gel electrophoresis and transferred to PVDF membranes. The membranes were blocked with 5% of milk in PBS for 2h and then incubated at 4°C overnight with the primary antibodies for HSP90 α/β (sc-13119 Santa Cruz Biotechnology) or CK10 (sc-53252 Santa Cruz Biotechnology); both diluted in TBS 1:250. After three washes in TBS, the membranes were incubated for 2h in HRP-conjugated goat anti-mouse antibody (sc-2005 Santa Cruz Biotechnology). The membranes were then washed three times and imaged with the Western sure premium chemiluminescent substrate (LICOR).
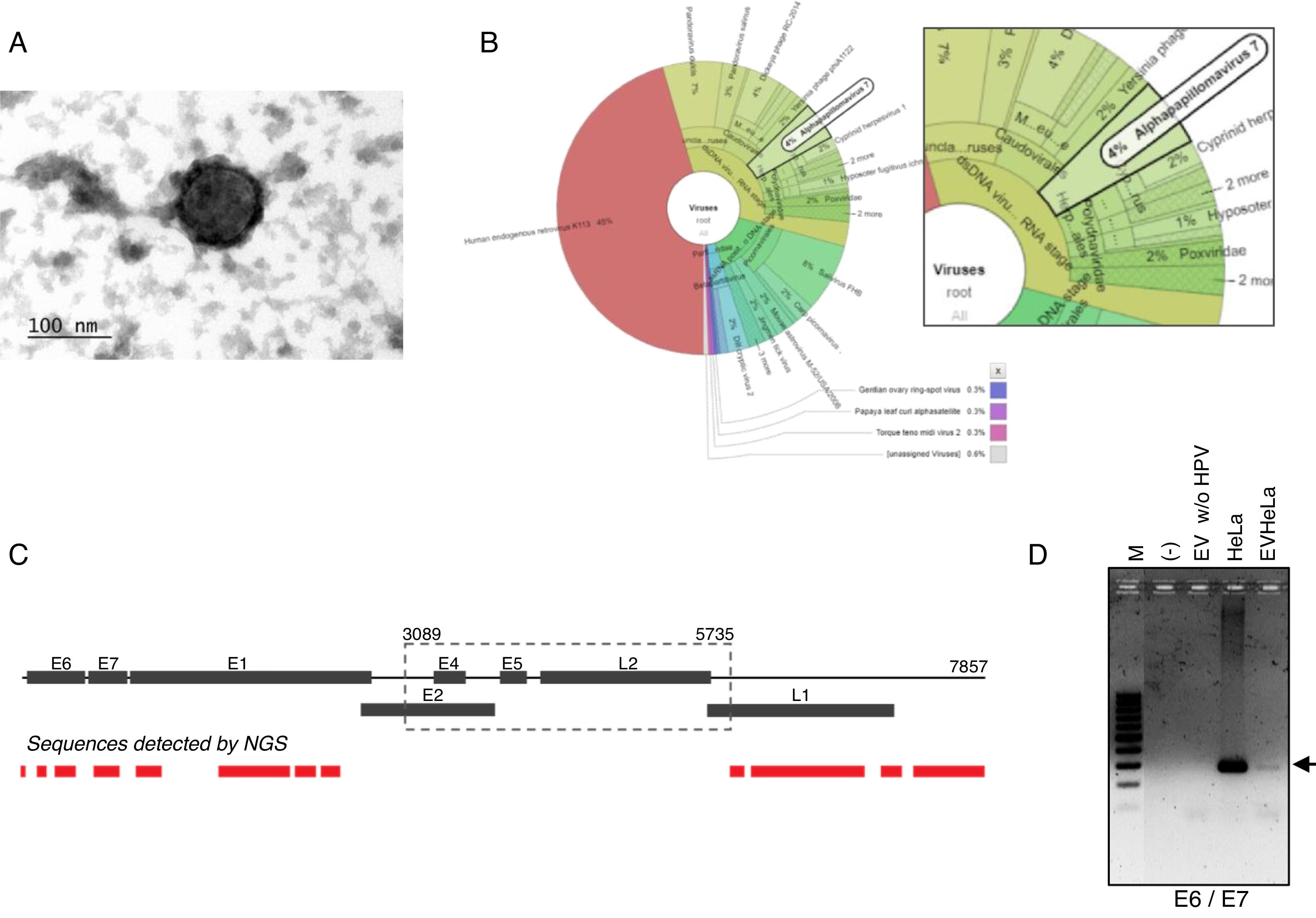
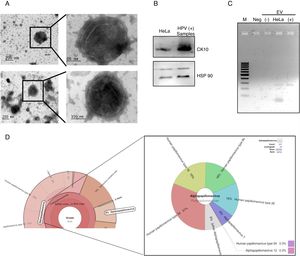
ResultsDetection of HPV18 DNA in extracellular vesicles from HeLa cells by NGS data analysisIn order to evaluate the presence of DNA from HPV in extracellular vesicles (EV), we obtained these from the supernatant of HeLa cell line by ultracentrifugation. We first confirmed the presence of exosomes from HeLa cells through TEM. The samples, enriched in exosomes, shown the previously reported size and structural characteristics as it could be seen in Fig. 1A. The DNA from these vesicles was analyzed through NGS. We performed the DNA extraction in duplicate and analyzed the two samples at different times. The NGS-22-DNA and NGS-11-DNA included 1,677,715 and 4,868,000 reads, respectively. We could detect the HPV in the two samples, and the NGS-22-DNA had until 40 mapped reads. These sequences corresponding to alpha papillomavirus 7, which type specie is the HPV18 (Fig. 1B). The sequence data revealed the presence of the LCR and L1, E6, E7, E1 and E2 genes (Fig. 1C). The specific sequences of the fragments depicted in Fig. 1C are shown in Table 1. The presence of the E6–E7 fragments detected in extracellular vesicles was confirmed by PCR with DNA of EV freshly isolated from HeLa cells (Fig. 1D) and the identity of this fragment was confirmed by Sanger sequencing.
HPV18 DNA in EV derived from HeLa cell line. (A) Electron microscopy of extracellular vesicles from HeLa cells showing the typical morphology. (B) Graphical representation of the unnormalized abundance of VPH in terms of number of reads. (C) Sequences of HPV detected in EV by NGS are indicated in the lower graph. The ORFs are indicated in the whole genome HPV-18 map. The dashed square comprises the region deleted in the inserted genome. (D) Detection of the region E6/E7 of HPV18 in HeLa cells and EV derived from this cell line but not in those derived from HPV-negative cells. M, 100bp DNA ladder.
Sequences of HPV from EV detected by NGS.
| Sequence | Gen | Location |
|---|---|---|
| CTGCAAGACATAGAAATAACCTGTGTATATTGCAAGACAGTATTGGAACTTACAGAGGTATTTGAA | E6 | 177–242 |
| ACTCTGTGTATGGAGACACATTGGAAAAACTAACTAACACTGGGTTATACAATTTATTAATAAGGTGCCTGCGGTGCCAGAAACCGTTGAATCCAGCAGAAAAACTTAGACACCTTAATGAAAAACGACGATTTCACAACATAGCTGGGCACTATAGAGG | E6 | 355–511 |
| GAGGAAGAAAACGATGAAATAGATGGAGTTAATCATCAACATTTACCAGCCCGACGAGCCGAACCACAACGTCACACAATGTTGTGTATGTGTTGTAAGTGTGAAGCCAGAATTGAGCTAGTAGTAGAAAGCTCAGCAGACGACCTTCGAGCATTCCAGCAGCTGTTTCTGAACACCCTGTCCTTTGTGTGTCCGTGGTGTGC | E7 | 692–894 |
| GGTACAGACGGGGAGGGCACGGGTTGTAACGGCTGGTTTTATGTACAAGCTATTGTAGACAAAAAAACAGGAGATGTAATATCAGATGACGAGGACGAAAATGCAACAGACACAGGGTCGGATATGGTAGATTTTATTGATACACAAGGAACATTTTGTGAACAGGCAGAGCTAGAGACAGCACAGGCAT | E1 | 929–1118 |
| CATATGGGCTATCATTTACAGATTTAGTTAGAAATTTTAAAAGTGATAAAACCACGTGTACAGATTGGGTTACAGCTATATTTGGAGTAAACCCAACAATAGCAGAAGGATTTAAAACACTAATACAGCCATTTATATTATATGCCCATATTCAATGTCTAGACTGTAAATGGGGAGTATTAATATTAGCCCTGTTGCGTTACAAATGTGGTAAGAGTAGACTAACAGTTGCTAAAGGTTTAAGTACGTTGTTACACGTACCTGAAACTTGTATGTTAATTCAACCACCAAAATTGCGAAGTAGTGTTGCAGCACTATATTGGTATAGAACAGGAATATCAAATATTAGTGAAGTAATGGGAGACACACCTGAGTGGATACAAAGACTTACTATTATACAACATGGAATAGATGATAGCAATTTTGATTTGTCAGAAATGGTACAATGGGCATTTGATAATGAGCTGACAGATGAAAGCGATATGGCATTTGAATATGCCTTATTAGCAGACAGCAACAGCAATGCAGCTGCCTTTTTAAAAAGCAATTGCCAAGCTAAATATTTAAAAGATTGTGCCA | E1 | 1557–2153 |
| ATATGTCACAGTGGATACGATTTAGATGTTCAAAAATAGATGAAGGGGGAGATTGGAGACCAATAGTGCAATTCCTGCGATACCAACAAATAGAGTTTATAACATTTTTAGGAGCCTTAAAATCATTTTTAAAAGGAACCCCCAAAAAAAA | E1 | 2196–2346 |
| ATTTTTGGTTGGAACCGTTAACAGATACTAAGGTGGCCATGTTAGATGATGCAACGACCACGTGTTGGACATACTTTGATACCTATATGAGAAATGCGTTAGATGGCAATCCAATAAGTATTGATAGAAAGCACAAACCATTAATA | E1 | 2454–2599 |
| TTATTAACTGTTGGTAATCCATATTTTAGGGTTCCTGCAGGTGGTGGCAATAAGCAGGATATTCCTAAGGTTTCTGCATACCAATATAGAGTATTTAGGG | L1 | 5736–5835 |
| TTATAATCCTGAAACACAACGTTTAGTGTGGGCCTGTGCTGGAGTGGAAATTGGCCGTGGTCAGCCTTTAGGTGTTGGCCTTAGTGGGCATCCATTTTATAATAAATTAGATGACACTGAAAGTTCCCATGCCGCCACGTCTAATGTTTCTGAGGACGTTAGGGACAATGTGTCTGTAGATTATAAGCAGACACAGTTATGTATTTTGGGCTGTGCCCCTGCTATTGGGGAACACTGGGCTAAAGGCACTGCTTGTAAATCGCGTCCTTTATCACAGGGCGATTGCCCCCCTTTAGAACTTAAAAACACAGTTTTGGAAGATGGTGATATGGTAGATACTGGATATGGTGCCATGGACTTTAGTACATTGCAAGATACTAAATGTGAGGTACCATTGGATATTTGTCAGTCTATTTGTAAATATCCTGATTATTTACAAATGTCTGCAGATCCTTATGGGGATTCCATGTTTTTTTGCTTACGGCGTGAGCAGCTTTTTGCTAGGCATTTTTGGAATAGAGCAGGTACTATGGGTGACACTGTGCCTCAATCCTTATATATTAAAGGCACAGGTATGCCTGCTTCACCTGGCAGCTGTGTGTATTCTCCCTCTCCAAGTGGCTCTATTGTTACCTCTGACTCCCAGTTGTTTAATAAACCATATTGGTTACATAAGGCACAGGGTCATAACAATGGTGTTTGCTGGCATAATCAATTATTTGTTACTGTGGTAGATACCACTCCCAGTACCAATTTAACAATATGTGCTTCTACACAGTCTCCTGTACCTGGGCAATATGATGCTACCAAATTTAAGCAGTATAGCAGACATGTTGAGGAATATGATTTGCAGTTTATTTTTCAGTTGTGTACTATTACTTTAACTGCAGATGTTATGTCCTATATTCATAG | L1 | 5882–6793 |
| TTGCGTCGCAAGCCCACCATAGGCCCTCGCAAACGTTCTGCTCCATCTGCCACTACGTCTTCTAAACCTGCCAAGCGTGTGCGTGTACGTGCCAGGAAGTAA | L1 | 7035–7136 |
| TATGTGTGTGTGTATATATATATACATCTATTGTTGTGTTTGTATGTCC | LCR | 7137–7185 |
| ATGTTTTGTGGTTCTGTGTGTTATGTGGTTGCGCCCTAGTGAGTAACAACTGTATTTGTGTTTGTGGTATGGGTGTTGCTTGTTGGGCTATATATTGTCCTGTATTTCAAGTTATAAAACTGCACACCTTACAGCATCCATTTTATCCTACAATCCTCCATTTTGCTGTGCAACCGATTTCGGTTGCCTTTGGCTTATGTCTGTGGTTTTCTGCACAATACAGTACGCTGGCACTATTGCAAACTTTAATCTTTTGGGCACTGCTCCTACATATTTTGAACAATTGGCGCGCCTCTTTGGCGCATATAAGGCGCACCTGGTATTAGTCATTTTCCTGTCCAGGTGCGCTACAACAATTGCTTGCATAACTATATCCACTCCCTAAGTAATAAAACTGCTTTTAGGCACATATTTTAGTTTGTTTTTACTTAAGCTAATTGCATACTTGGCTTGTACAACTACTTTCATGTCCAACATTCTGTCTACCCTTAACATGAACTATAATATGACTAAGCTGTGCATACATAGTTTATGCAACCGAAATAGGTTGGGCAGCACATACTATACTTTTC | LCR | 7286–7857 |
| ATTAATACTTTTAACAATTGTAGTA | LCR | 1–25 |
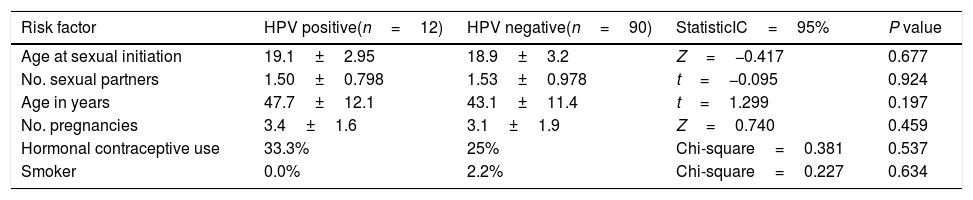
The episomal state of the virus is common in infected patients without lesions or with LSIL. Thus, we also evaluated if the EV from cervical samples of patients with HPV infection but without lesions or with LSIL could contain DNA of E6/E7 HPV, in spite of the expected episomal status. For this purpose, we obtained the DNA from cervical samples of voluntary donors, the status of HPV infection was established by PCR and the products were sequenced to identify the type of virus. We evaluate 106 cervical samples and found 16 positives for HPV (15.7%). Four samples were excluded due to the viral type was undetermined in three of them and the type 6 (low-risk) was detected in the other. Twelve cervical samples (11.8%), of the 102 included, were positive for some high-risk type of HPV. Three of them were positive for HPV 16, four for HPV 18, and the other high-risk types detected were 56, 69, 30, 71 and 85. Three of this samples had infection for more than one viral type (16 and 11, 16 and 71 or 16, 30 and 71). The relation between the positive status with different risk factor was analyzed but none of the variables had significant differences between the positive samples and those negative for HPV (Table 2).
Risk factors associated with HPV infection.
| Risk factor | HPV positive(n=12) | HPV negative(n=90) | StatisticIC=95% | P value |
|---|---|---|---|---|
| Age at sexual initiation | 19.1±2.95 | 18.9±3.2 | Z=−0.417 | 0.677 |
| No. sexual partners | 1.50±0.798 | 1.53±0.978 | t=−0.095 | 0.924 |
| Age in years | 47.7±12.1 | 43.1±11.4 | t=1.299 | 0.197 |
| No. pregnancies | 3.4±1.6 | 3.1±1.9 | Z=0.740 | 0.459 |
| Hormonal contraceptive use | 33.3% | 25% | Chi-square=0.381 | 0.537 |
| Smoker | 0.0% | 2.2% | Chi-square=0.227 | 0.634 |
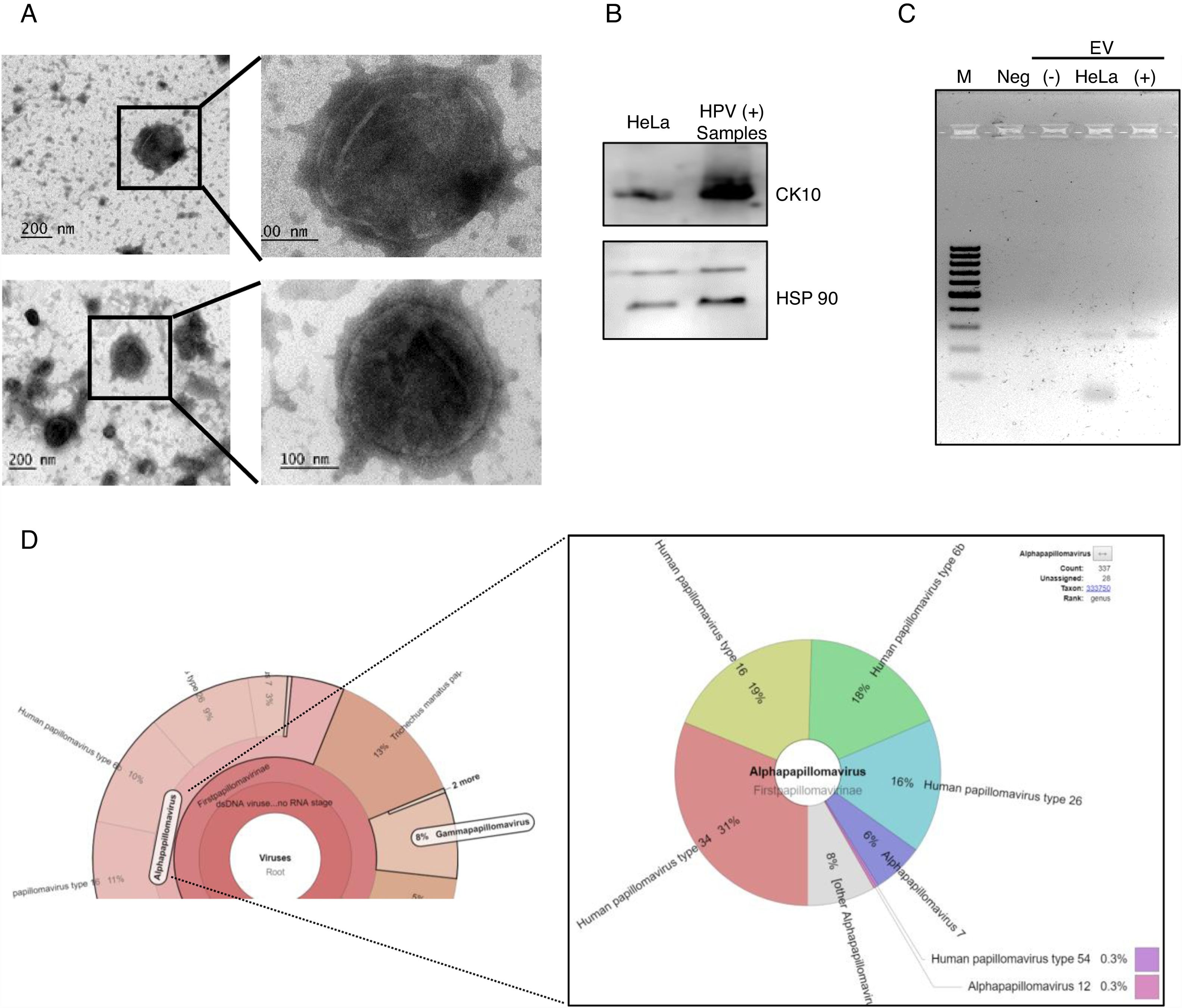
After the analysis of HPV status, we isolated EV from the supernatant of positive and negative samples. The presence of extracellular vesicles was confirmed by TEM (Fig. 2A). Due to the samples could contain EV from different cellular types, we evaluated the presence of cytokeratin 10 (CK10) in the EV derived from HPV-positive samples and HeLa cell line, which is expressed in keratinocytes from the parabasal layer and it was positive in both. The HPS90 protein was also evaluated and found positive in EV derived from HPV-positive cervical samples and cell line (Fig. 2B). Finally, we evaluated the presence of DNA of E6/E7 HPV from HPV in the EV from cervical samples by PCR. We detected the fragment corresponding to E6/E7 expected in the EV derived from HPV-positive samples but not in those from negative samples (Fig. 2C). To further confirm the presence of DNA from HPV in EV and to evaluate if the viral type detected in the EV correspond to that in the cellular fraction of the cervical samples; the DNA obtained from EV of four separate samples with LSIL and HPV positive was analyzed through NGS. The analysis confirmed the presence of HPV in all the samples. The number of mapped reads of HPV was 3,385,430 and 25,499. Even if only one viral type was detected in the cells of cervical samples, the NGS detect several other in EV (Fig. 2D) due to the necessary fragmented status of the DNA in the technique. In the four samples analyzed the viral type detected in cells was included in that reported in the EV.
DNA of HPV in EV derived from HPV-positive cervical samples. (A) Electron microscopy of extracellular vesicles from positive (top panel) and negative (lower panel) samples. (B) Detection of CK10 and HSP90 proteins in EV from HPV-positive cervical samples by immunoblot. (C) Detection of the region E6/E7 of HPV18 in EV derived from HPV-positive samples but not in those derived from HPV-negative cells. The EV of HeLa cells was employed as a positive control. Neg, negative control without template. M, 100bp DNA ladder. (D) Graphical representation of the unnormalized abundance of VPH in terms of number of reads and the viral types detected, in EV from a representative sample.
We show the presence of DNA-HPV in extracellular vesicles enriched with exosomes regardless of the integration status in the cell. The ultracentrifugation isolation is the gold standard to obtaining EV. We used this methodology with the recommended conditions to the obtention of EV enriched in exosomes. The visualization with TEM confirmed the obtention of the expected EV8,11 when obtained from supernatants of culture medium as well as cervical samples.
Fragments fused with chromosome 8 were detected (data not shown), but in a low frequency due to the astringency necessary to eliminate the human DNA for detection of other organisms.16,17 But the fragments of HPV detected in EV from HeLa cell line and the absence of the region 3089 to 5735nt not only confirmed that they constitute some of the genome of this cell line, but also is congruent with the previously published reports. These indicate the presence of DNA fragments in exosomes reflecting the completeness of the genome in the cells of origin13,20 and HeLa cell line with HPV 18 integrated with the loss of the genes E4, E5, L2, and partial sequence of E2 and L1.21 The absence of fragments in regions other than between bases 3089 and 5735 of the viral genome could be attributable to the astringency in the analysis, since the modified PU1M/2R allowed us to amplify the region between nucleotides 426 and 690 even if only the sense primer hybridizes in a region detected by NGS.
In cervical cancer and high-grade lesions viral genomes are usually integrated, but in HPV infections and low-grade lesions the genome is found in episomal state.22 In all the EV of cervical samples evaluated, we found the DNA of HPV despite the expected episomal status. This finding agrees with previous reports indicating the presence of circulating DNA of HPV in serum of patients with cancer3,22 and exosomes of nipple discharges4; however, most authors do not evaluate the origin of circulating DNA in serum. The vesicles obtained from cervical samples allow us to identify the HSP90 protein, which is a constant component of extracellular vesicles but is also enriched in those derived from cancer cells.23,24 The HSP90 has been previously reported in HeLa cells regulating the function of survivin through their interaction 25; and also has been reported that the presence of survivin in EV is regulated by E6/E7 expression.12 Another molecule identified in total protein from EV was CK10, which is expressed in the less differentiated layers of the epithelium. This helped us to confirm the presence of EV derived from keratinocytes in the obtained cervical scrapples.
The content of viral genomes in EV previously observed for other viruses14 and the positive results for the detection of exosomal viral DNA of HPV, allows us to focus on exosomes not only as carriers of molecules that would contribute to the transformation of nearby cells but also as a source of DNA for the detection of an HPV infection. This possibility increases in the infection and transformation by HPV due to this finding and the recent reports about the extracellular location of their DNA, and their possible distribution through EV in fluids. The detection of viral DNA in EV and their coincidence with the viral type detected in the cells, made them useful for the diagnosis of the infection by direct or nested PCR if the EV could be isolated from different fluids. Several additional types of virus were detected through NGS, this occur due to the fragmented regions of the viral gene L1 and the other different to this. For this reason, a greater number of cervical samples along with their EV need to be analyzed to support the correspondence between the viral types detected. On the other hand, episomal DNA contained in EV from keratinocytes had not been previously reported and suggests the possibility of viral DNA transmission between cells through exosomes. This transmission amplifies the possibility of integration of DNA coding for viral oncogenes or their expression in non-infected keratinocytes, but this hypothesis requires a further detailed study. This work exposes the presence of HPV DNA including the viral oncogenes into EV independent of their integration status so as the correspondence between the viral type in the vesicles and their origin cells.
FundingThis research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflicts of interestThe authors declare no conflict of interest.
The authors acknowledge the Sequencing Laboratory, Instrument Center of National Medical Center “Siglo XXI”, IMSS, Mexico City, for their technical assistance and support. We also acknowledge to the Electron Microscopy Unit at the National Laboratory of Experimental Services (LaNSE), CINVESTAV, Mexico City for their technical assistance.