This study aimed to collect and summarize test data and conduct a meta-analysis, with respect to the Multitarget Stool DNA test sensitivity and specificity, compared to colonoscopy.
Material and methodsAll manuscripts were screened for eligibility according to inclusion criteria. Participants were a normal population at an average risk of developing CRC. Intervention was Stool based and DNA panel tests compared with colonoscopy, and outcome was detection of CRC and any pre-cancerous lesions. Inter-study and inconsistency (using the I-squared test) were assessed.
ResultsMeta-analyses of the Mt-sDNA test showed a combined sensitivity of 89%, 51%, and 76% for the detection of CRC, advanced adenoma (AA), and combined CRC and AA, respectively. The overall specificity was 91%, 89%, and 90% for the detection of CRC, AA, and combined CRC and AA, respectively.
ConclusionMt-sDNA had significantly acceptable diagnostic accuracy for CRC and AA diagnosis, but still has lower sensitivity and specificity than colonoscopy.
Este estudio tuvo como objetivo recopilar y resumir los datos de las pruebas y realizar un metaanálisis con respecto a la sensibilidad y especificidad de la prueba de DNA en heces multiobjetivo, en comparación con la colonoscopia.
Material y métodosTodos los manuscritos fueron examinados para determinar su elegibilidad de acuerdo con los criterios de inclusión. Los participantes eran una población normal con un riesgo promedio de desarrollar CRC. La intervención se basó en heces y pruebas de panel de DNA en comparación con la colonoscopia, y el resultado fue la detección de CRC y cualquier lesión precancerosa. Se evaluaron la inconsistencia entre estudios y la inconsistencia (mediante la prueba de I cuadrado).
ResultadosLos metaanálisis de la prueba Mt-sDNA mostraron una sensibilidad combinada del 89%, 51% y 76% para la detección de CRC, adenoma avanzado (AA) y CRC y AA combinados, respectivamente. La especificidad general fue del 91%, 89% y 90% para la detección de CRC, AA y CRC y AA combinados, respectivamente.
ConclusiónMt-sDNA tuvo una precisión diagnóstica significativamente aceptable para el diagnóstico de CRC y AA, pero aún tiene una sensibilidad y especificidad más bajas que la colonoscopia.
According to the latest Global Cancer Statistics, colorectal cancer (CRC) is the third most common cancer, but is the second leading cause of cancer-related death, worldwide.1 The incidence of colorectal cancer vary widely across different regions of the world, as it was about 9-fold variation, and 4-fold higher in transitioned countries as well.1,2 Socioeconomic development, lack of CRC screening and early diagnosis programs, and westernization of most developed countries recently leaded to an increasing trends of incidence and mortality in low and lower-middle income countries.1–3 However there was declines in both CRC incidence and mortality in most developed countries because of implementation of screening programs including colonoscopy and removal of precancerous lesions, healthy lifestyle choices, declines in smoking, and best practices in early detection and treatment of CRC.1–3
CRC screening programs, which lead to early detection, can reduce approximately 60% of CRC-related deaths. Recent studies have shown that 60–70% of CRC cases can be diagnosed at an early stage of the disease by screening and early diagnostic techniques, including colonoscopy.4,5 This allows the diagnosis of cancer in the curable stage and also decreases the CRC incidence in average-risk individuals.6 Screening strategies should therefore include a standard screening method to detect precancerous lesions in asymptomatic individuals, identifying those eligible for screening by risk assessment.6,7
Increased knowledge on specific changes and molecular pathways underlying CRC malignancy has led to the development of new laboratory-based diagnostic tests, including molecular and immunologic methods, in recent years. Compared with colonoscopy as an approved gold standard method for CRC screening in many centers, new techniques are non-invasive, simple, and patient friendly. According to the last released guidelines of American Cancer Society's in 2018, U.S. Preventive Services Task Force in 2016, and the National Comprehensive Cancer Network in 2016, multi-target stool DNA (Mt-sDNA) tests (combined Fecal Immunochemical Test (FIT)-DNA stool tests) every 3-years have been recommended as a potential screening method in average-risk populations, with almost the same results in CRC incidence reduction compared with 10-year colonoscopy (63% vs. 65%).6,8,9 Most recently United State (US) Preventative Services Task Force has been endorsed Mt-sDNA test as first-line CRC screening test, as this test is cost effective compared with no screening, and is enough efficient compared with other screening modalities.10 Therefore, Mt-sDNA-based testing may be a good substitute for individuals unable to undergo colonoscopy because of possible harm, unavailability, and unpleasant and time-consuming cathartic bowel preparation.11 However, colonoscopy is strongly recommended for individuals with abnormal findings in any other screening method, including sigmoidoscopy, CT colonoscopy, and stool-based methods including Guaiac Fecal Occult Blood Test (FOBT), Fecal Immunochemical Test (FIT), and stool DNA test. Although different screening modalities were presented in majority of guideline, however there is still discrepancy with regards to the respective role, ranking and cost effectiveness of these tests.8,9,12
Therefore, we performed a systematic review and meta-analysis of the most reliable published evidence on the diagnostic accuracy of Mt-sDNA tests compared with colonoscopy during the last 20 years. This information will aid clinical decision-making.
Material and methodsWe designed and performed this study as a systematic review and meta-analysis following the PRISMA guidelines.13
Eligibility criteriaThe systematic review question was the effectiveness of Stool-based DNA tests for detecting CRC and any other pre-cancerous lesions (advanced adenoma) compared with colonoscopy as a gold standard method in a normal population (asymptomatic persons with an average risk of developing CRC).
Information sourcesWe searched all available databases including PubMed, MEDLINE, CINAHL Plus with full text (EBSCO), Web of Science Core Collection, EMBASE (Ovid), SCOPUS, ProQuest Central, Joanna Briggs Institute EBP database, and Google Scholar, as well as SID, Magiran, Medlib, and Irandoc as Iranian databases.
Search strategyWe used Boolean Logics, including the following key words:
“Colorectal”, “Cancer”, “Malignancy”, “Neoplasm”, “Screening”, “Early Detection”, “Colonoscopy”, “Stool based tests”, “Stool DNA Test”, “Mt-sDNA-based testing”.
In addition, we used the methods “snowballing” (search for related articles or similar results), the “ancestry” method (follow up on citations in selected publications), and use the “cited references”. In some cases, we contacted the study authors to identify additional studies.
Selection processWe selected studies according to our defined PICOS items, first from Title and Abstracts, and then from the text of the manuscripts. All manuscripts were screened for eligibility according to PICOS criteria, time of publication, and type of study. Studies with reliable and available end points (sensitivity (SE), specificity (SP), true positives (tp), false positives (fp), false negatives (fn), true negatives (tn), number of individuals with disease (CRC and AA), and number of healthy controls were included in the meta-analysis.
Criteria for considering studies for this review were as follows:
PICOSParticipantsNormal population (average risk of developing CRC).
InterventionStool based DNA test panel compared with colonoscopy.
Panel includes mutational aberrations including Kirsten rat sarcoma viral oncogene homolog (KRAS) and v-raf murine sarcoma viral oncogene homolog B1 (BRAF) and/or hyper methylated promoter genes including Secreted Frizzled-Related Protein-2 (SFRP2), Tissue factor pathway inhibitor 2 (TFPI2), Bone morphogenetic protein 3 (BMP3), N-Myc downstream regulated gene 4 (NDRG4), Tissue factor pathway inhibitor 2 (TFPI2), and vimentin) as well as Fecal Immunochemical Test (FIT, any brand)
Control/compareOptical colonoscopy (gold standard method).
OutcomeDetecting any
- 1)
Precancerous lesions (AA)
AA:
An AA was defined as
- -
an adenoma 10mm or
- -
an adenoma with villous histology (25% villous)
- -
and/or high-grade dysplasia of any size.
- 2)
Neoplastic lesions (including carcinoma in situ early stage or advanced CRC)
Test accuracy and reliability in detecting CRC and/or AA (Clinical sensitivity and specificity; predictive value)
Publication datePublished in the last 20 years
Publication language: English
We did not consider any geographic restraints in the search strategy.
Data collection processTwo main investigators (RD, SD) performed the appraisal of the included manuscript and data collection independently, according to our eligibility criteria.
Data itemsStudies with reliable and available end points (SE, SP, true positives (tp), false positives (fp), false negatives (fn), true negatives (tn), number of individuals with disease (CRC and AA), and number of healthy controls were included in the meta-analysis.
Study risk of bias assessmentThe risk of bias was assessed using the Cochrane Collaboration tool,14 which included six domains, including reporting bias, attrition bias, performance bias, detection bias, selection bias, and other bias. Accordingly, each domain was assessed as having a low, unclear, or high risk of bias.
Effect measuresFor the meta-analysis of the diagnostic accuracy of Mt-sDNA test for detecting CRC and AA compared with colonoscopy, hierarchical logistic regression and the HSROC (hierarchical summary receiver operating characteristic) model were applied.15 The summary measures were sensitivity and specificity, positive and negative likelihood ratios, and diagnostic odds ratios.
The primary dataset was designed for analysis including true positives (tp), false positives (fp), false negatives (fn), true negatives (tn), number of individuals with disease (CRC and AA), and number of healthy controls.
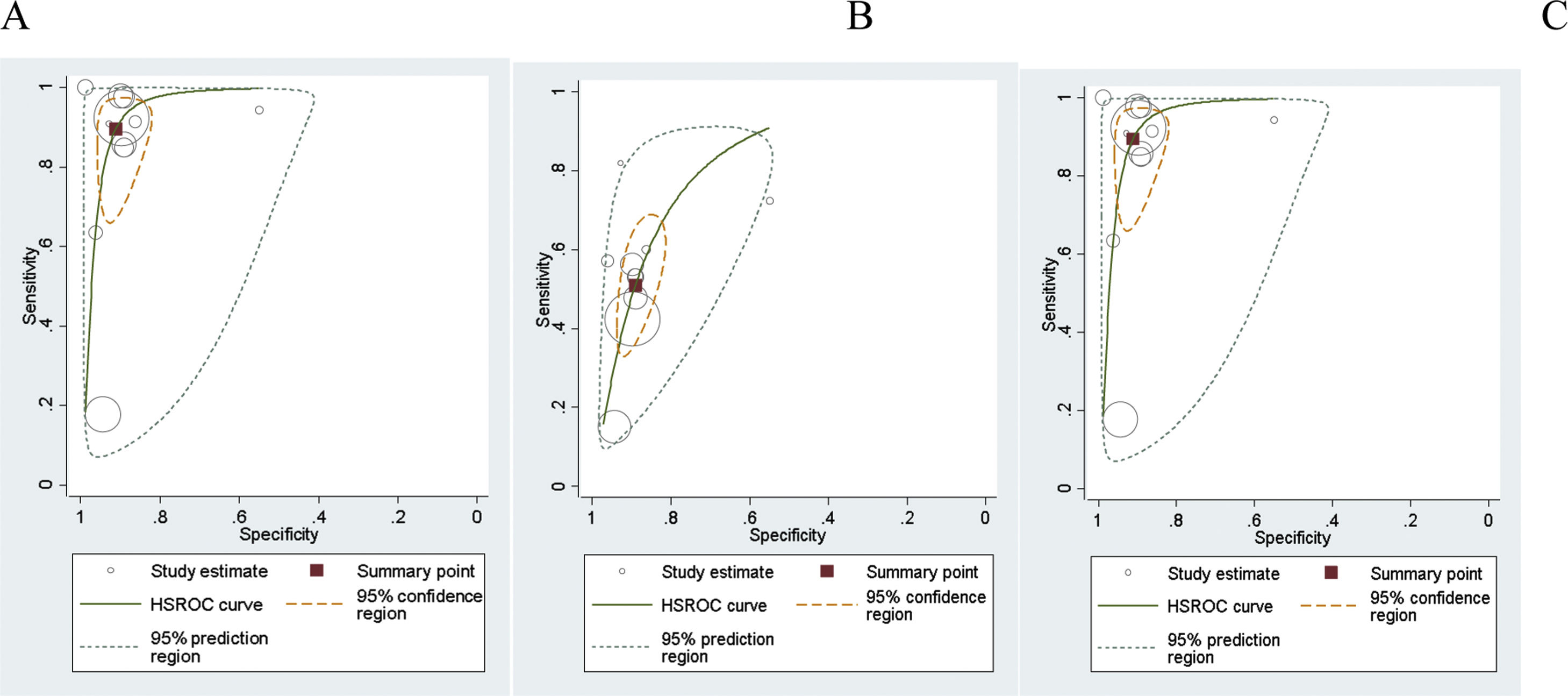
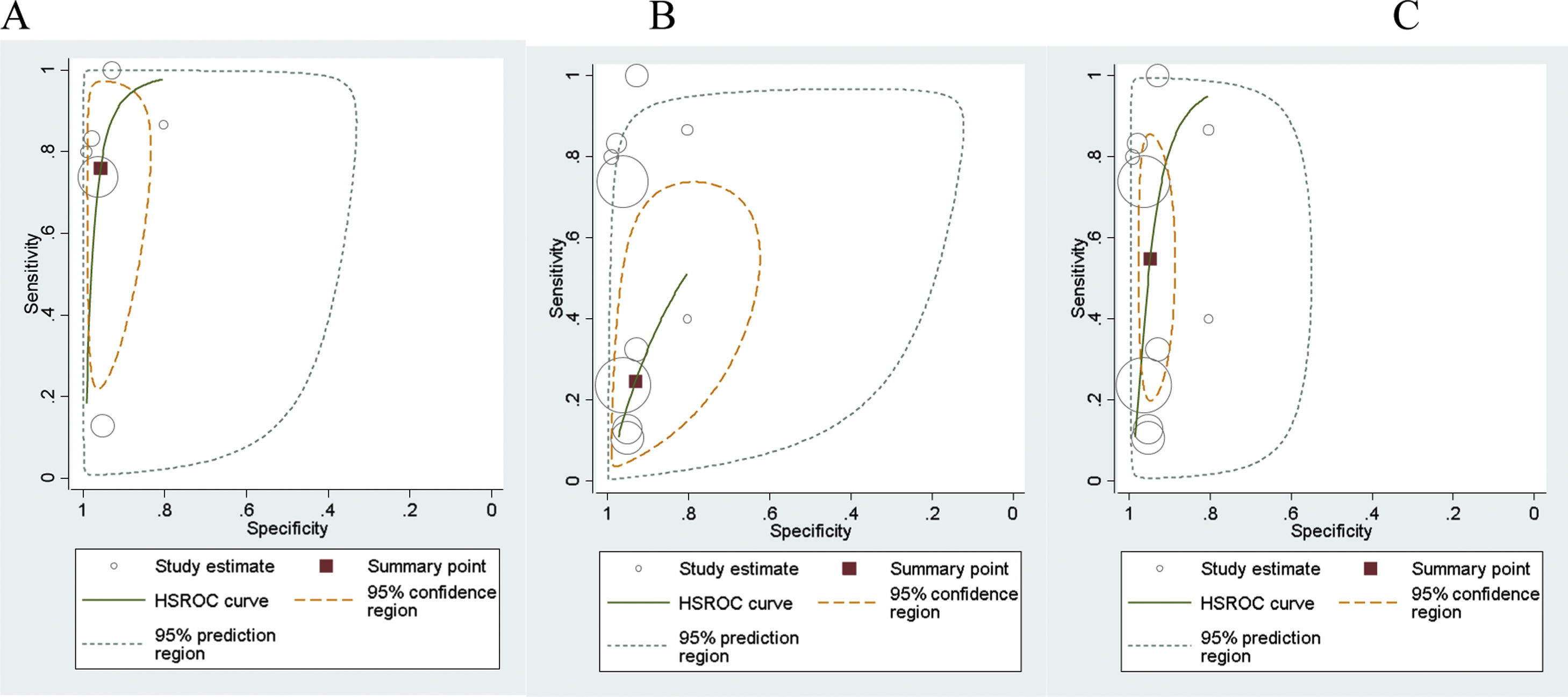
The summary receiver operating characteristic curves (SROC) were applied to simplify the graphical summaries for the fitted model, summary points and confidence regions. The size of each circle shows the total number of cases in each study. The size of each point was scaled according to the precision of sensitivity and specificity in the study. The solid circle (summary point) represents the summary estimate of the sensitivity and specificity of the related test. The summary point is surrounded by a dotted line representing the 95% confidence region and a dashed line representing the 95% prediction region. Finally, a summary curve from the HSROC model was obtained.
Synthesis methodsThe quantity I2 was used to assess the impact of unobserved heterogeneity across studies that are attributed to the heterogeneity between studies rather than to chance.16 A mixed model was used to express both the intra-class correlation coefficient (ICC) and Midas (MED) sensitivity and specificity separately as percentages with 95% confidence intervals (CIs). In addition, the I2value ranged from 0 to 100, while 0% indicated no observed heterogeneity and an increase in the value greater than 75% was considered as substantial heterogeneity between studies. All analyses were performed using STATA MP 14.2 (StataCorp LP, College Station, TX 77845, USA) software. Inter-study variation in sensitivity and specificity, receiver operating characteristic (ROC) Area with 95% CI, heterogeneity (using Chi-squared test), and inconsistency (using the I-squared test) were performed using the “midas” method. The reference test was colonoscopy and the index tests were Mt-sDNA and FIT tests.17
Reporting bias assessmentPublication bias across the studies was assessed with Deek's asymmetry test, which performs a linear regression of the log odds ratios on the inverse root of effective sample sizes as a test for funnel plot asymmetry in the diagnostic meta-analysis.18 A non-zero slope coefficient is suggestive of a significantly small study bias (p value<0.10).
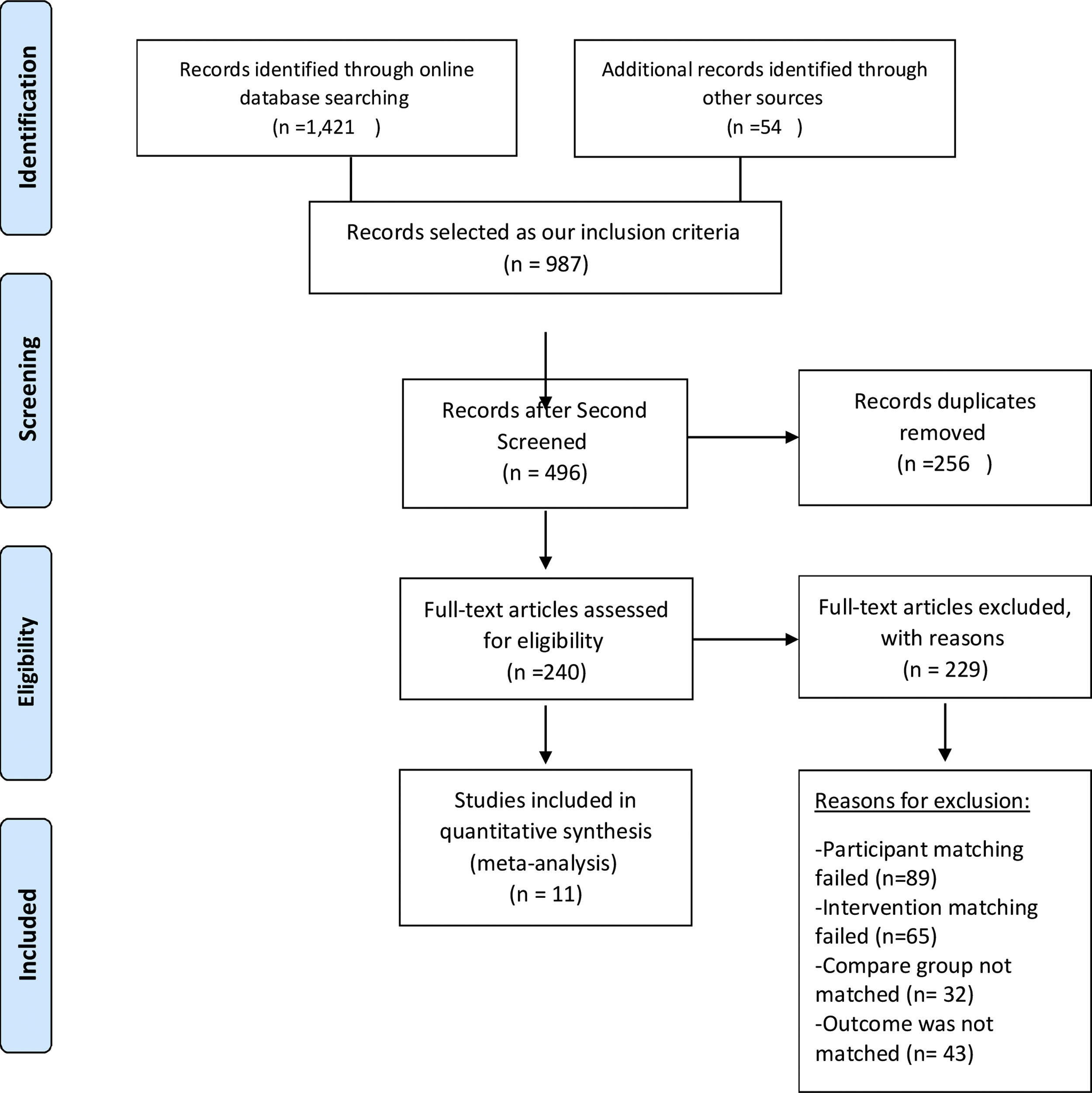
ResultsStudy selectionAn advanced search in all available databases using intended keywords found 450 published manuscripts in the PubMed database, 89 in EBSCO, 243 in Web of Science Core Collection, 432 in EMBASE (Ovid), 87 in SCOPUS, 80 in Google Scholar, and 40 in Iranian databases (SID, Magiran, Medlib, and Irandoc). Meanwhile, 54 studies were selected by a search of manuscript bibliographies. A total of 1475 published studies were screened and assessed for eligibility. Of these studies, 987 were selected based on our inclusion criteria. Records were screened independently by two co-investigators for reliability and data quality, and after excluding 256 duplicates, finally, 11 full text articles were included in the review and quantitative analysis (Fig. 1).
Study characteristicsThe information of included studies is summarized in Table 1 (Table 1).
Summary information of 11 included studies in systematic review and meta-analysis.
| No | Author/country (refs) | Study type/design | Number of case/control in each group | AgeMedian/mean (range) | Intervention tests | Length of follow-up |
|---|---|---|---|---|---|---|
| 1 | Mu J et al., 2020China21 | Blinded case control | Total evaluated 839Case: 252CRC=203Advanced colorectal neoplasia=49Control: 587Non-advanced neoplasia=156Normal Subjects=431 | 40–85 | Multitarget stool DNA panel:-KRAS-BMP3-NDRG4-β actin- methylated B2M quantitation“ColoClear” similar to ColoGuard and FIT (Hemmocult II) | 1st May 2015 to 30th October 2017 |
| 2 | Bosch LJW et al., 2019Netherlands23 | Randomized control trial (screening) | Total evaluated 1014- CRC: 7- Advanced precancerous lesions: 119- Advanced Adenoma: 92- Advanced Serrated Polyp: 27- High-grade Dysplasia: 22 | 50–75 | Multitarget stool DNA panel:-KRAS-BMP3-NDRG4-β actinand FIT (OC-Sensor; Eiken Chemical,Tokyo, Japan) | From June 2009 and July 2010 |
| 3 | Sun M et al., 2019China24 | Case–control | Total evaluated 233Cases:CRC: 105AA: 20Polyps: 6Controls:102 (Colonoscopy negative) | Not assessed | Multitarget stool DNA panel:-KRAS-BMP3-NDRG4-SDC2-SFRP2and FIT(Fecal Occult Blood Test Kit (colloidal gold) | Not assessed |
| 4 | Park SK et al., 2017Korea22 | Randomized control trial (screening) | Total evaluated 111- CRC: 35- AA: 36- Normal: 40 | Mean: 55.7 | Stool-based DNA methylation markers:-SFRP2-TFPI2-NDRG4-BMP3 | Between August 2012 and March 2014 |
| 5 | Redwood DG et al., 2016Alaska19 | Prospective, cross-sectional | Total evaluated 661Screening Relevant Colorectal Neoplasia (SRN): 92CRC: 10Villous Adenoma: 28Sessile serrated polyps ≥1cm: 16Tubular adenoma: 38Non-SRN: 569 | 40–85 | Multitarget stool DNA panel:-KRAS-BMP3-NDRG4-β actin(Cologuard)and FIT (OCSensor Diana, PolyMedco, Inc, Portlandt, NY) | From February 6, 2012, through August 7, 2014 |
| 6 | Imperial TF et al., 2014USA31 | Cross sectional | Total evaluated 9989CRC: 65Advanced precancerous lesions: 757High Grade dysplasia: 39Sessile serrated polyps ≥1cm: 99 | 50–84 | Multitarget stool DNA panel:-KRAS-BMP3-NDRG4-β actin(Cologuard)and FIT(OC FIT-CHEK, Polymedco) | From June 2011 to November 2012 |
| 7 | Lidgard GP et al., 2013USA11 | Blinded, multicenter, case–control | Total evaluated 1003 (459 asymptomatic patients and 544 referred patients)CRC: 93Advanced Adenoma: 84Sessile Serrated Adenoma ≥1cm: 30 | Median 65 years (range 38–87) | Multitarget stool DNA panel:-KRAS-BMP3-NDRG4-β actinand FIT(OC-FIT CHEK, Polymedco, Cortlandt Manor, NY) | Not assessed |
| 8 | Ahlquist DA et al., 2012USA, Canada, Denmark32 | Blinded multicenter case–control | Total evaluated 678Cases:385252 CRC, 133 adenoma≥1cmControls: 293 (normal colonoscopy) | 60 (39–92) | Multitarget stool DNA panel:-KRAS-NDRG4-BMP3-TFPI2-Vimentin-β-actinHemoglobin (HemoQuant) | Not assessed |
| 9 | Imperial TF et al., 2004USA20 | Randomized control trial | Total evaluated 4404CRC: 31CRC plus adenomas with High grade dysplasia: 71Advanced adenoma: 403Minor polyp: 648Normal: 1423 | ≥50 (68.6) | Multitarget stool DNA panel:-KRAS-APC-P53-BAT-26-Long DNAHemoccult II (guaiac-based tests) | From August 2001 and March 2003 |
| 10 | Tagore KS et al., 2003USA25 | Case control | Total evaluated 212Cases: 8052 invasive CRC, 28 advanced adenomaControls: 212113: Negative colonoscopy99: minor polyps | ≥50 | Multitarget stool DNA panel:-MTAP-KRAS-P53-APC-BAT-26 | not assessed |
| 11 | Ahlquist DA et al., 2000USA30 | Feasibility StudyIncluding 2 clinical pilot clinical trials | Total evaluated 61CRC: 22Adenoma≥1cm: 11Normal: 28 | Median 70 (38–88) | Multitarget stool DNA panel:-KRAS-APC-P53-Bat-26-L-DNA | Not assessed |
The risk of bias was assessed in the clinical trials. An unclear risk of selection bias (due to lack of information on the method of randomization and concealment), performance bias (due to lack of information on blind assessments), and detection bias (blinded outcome assessment) was observed. The risk of bias for the included studies was low for reporting bias, attrition bias, and other sources of bias (Fig. 2).
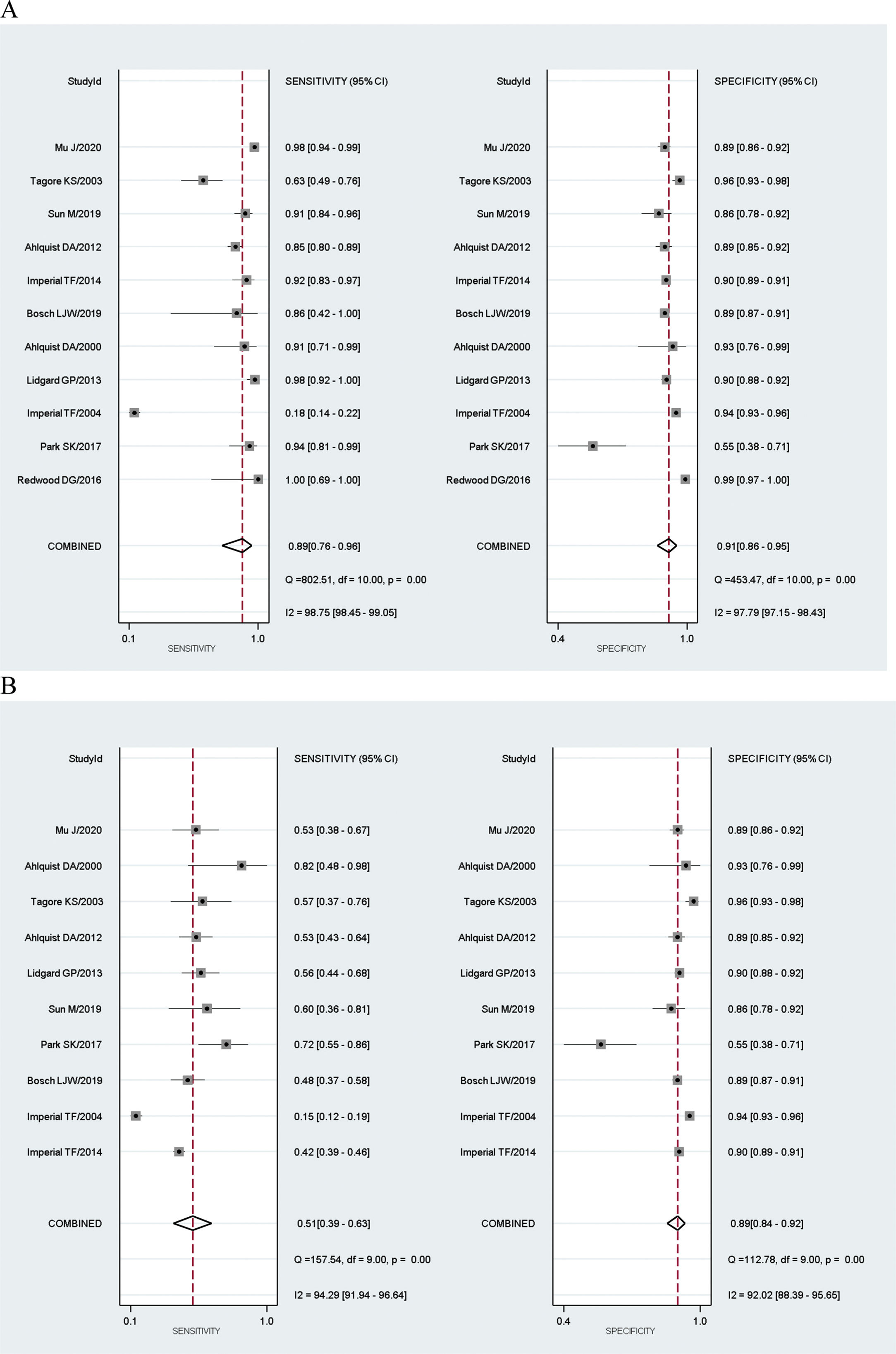
Results of individual studiesFor the Mt-sDNA diagnostic test accuracy, 11 studies were included in the meta-analysis. For the FIT diagnostic accuracy test, 6 studies were included. Results of Mt-SDNA and FIT tests underwent total and subgroup analysis for CRC and AA and for combined CRC and AA diagnostic accuracy compared with colonoscopy (Table 2).
Comparing multitarget stool DNA and FIT tests, advantages and disadvantages.6,35.
| Recommended testing interval | Sensitivity6 | Specificity6 | Procedure | Harms | Cost | Availability | Diet and/or medication restriction | Patient compliance rate | Side-affecting detection biases | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Colorectal cancer | Advanced adenoma | ||||||||||
| MT-sDNA test | Every 3 years | 92% | 24–42% | 87% | Simple | Non-invasive | Costly | Less available | No | Nearly 70% | Unaffected |
| FIT | Annually | 76–95% | 27–47% | 89–96% | Simple | Non-invasive | More cost-effective | More available | No | Nearly 14% | Affected |
Eleven studies achieved the eligibility to be included in the diagnostic accuracy meta-analysis. The total number of CRC patients in the 11 studies ranged from 7 to 435 cases, and AA patients from 11 to 757 cases, while the control groups were between 28 and 4457 people. There were two outcome analyses for ten studies (CRC and AA), and one study only had diagnostic tests for CRC cases in the dataset.
The results showed that the sensitivity of Mt-sDNA test for CRC diagnosis ranged between 100%,19 and 18%.20 The specificity was at its highest in the Redwood et al., 2020 study (99%),21 and lowest in the Park et al., 2017 study (55%).22
FIT test subgroupSix studies were eligible for inclusion in the diagnostic accuracy meta-analysis. The total number of CRC patients in the six studies ranged from 7 to 252, AA from 20 to 757, and healthy controls from 102 to 4454.
There were two outcome analyses for four studies (CRC and AA), and two studies only had diagnostic tests for CRC cases in the dataset. The sensitivity of the FIT test for CRC diagnosis ranged from 100%,23 to 13%.20 The specificity was the highest in the Redwood et al. (2016) study (99%),19 and lowest in the Sun et al. (2019) (80%) study.24
Results of synthesisMeta-analysis results of the Mt-sDNA test showed that the combined sensitivity for diagnosis was 89% for CRC (95% CI: 0.76–0.96), 51% for AA (95% CI: 0.39–0.63), and 76% for combined CRC and AA (95% CI: 0.61–0.86). Combined specificity was 91% (95% CI: 0.86–0.95) for CRC, 89% (95% CI: 0.84–0.92) for AA, and 90% (95% CI: 0.87–0.93) for combined CRC and AA.
The diagnostic odds ratio of the Mt-sDNA test was 87.42 for CRC (95% CI: 31.79–240.43), 8.46 for AA (95% CI: 5.59–12.81), and 87.42 for combined CRC and AA (95% CI: 31.79–240.43).
The results are presented in Figs. 3 and 4.
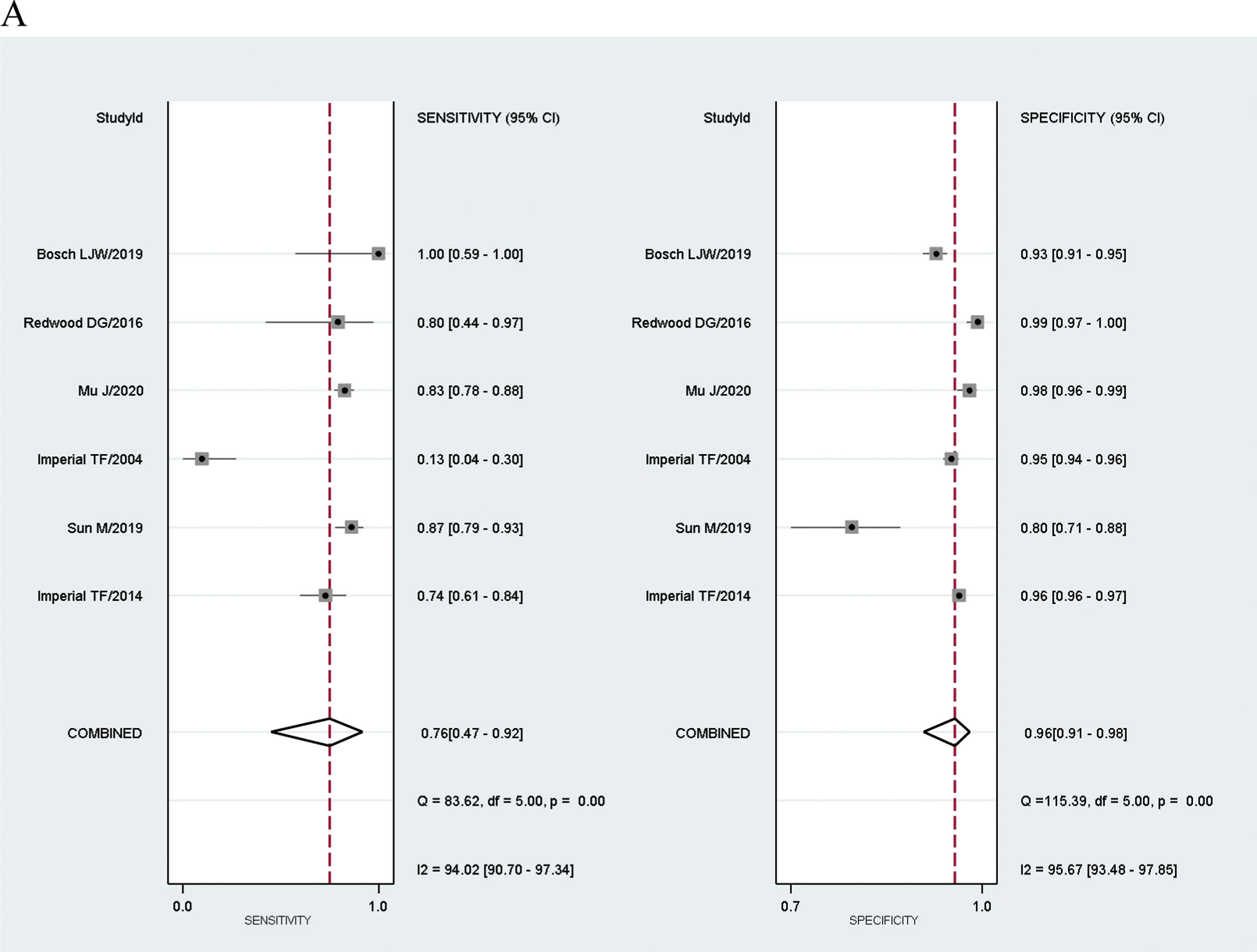
Meta-analysis results of FIT showed that the combined sensitivity for diagnosis was 76% for CRC (95% CI: 0.47–0.92), 25% for AA (95% CI: 0.14–0.39), and 55% for combined CRC and AA (95% CI: 0.29–0.77).
The diagnostic odds ratio of the FIT test was 70.72 for CRC (95% CI: 17.04–293.54), 4.46 for AA (95% CI: 2.47–8.05) and 21.84 for combined CRC and AA (95% CI: 6.65–71.70). The results are presented in Fig. 5 and 6.
Deek's asymmetry test was used to assess publication bias across the studies.
The statistically non-significant p-value for the slope coefficient suggests symmetry in the data and a low likelihood of publication bias across the studies regarding the diagnostic accuracy of Mt-sDNA testing for CRC (P=0.13) and AA (P=0.13) separately.
There was a significant study bias across the studies on Mt-sDNA testing for combined CRC and AA (P=0.03).
Deek's asymmetry test was used to assess publication bias across the studies. There was a significant study bias across the studies on Mt-sDNA testing for combined CRC and AA (P=0.03), but this was not significant for CRC (P=0.13) and AA (P=0.13) separately.
The statistically non-significant p-value for the slope coefficient suggests symmetry in the data and a low likelihood of publication bias across the studies regarding the diagnostic accuracy of FIT tests for CRC (P=0.51), AA (P=0.67), and combined CRC and AA (P=0.36).
The results with regression lines superimposed on a funnel plot are presented in Figs. 7 and 8.
Certainly of evidencesHeterogeneity analysis using the “midas” model showed that there was substantial heterogeneity between the 11 studies with Mt-sDNA results compared with colonoscopy, and the I2 was 100 (95% CI: 100–100) for the CRC subgroup, I2=99 (95% CI: 98–99) for AA, and I2=100 (95% CI: 100–100) for the diagnosis of combined CRC and AA.
Heterogeneity between the six studies with FIT tests and colonoscopy showed that I2=97 (95% CI: 95–99) for CRC diagnosis, I2=94 (95% CI: 90–99) for AA diagnosis, and I2 was 99 (95% CI: 99–100) for the diagnosis of combined CRC and AA.
DiscussionMeta-analyses of diagnostic accuracy studies of Mt-sDNA test showed a combined sensitivity of 89%, 51%, and 76% for the detection of CRC, AA, and combined CRC and AA, respectively. The overall specificity was 91%, 89%, and 90% for the detection of CRC, AA, and combined CRC and AA, respectively. Meta-analyses of diagnostic accuracy studies of the FIT test showed a combined sensitivity of 76%, 25% and 55% for the detection of CRC, AA, and combined CRC and AA, respectively. The combined specificity was 96%, 93%, and 95% for the detection of CRC, AA, and combined CRC and AA, respectively. Therefore, Mt-sDNA had a higher sensitivity for CRC and AA diagnosis, with almost the same specificity rates as FIT. The Mt-sDNA test is easy, safe, and convenient, does not require any dietary restrictions or bowel preparations, and should be performed every three years. However, using this test for CRC screening is still a challenge in terms of cost and accuracy.
Despite reliable and comprehensive studies in the last two decades, additional evidence-based analysis is needed to prove the diagnostic accuracy and specificity of the multi-target stool DNA (Mt-sDNA) test. Therefore, this study aimed to collect and summarize test data during last 20 years, and conduct a meta-analysis, with respect to the Mt-sDNA test sensitivity and specificity, compared to colonoscopy.
Colonoscopy is the most widely used and accepted CRC screening method in many countries, and is presented as the “gold standard” test due to being a sensitive and specific test. However, colonoscopy has some major barriers and limitations for implementation on a large scale in different populations.25 Stool-based screening modalities are patient-friendly, non-invasive, easy, quick, and can be performed at home by individuals. Historically, guaiac-based fecal occult blood test (gFOBT) was the most available and common test for CRC screening, but some of its disadvantages led to a decline in its popularity. Individuals must follow diet and drug restrictions and obtain three stool samples. Meanwhile, few evidence-based studies have revealed that the new stool based test and fecal immunochemical test (FIT) were significantly more sensitive and specific than other methods.26–29
Recently, Mt-sDNA testing has emerged as a reliable, feasible, non-invasive, and patient-friendly modality for CRC screening, and has the potential to enhance screening and compliance. Previous screening modalities, including fecal occult blood, colonoscopy, and sigmoidoscopy, have side-affecting detection biases (right- or left-side), but the detection accuracy of the Mt-sDNA panel is unaffected by the site of cancer and/or adenoma.30 The diagnostic accuracy of Mt-sDNA test is increased and strengthened by improving the included targeted DNA alterations and hyper-methylations.
The final established Mt-sDNA test panel, named “Cologuard” (Exact Sciences, Madison, WI, USA), was approved by the U.S. Food and Drug Administration (FDA) in August 2014 based on published results by Imperial et al. in 2014.31 Cologuard was approved as a sensitive, non-invasive, and acceptable screening test for eligible average-risk individuals. Cologuard was included in the American Cancer Society's (2018) CRC screening guidelines and as a recommended option for adults over the age of 50 by the U.S. Preventive Services Task Force (2016) and the National Comprehensive Cancer Network (2016).
Timeline of major milestones of Mt-sDNA test started by, Ahlquist et al., in 2000. They performed a feasibility study of an Mt-sDNA assay panel including point mutation detection of KRAS, p53, and Adenomatous polyposis coli (APC) genes, and a Microsatellite instability (MSI) assay of Bat-26. Their results showed that the Mt-sDNA panel had a sensitivity of 91% for CRC and 82% for AA, with a specificity of 93%. Meanwhile, they revealed that the Mt-sDNA assay could detect AA with a significantly higher sensitivity than the FIT (Hemoccult) test.30 Then in 2003, Tagore et al. designed and performed a large prospective study using the same stool DNA panel, supporting the feasibility of this test for CRC screening. However, the stool DNA panel was more sensitive to early stage lesions and AA in curable status compared to historical stool tests, including FOBT and/or FIT.25 The preliminary study report by Imperial et al. in 2004 revealed that although the diagnostic accuracy of Mt-sDNA is lower than that of colonoscopy for CRC, it had four times higher sensitivity and over two times higher sensitivity for invasive CRC and AA, respectively, compared to Hemoccult II.20 In 2012, Ahlquist et al., designed and incorporated the first clinical implementation of a novel Mt-sDNA panel assessed by a next-generation test. This novel prototype multi-marker panel included four gene methylation assays (NDRG4, BMP3, TFPI2, and vimentin), mutations in the K-ras gene, and ACTB (β-actin gene) as a reference gene, as well as FIT. The overall sensitivity for CRC detection was 85% and 63% for AA, with 90% specificity.32
The “DeeP-C” study started in middle of 2011 by Imperial et al., and their first results published in NEJM in 2014 (ClinicalTrials.gov identifier NCT01397747). They compared the commercial FIT test with the new Mt-sDNA panel including quantitative molecular assays of K-ras mutation, methylation aberrations in the NDRG4 and BMP3 promoter regions, and β-actin, as well as the FIT test, among 9989 asymptomatic persons and 65 CRCs and 757 advanced precancerous lesions (APLs) were detected. This Mt-sDNA panel had significantly higher sensitivity for the detection of any CRC (92.3% vs. 73.8%) and advanced precancerous lesions (42.4% vs. 23.8%) compared with FIT alone. Although FIT was still more specific for both CRC and AA in this study, we know that sensitivity is the most important characteristic of any screening modality.31
Another major study provided better results using this Mt-sDNA panel in early 2016. Redwood et al. performed a prospective cross-sectional surveillance study among 661 asymptomatic Alaska Native adults (435 in the screening group and 226 in the surveillance group). Interestingly, their study revealed a sensitivity of 100% for CRC detection, and 75% for large adenomas in the screening group, with a specificity of 93%. Their results once again emphasized that Mt-sDNA has a significantly higher sensitivity for CRC and AA detection than FIT alone.19 However, Bosh et al., revealed lower CRC and AA detection rates among 6600 asymptomatic individuals in the Netherlands (Amsterdam and Rotterdam), with a sensitivity of 85.7% for CRC and 47.8% for AA; this study was performed between 2009 and 2010, before Food and Drug Administration (FDA) approval of Cologuard.23
In line with these studies, a few studies have tried to adjust the gene panel and modify the techniques used. For example, in 2017, Park et al., attempted to validate aberrantly hypermethylated promoter genes (SFRP2, TFPI2, NDRGA, and BMP3) as stool-based DNA markers for CRC and AA detection in the Korean population.22 Using these four markers, the sensitivity was lower than that of the Redwood trial (94.3% for CRC and 72.2% for AA) with a specificity of 55%. Thus, using only four methylated markers is not reliable enough for CRC screening compared with the Mt-sDNA panel,22 Sun et al., included the syndecan-2 protein (SDC2) and secreted frizzled-related protein 2 (SFRP2) methylation assessment in addition to the Cologuard panel for CRC screening in a Chinese population.24 Next-generation sequencing analysis provided a larger and more comprehensive detection ability of most gene alterations and DNA methylation assays.
The superior sensitivity of Mt-sDNA tests is still under fire because of its high cost and poor availability, and remains a challenge in deciding whether this screening strategy should be used at the population level. However US Preventative Services Task Force has been endorsed Mt-sDNA test as first-line CRC screening test, with about 70% adherence rate.10 Mt-sDNA test is cost effective compared with no screening, and is enough efficient compared with other screening modalities.10 Meanwhile, the cost effectiveness of Mt-sDNA test has been approved by a few microsimulation models compared to no screening, however Medicare beneficiaries study revealed acceptable cost-effectiveness threshold.12,33
While sessile serrated polyp (SSP) is a specific type of hyperplastic colonic polyp, and paly as a precursor of about 30% of all colorectal cancers, it is worth mentioning that we had limited evidence about including and detection of SSPs.34 Based on earlier research studies, FIT test was obviously undetectable for sessile serrated adenomas/polyps, while MT-sDNA had larger diagnostic accuracy.19 Also the analytic results of Bosch et al, revealed that advanced serrated polyp(ASP) detection by MT-sDNA test was significantly superior to that by FIT, with higher sensitivity (P=0.02), as the sensitivity was 40.7%(95% CI: 22.4–61.2) for MT-sDNA compared with sensitivity of 7.4%(95% CI: 0.9–24.3) for FIT test including 3 threshold (50, 75, and 100).23 Also Redwood et al study showed the detection accuracy of 67% by MT-sDNA vs 11% by FIT (P=0.07) for sessile serrated polyps.19 Despite the superior diagnostic accuracy of MT-sDNA panel for SSPs, and regarding the importance of these types of polyps as precancerous lesion, we just find only 4 relevant studies, that reported data for serrated lesions, so due to lack of data, we didn’t included this option as our aims and in analysis.
The reluctance to switch from FIT to Mt-sDNA test for screening programs is the high cost of Mt-sDNA test resulting in lower overall cost-effectiveness and availability of this test compared to FIT or colonoscopy. Despite the approved advantages of Mt-sDNA test, more evidences need to focus more on the barriers to the widespread use of Mt-sDNA in screening programs.
However, ensuring the clinical diagnostic accuracy of the Mt-sDNA test for any CRC and precancerous lesions is still one of the main concerns of health care providers.
LimitationsAs the Mt-sDNA test has been recently approved by the FDA for routine use in CRC screening, we found limited studies about the accuracy of this test. Although molecular tests and techniques have been developed over the last two decades, there is a discrepancy in the results of sensitivity and specificity of this test, largely due to study sample size and included gene alterations and DNA methylation tests. Therefore, there was a significant study bias across studies regarding the diagnostic accuracy of Mt-sDNA testing for CRC and AA. However the studies included some kind of multitarget testing in stool, but with different markers. Also some recent trials included additional target tests and updated the “Cologuard” panel, for achieving better accuracy results. Then, the pooled performance is not referred only to Cologuard, but the combination of many studies.
ConclusionsThe superior sensitivity of Mt-sDNA tests is still under fire because of its high cost and poor availability, and it is still unclear how test results could be verified independently by other tests. However, our study results show that Mt-sDNA assays have a higher sensitivity for CRC and AA diagnosis. Mt-sDNA tests have major advantages over previously commonly used stool based tests (gFOBT and FIT) in average-risk populations. This evidence indicates the reliability and accuracy of Mt-sDNA tests in population-based CRC screening programs. However, larger population-based screening studies and clinical trials using the Mt-sDNA tests are needed in the future.
Authors’ contributionsRD, SD, MAJ: Performed substantial contributions to the conception and design of the work; the acquisition, analysis, and interpretation of data for the work; AND HMA, MHS: Performed drafting the work and revising it critically for important intellectual content; AND MHS: Revised and approved the version to be published; AND All the authors confirmed agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Availability of data and materialData are openly available in a public repository that issues datasets with the responsibility of the corresponding author.
Statement of ethicsCurrent study was reviewed and corroborated by the ethics committee of Tabriz University of Medical Sciences (ID: IR.TBZMED.REC.1395.1333).
Consent to participantsN/A.
Consent for publicationThe manuscript has been read and approved by all the authors, that the requirements for authorship as stated earlier in this document have been met, and that each author believes that the manuscript represents honest work, if that information is not provided in another form. All authors accepted the consent for publication.
FundingThe study was approved and supported as a research grant, by Ministry of Health and Medical Education, Deputy of Research and Technology (Grant number: 700/98, 2015.03.14 [1394/12/24]) from Iran Ministry of Health.
Conflicts of interestThe author reports no conflicts of interest in this work.
We would like to thanks to all research team and students.