IDENTIFICATION OF NEW GENES INVOLVED IN GERMLINE PREDISPOSITION TO EARLY-ONSET GASTRIC CANCER
1Gastroenterology Department, Hospital Clínic, Institut d'Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas (CIBEREHD), University of Barcelona. 2Bioinformatics Platform, Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas (CIBEREHD), Barcelona. 3Gastroenterology Department, Hospital Donostia-Instituto Biodonostia, Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas (CIBEREHD), Basque Country University (UPV/EHU), San Sebastián.
Introduction: Gastric cancer (GC) is the fifth most common cancer worldwide, showing a high mortality rate. Up to 10% of GC cases show familial aggregation and a genetic cause is present in 5% of all cases. The main associated syndrome is hereditary diffuse GC, which is mainly caused by CDH1 mutations. However, the genetic cause for a high number of families with GC aggregation is unclear, especially in early-onset patients. The aim was to identify new candidate genes involved in germline predisposition to GC.
Methods: Whole-exome sequencing (WES) of germline samples was performed in 20 patients with GC before the age of 50 and without CDH1 mutations. Nine tumor samples were also available and WES was performed. Sequencing data of germline samples were filtered by a pipeline to select variants with plausible pathogenicity, rare frequency in the general population and involved in cancer. After that, a manual filtering was performed to prioritize those genes involved in germline predisposition to any type of cancer and genes according to gene function. These genetic variants were prevalidated with Integrative Genomics Viewer (IGV). In addition, data of tumor samples was used to analyze its tumor profile using SigProfilerWeb. Subsequently, a selection step was performed according to gene function and tumor profile. Selected variants were validated by Sanger sequencing.
Results: After manual filtering, 274 genetic variants located on 205 genes were prioritized. Using IGV, 86 genetic variants were discarded. Among the remaining, 60 variants (54 genes) were selected and Sanger sequencing was performed. Finally, 58 final genetic variants located on 52 candidate genes were validated.
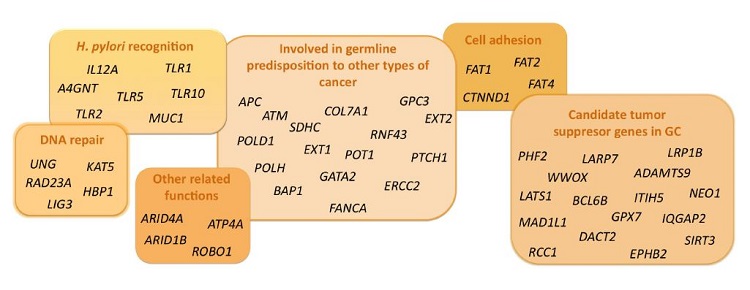
Conclusions: Among 52 candidate genes, APC, SDHC, FAT4, CTNND1 and TLR2 seem to be the most promising ones. This project will be helpful to increase the knowledge of predisposition to early-onset GC, although replication in a different cohort and functional studies will be needed.








