It has been reported that ADAM33 (a disintegrin and metalloproteinase domain 33) polymorphisms might be associated with susceptibility to allergic rhinitis (AR).
ObjectiveOwing to mixed and inconclusive results, we conducted a meta-analysis to systematically summarise and clarify the association between ADAM33 S2, V4, T1, T2 and T+1 polymorphisms and AR risk.
Methods/resultsA systematic search of studies on the association of ADAM33 polymorphisms with susceptibility to AR was conducted in Pubmed and Embase. A total of five case–control studies with 1251 patients and 1634 controls were included. Meta-analysis indicated an association between the ADAM33 S2 and AR in allele comparison (G/C:OR=1.40, 95% CI 1.08–1.82, P=0.012), heterozygote comparison (CG/CC:OR=1.24, 95% CI 1.04–1.48, P=0.015), and dominant comparison (CG+GG/CC:OR=1.39, 95% CI 1.05–1.85, P=0.023). The meta-analysis also revealed an association between the ADAM33 V4 and AR in allele comparison (G/C:OR=1.67, 95% CI 1.01–2.75, P=0.044). However, no association was found between AR and the ADAM33 T1, T2 and T+1 polymorphisms in any gene model comparison.
ConclusionsThis meta-analysis demonstrates that the ADAM33 S2 and V4 polymorphisms confer susceptibility to AR. However, these results should be interpreted with caution due to limited sample and heterogeneity. Large-scale and well-designed studies are needed to validate our findings.
Allergic rhinitis (AR) is defined as a nasal disorder induced by immunoglobulin E (IgE)-mediated immune response and characterised by nasal symptoms including rhinorrhoea, sneezing, nasal obstruction, and itching.1 It is one of the most common atopic diseases throughout the world. The development of AR entails a complex interaction between genetic predisposition and environmental exposure to different factors, of which the most important is the implicated allergen. Despite environmental exposures playing important roles in the development of atopy, there is a strong inherited susceptibility in the aetiology of AR. Variants in several genes have been identified as biologically plausible candidates for effects on AR, such as ADAM33. ADAM33 (a disintegrin and metalloproteinase domain 33), a gene located on chromosome 20p13 and a member of the ADAM family, is a complex molecule whose expression is largely restricted to mesenchymal cells, including fibroblast and smooth muscle, and codes for a protein important for cell fusion, cell adhesion, cell signalling and proteolysis.2 It has been illustrated that the alteration of this protein function might be involved in airway remodelling as ADAM33 is expressed in the smooth muscle, myofibroblasts, and fibroblasts of asthmatic airways.3 As a result, the variation in ADAM33 polymorphisms may induce the development of some diseases, including asthma and AR.4,5
ADAM33 is highly polymorphic and several of its polymorphisms have been studied in AR. However, the results in these studies were mixed and inconclusive.6–10 The numbers of studies on the ADAM33 polymorphisms were various and some polymorphisms were not possible for meta-analysis due to the limited data. We selected five ADAM33 polymorphisms for meta-analysis based on the number of studies published. The aim of the present study was to use meta-analysis to determine whether the ADAM33 S2 (rs528557 C>G Exon 19), V4 (rs2787094 C>G 3′ UTR), T1 (rs2280091 A>G Exon 20), T2 (rs2280090 G>A Exon 20) and T+1 (rs2280089 G>A Intron 20) polymorphisms confer susceptibility to AR.
Methods and materialsLiterature search strategyTwo independent investigators carried out a systematic search in PubMed and Embase databases, with the last search update on December 1, 2014. The following terms were used: “polymorphism OR SNP OR variant”, “allergic rhinitis OR rhinallergosis” and “ADAM33 OR metallopeptidase domain 33”, without any limitation applied. The reference lists of retrieved studies and recent reviews were also manually searched for further relevant studies.
Inclusion and exclusion criteriaStudies in this meta-analysis must meet the following inclusion criteria: (1) evaluation of the association between ADAM33 polymorphisms and AR; (2) case–control study; (3) Frequencies of ADAM33 polymorphisms in the controls must be in the Hardy-Weinberg equilibrium (HWE); (4) detailed genotype data could be acquired to calculate the odds ratios (ORs) and 95% confidence intervals (CI); exclusion criteria: (1) duplication of previous publications; (2) comment, review and editorial; (3) family-based studies of pedigrees; (4) study with no detailed genotype data. When there were multiple publications from the same population, only the largest study was included. The study selection was achieved by two investigators independently, according to the inclusion and exclusion criteria by screening the title, abstract and full-text. Any dispute was solved by discussion.
Data extractionThe data of the eligible studies were extracted in duplicate by two investigators independently. The following contents were collected: the first author, year of publication, country, ethnicity, genotyping methods, total numbers of cases and controls, genotype frequencies in cases and controls for ADAM33 S2, V4, T1, T2 and T+1 genotypes, respectively. Two authors checked the extracted data and reached a consensus on all the data. If dissent existed, they would recheck the original data of the included studies and have a discussion to reach consensus. If the dissent still existed, the third investigator would be involved to adjudicate the disagreements.
Statistics analysisWe conducted our meta-analysis according to the PRISMA checklists and followed the guidelines.11 HWE was evaluated for each study by Chi-square test in control groups, and P<0.05 was considered a significant departure from HWE. Variant genotype frequencies were compared between cases and controls. The OR and 95% CI were calculated to evaluate the strength of the association between ADAM33 SNPs and susceptibility to AR. Between-study heterogeneity was estimated using the χ2-based Q statistic and I2 test. When I2>50% and P<0.1, heterogeneity was considered statistically significant, and random effects model was used to analyse the data subsequently. On the contrary, the fixed effects model was chosen. Sensitivity analysis was also performed to evaluate the effect of each study on the combined ORs by omitting each study in each turn. Potential publication bias was checked by Harbord's test. An asymmetric plot and P<0.05 was considered a significant publication bias. All statistical analyses were performed with Stata 13.1 software (StataCorp, College Station, TX, USA).
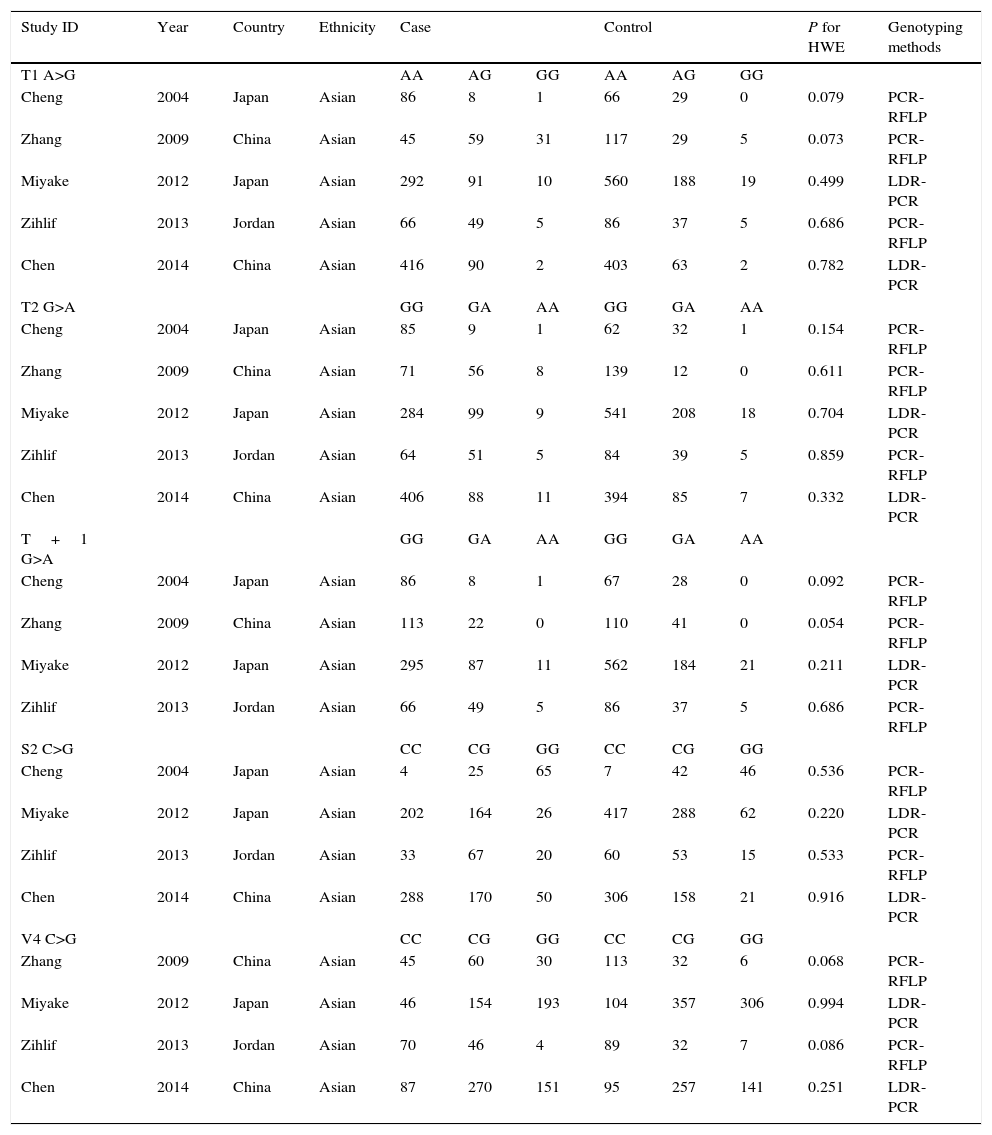
ResultsCharacteristics of studiesFollowing the search strategy, 21 potentially relevant studies were retrieved. According to the inclusion criteria, five studies6–10 with full-text were included into this meta-analysis and 16 studies were excluded. These studies included 1251 AR cases and 1634 healthy controls. Different genotyping methods were utilised including polymerase chain reaction–restriction fragment length polymorphism (PCR–RFLP) and TaqMan, polymerase chain reaction–ligation detection reaction (LDR–PCR). The genotyping distribution was in agreement with HWE in all studies. The characteristics of each study are shown in Table 1.
Characteristics of studies included in the meta-analysis.
| Study ID | Year | Country | Ethnicity | Case | Control | P for HWE | Genotyping methods | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| T1 A>G | AA | AG | GG | AA | AG | GG | |||||
| Cheng | 2004 | Japan | Asian | 86 | 8 | 1 | 66 | 29 | 0 | 0.079 | PCR-RFLP |
| Zhang | 2009 | China | Asian | 45 | 59 | 31 | 117 | 29 | 5 | 0.073 | PCR-RFLP |
| Miyake | 2012 | Japan | Asian | 292 | 91 | 10 | 560 | 188 | 19 | 0.499 | LDR-PCR |
| Zihlif | 2013 | Jordan | Asian | 66 | 49 | 5 | 86 | 37 | 5 | 0.686 | PCR-RFLP |
| Chen | 2014 | China | Asian | 416 | 90 | 2 | 403 | 63 | 2 | 0.782 | LDR-PCR |
| T2 G>A | GG | GA | AA | GG | GA | AA | |||||
| Cheng | 2004 | Japan | Asian | 85 | 9 | 1 | 62 | 32 | 1 | 0.154 | PCR-RFLP |
| Zhang | 2009 | China | Asian | 71 | 56 | 8 | 139 | 12 | 0 | 0.611 | PCR-RFLP |
| Miyake | 2012 | Japan | Asian | 284 | 99 | 9 | 541 | 208 | 18 | 0.704 | LDR-PCR |
| Zihlif | 2013 | Jordan | Asian | 64 | 51 | 5 | 84 | 39 | 5 | 0.859 | PCR-RFLP |
| Chen | 2014 | China | Asian | 406 | 88 | 11 | 394 | 85 | 7 | 0.332 | LDR-PCR |
| T+1 G>A | GG | GA | AA | GG | GA | AA | |||||
| Cheng | 2004 | Japan | Asian | 86 | 8 | 1 | 67 | 28 | 0 | 0.092 | PCR-RFLP |
| Zhang | 2009 | China | Asian | 113 | 22 | 0 | 110 | 41 | 0 | 0.054 | PCR-RFLP |
| Miyake | 2012 | Japan | Asian | 295 | 87 | 11 | 562 | 184 | 21 | 0.211 | LDR-PCR |
| Zihlif | 2013 | Jordan | Asian | 66 | 49 | 5 | 86 | 37 | 5 | 0.686 | PCR-RFLP |
| S2 C>G | CC | CG | GG | CC | CG | GG | |||||
| Cheng | 2004 | Japan | Asian | 4 | 25 | 65 | 7 | 42 | 46 | 0.536 | PCR-RFLP |
| Miyake | 2012 | Japan | Asian | 202 | 164 | 26 | 417 | 288 | 62 | 0.220 | LDR-PCR |
| Zihlif | 2013 | Jordan | Asian | 33 | 67 | 20 | 60 | 53 | 15 | 0.533 | PCR-RFLP |
| Chen | 2014 | China | Asian | 288 | 170 | 50 | 306 | 158 | 21 | 0.916 | LDR-PCR |
| V4 C>G | CC | CG | GG | CC | CG | GG | |||||
| Zhang | 2009 | China | Asian | 45 | 60 | 30 | 113 | 32 | 6 | 0.068 | PCR-RFLP |
| Miyake | 2012 | Japan | Asian | 46 | 154 | 193 | 104 | 357 | 306 | 0.994 | LDR-PCR |
| Zihlif | 2013 | Jordan | Asian | 70 | 46 | 4 | 89 | 32 | 7 | 0.086 | PCR-RFLP |
| Chen | 2014 | China | Asian | 87 | 270 | 151 | 95 | 257 | 141 | 0.251 | LDR-PCR |
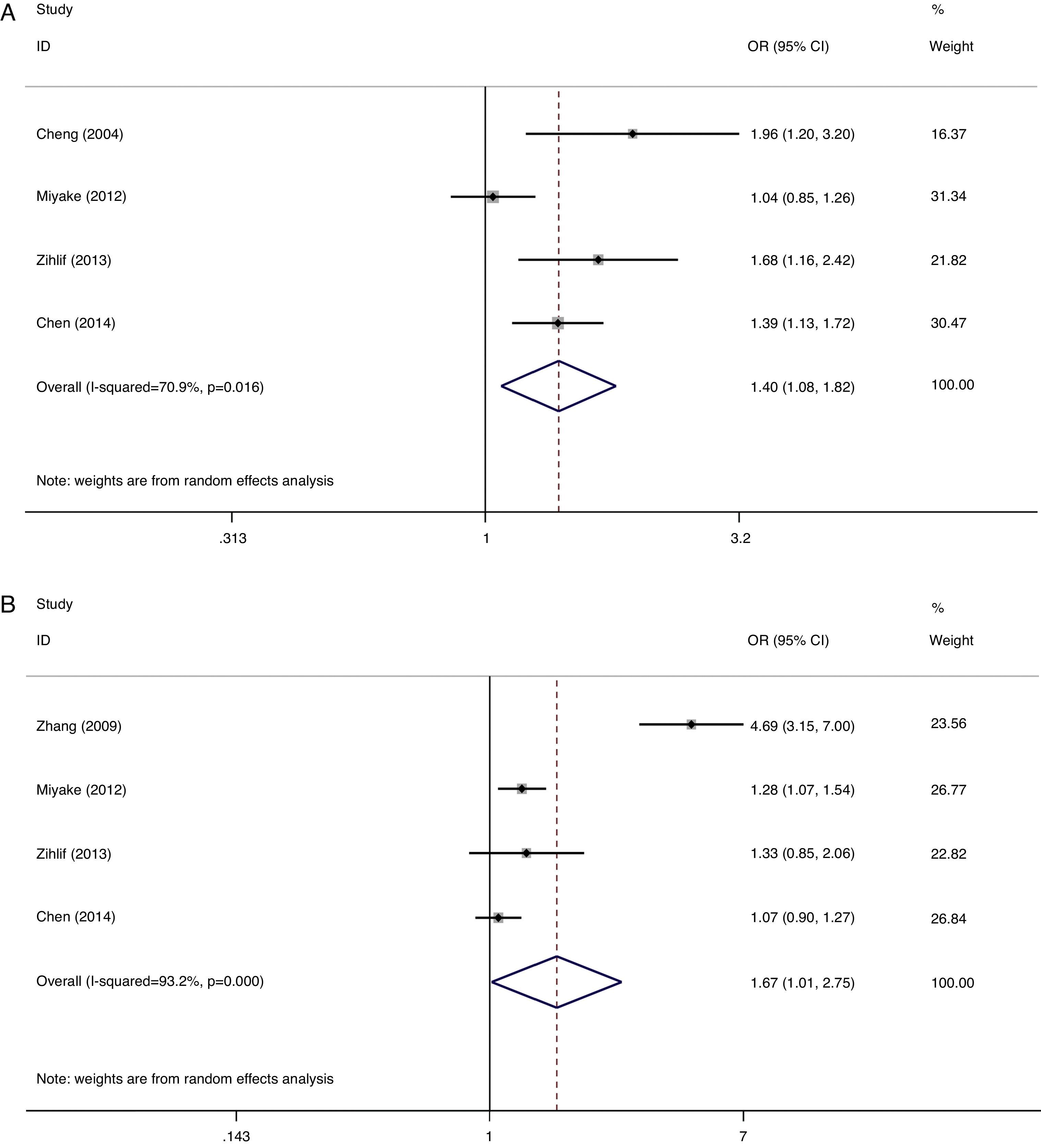
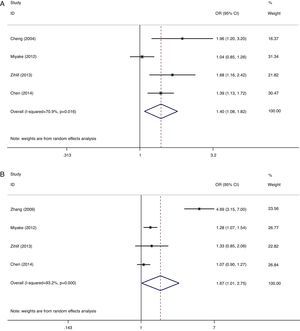
A summary of the association results between ADAM33 S2 and V4 polymorphisms and AR is provided in Table 2. OR is as effect estimate, OR=the ratio of the number of events and not occur in case/the ratio of the number of events and not occur in control. The statistical significant level was determined by Z-test with P value less than 0.05. Due to the limited studies and heterogeneity, these results should be treated with caution. Random-effects model was used in all models due to presence of heterogeneity except for ADAM33 S2 heterozygote model. A significant increased risk of AR was observed in ADAM33 S2 and V4 allele comparison (G vs. C:OR=1.40, 95% CI 1.08–1.82, P=0.012 and OR=1.67, 95% CI 1.01–2.75, P=0.044, Fig. 1); ADAM33 S2 heterozygote comparison (CG vs. CC:OR=1.24, 95% CI 1.04–1.48, P=0.015) and dominant comparison (CG+GG vs. CC:OR=1.39, 95% CI 1.05–1.85, P=0.023). No significant association was found in other models comparison. However, a trend of increased susceptibility still existed.
Analysis of the association between the ADAM33 S2 and V4 polymorphisms and AR.
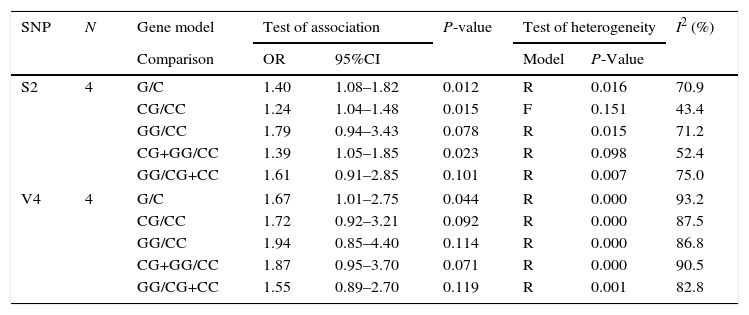
| SNP | N | Gene model | Test of association | P-value | Test of heterogeneity | I2 (%) | ||
|---|---|---|---|---|---|---|---|---|
| Comparison | OR | 95%CI | Model | P-Value | ||||
| S2 | 4 | G/C | 1.40 | 1.08–1.82 | 0.012 | R | 0.016 | 70.9 |
| CG/CC | 1.24 | 1.04–1.48 | 0.015 | F | 0.151 | 43.4 | ||
| GG/CC | 1.79 | 0.94–3.43 | 0.078 | R | 0.015 | 71.2 | ||
| CG+GG/CC | 1.39 | 1.05–1.85 | 0.023 | R | 0.098 | 52.4 | ||
| GG/CG+CC | 1.61 | 0.91–2.85 | 0.101 | R | 0.007 | 75.0 | ||
| V4 | 4 | G/C | 1.67 | 1.01–2.75 | 0.044 | R | 0.000 | 93.2 |
| CG/CC | 1.72 | 0.92–3.21 | 0.092 | R | 0.000 | 87.5 | ||
| GG/CC | 1.94 | 0.85–4.40 | 0.114 | R | 0.000 | 86.8 | ||
| CG+GG/CC | 1.87 | 0.95–3.70 | 0.071 | R | 0.000 | 90.5 | ||
| GG/CG+CC | 1.55 | 0.89–2.70 | 0.119 | R | 0.001 | 82.8 | ||
N=number of studies, F=fixed effects model, R=random effects model.
A summary of the association results between ADAM33 T+1, T1 and T2 polymorphisms and AR is shown in Table 3. No association was found between AR and ADAM33 T+1, T1 and T2 polymorphisms using the five models comparison.
Analysis of the association between the ADAM33 T+1, T1 and T2 polymorphisms and AR.
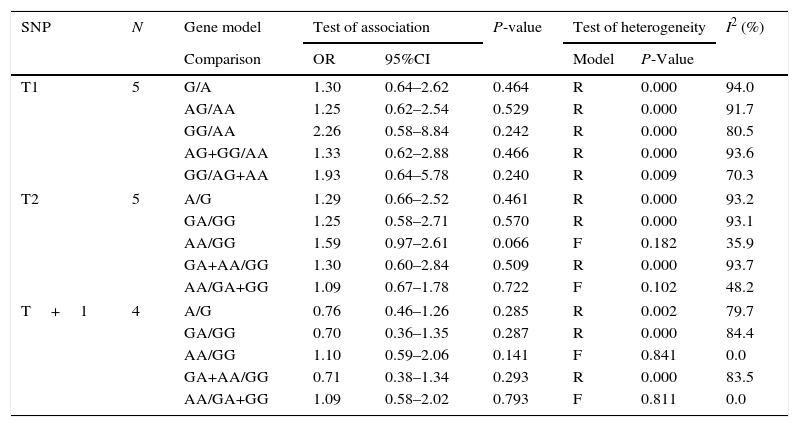
| SNP | N | Gene model | Test of association | P-value | Test of heterogeneity | I2 (%) | ||
|---|---|---|---|---|---|---|---|---|
| Comparison | OR | 95%CI | Model | P-Value | ||||
| T1 | 5 | G/A | 1.30 | 0.64–2.62 | 0.464 | R | 0.000 | 94.0 |
| AG/AA | 1.25 | 0.62–2.54 | 0.529 | R | 0.000 | 91.7 | ||
| GG/AA | 2.26 | 0.58–8.84 | 0.242 | R | 0.000 | 80.5 | ||
| AG+GG/AA | 1.33 | 0.62–2.88 | 0.466 | R | 0.000 | 93.6 | ||
| GG/AG+AA | 1.93 | 0.64–5.78 | 0.240 | R | 0.009 | 70.3 | ||
| T2 | 5 | A/G | 1.29 | 0.66–2.52 | 0.461 | R | 0.000 | 93.2 |
| GA/GG | 1.25 | 0.58–2.71 | 0.570 | R | 0.000 | 93.1 | ||
| AA/GG | 1.59 | 0.97–2.61 | 0.066 | F | 0.182 | 35.9 | ||
| GA+AA/GG | 1.30 | 0.60–2.84 | 0.509 | R | 0.000 | 93.7 | ||
| AA/GA+GG | 1.09 | 0.67–1.78 | 0.722 | F | 0.102 | 48.2 | ||
| T+1 | 4 | A/G | 0.76 | 0.46–1.26 | 0.285 | R | 0.002 | 79.7 |
| GA/GG | 0.70 | 0.36–1.35 | 0.287 | R | 0.000 | 84.4 | ||
| AA/GG | 1.10 | 0.59–2.06 | 0.141 | F | 0.841 | 0.0 | ||
| GA+AA/GG | 0.71 | 0.38–1.34 | 0.293 | R | 0.000 | 83.5 | ||
| AA/GA+GG | 1.09 | 0.58–2.02 | 0.793 | F | 0.811 | 0.0 | ||
N=number of studies, F=fixed effects model, R=random effects model.
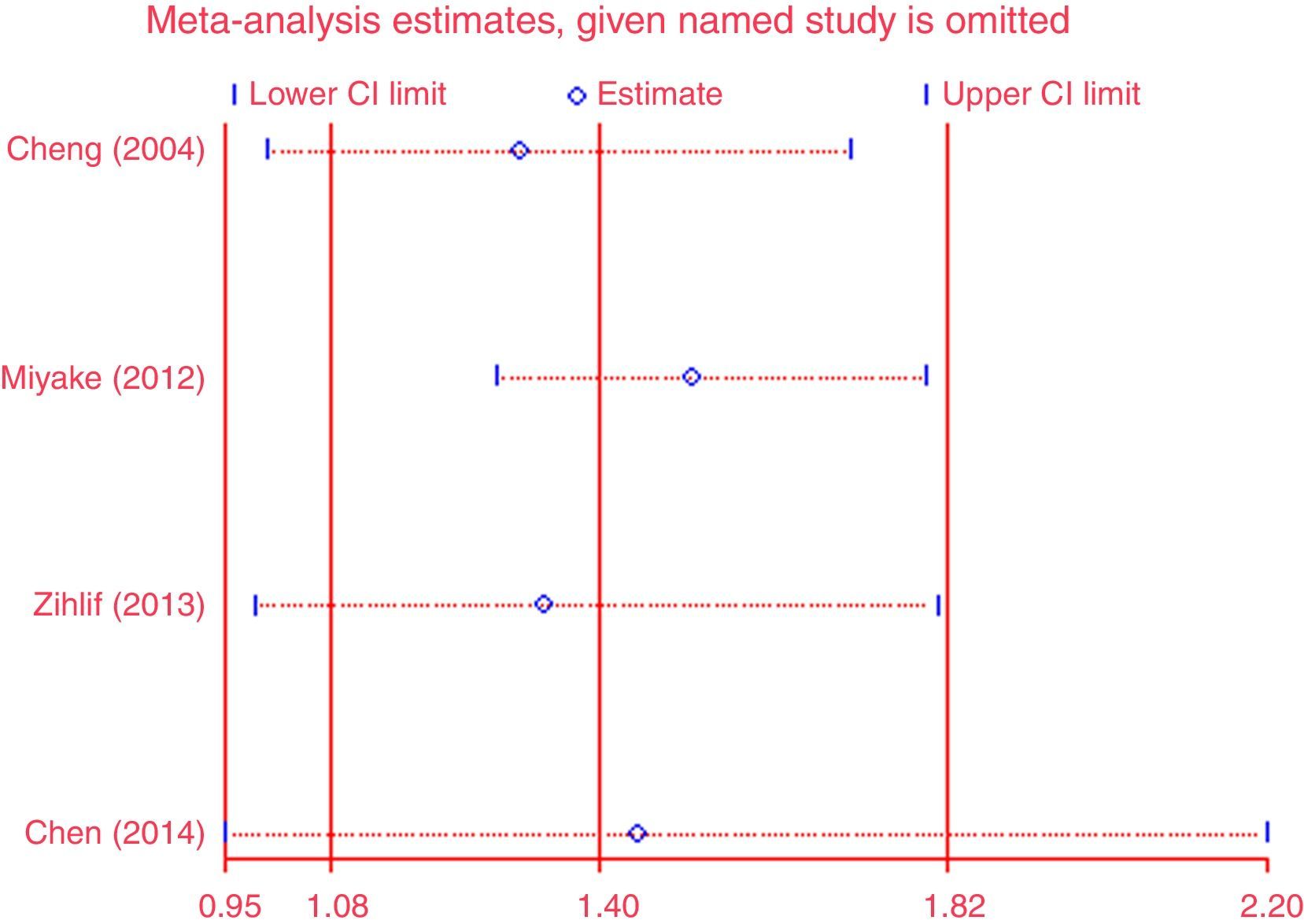
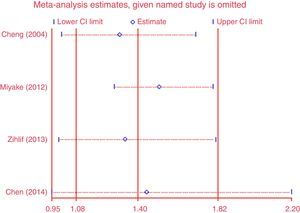
Except for CG vs. CC (I2=43.4%, P=0.151) for S2, AA vs. GG (I2=35.9%, P=0.182) and AA vs. GA+GG (I2=48.2%, P=0.102) for T2, AA vs. GG (I2=0%, P=0.841) and AA vs. GA+GG (I2=0%, P=0.811) for T+1, there was significant heterogeneity for all the remaining comparisons. Sensitivity analysis was performed to examine the influence set by the individual study on the pooled ORs for ADAM33 S2 (Fig. 2), V4, T1, T2 and T+1 by deleting each study once in every genetic model. As is shown Fig. 2, after deleting each study once, the point estimates of the combined effect was within 95% confidence interval of the total combined effect. So the magnitude of effect in overall analysis was not excessively influenced. Publication bias of the included studies was assessed by Harbord's test. Symmetrical plots were obtained in all the genetic models and there was no apparent evidence of publication bias. As is shown in Fig. 3 for allele comparison of ADAM33 S2 (G vs. C), the plot was approximately symmetrical and 95% CI for intercept included 0, in addition, the result of the quantitative analysis was P=0.189>0.05; these results indicated that there was no publication bias.
AR may be a multifactorial disease. Some studies have shown that a combination of genetic susceptibility and environmental factors can ultimately induce the occurrence of this disease.12,13 More than 100 SNPs including ADAM33 polymorphisms have been reported to be associated with the risk of AR.14 ADAM33 (a disintegrin and metalloproteinase domain 33), located on chromosome 20p13, was the first gene identified in asthma by positional cloning.5 ADAM33 is a member of the multifunctional ADAM family of genes that code for zinc-dependent metalloproteinases. The ADAM33 protein has several domains, including metalloprotease-like, disintegrin-like, cysteine-rich, epidermal growth factor-like, transmembrane, and cytoplasmic domains.15 This protein is important for cell fusion, cell adhesion, cell signalling, and proteolysis and has been suggested to play an important role in the activation of growth factors and Th2 cytokines.16 ADAM33 mRNA expression was found to be elevated in the lungs of a murine model of allergen-induced chronic airway inflammation.17 Angiogenesis is a fundamental process that supports many aspects of tissue inflammation and remodelling. Therefore, ADAM33 polymorphism may play a vital role in airway remodelling in AR and asthma.
Previous studies have investigated the association between ADAM33 polymorphisms and the risk of AR. Cheng et al.10 found that ADAM33 S2, T1, T2 and T+1 polymorphisms were significantly associated with AR susceptibility but not V4 in Japanese cedar pollinosis (a kind of seasonal allergic rhinitis). Miyake et al.6 reported that ADAM33 S2, V4, T1, T2 and T+1 polymorphisms conferred to AR susceptibility in Japanese adult women. A study conducted by Zhang et al.7 indicated that ADAM33 V4, T1, and T2 polymorphisms may be the causal variants but not T+1 in AR disease in Chinese Han population with asthma. Another research by Chen et al.8 revealed that ADAM33 S2 polymorphism was associated with persistent allergic rhinitis but not V4, T1 and T2 in Chinese population. However, Zihlif et al.9 reported that no significant difference in the allelic frequencies of ADAM33 S2, V4, T1, T2 and T+1 between AR patients and the control volunteers were found among Jordanians.
In the current study, we pooled and analysed these data including 1251 cases and 1634 controls from Asia. The results showed that a significant increased risk of AR was observed in ADAM33 S2 and V4 allele comparison, in ADAM33 S2 heterozygote comparison and dominant comparison. However, no association was found between AR and ADAM33 T+1, T1 and T2 polymorphisms in any gene model. Strangely, a recent similar meta-analysis showed different results from ours.18 Some reasons can contribute to the difference. First, two publications cited by the meta-analysis were from the same population and area, however we ruled out one of these articles. Second, when there were duplicated articles, we integrated the latest study. There were some different data between these repeated studies.
The association between ADAM33 polymorphisms and asthma has been reported previously. Recent several meta-analysis showed that polymorphisms in the ADAM33 gene are risk factors for asthma in the Asian (T1, V4, F+1, T2, and T+1)19,20 and European (S2)21 population. However, these results were not fully consistent with our study. Therefore, although asthma and AR have been considered to be atopic diseases, the two kinds of disease may have different aetiologies and genetic basis.22
Except for CG vs. CC for S2, AA vs. GG and AA vs. GA+GG for T2, AA vs. GG and AA vs. GA+GG for T+1, there was significant heterogeneity for all the remaining comparisons. Although heterogeneity existed, the results from our sensitivity analyses were consistent and robust.
In spite of the considerable efforts to explore the possible relationship between ADAM33 SNPs and AR risk, some limitations should be considered. First, heterogeneity and confounding factors may have distorted the analysis, and potential publication bias may have further affected the findings. Second, the numbers of subjects and studies included in the meta-analysis was small, and may not have been sufficient to reveal the associations between ADAM33 polymorphisms and AR. Third, our ethnic-specific analysis only included data on Asian patients, thus, our results are applicable to only this ethnicity, and further studies are required in other ethnic populations. Fourth, data were not stratified by sex, disease severity, or clinical or environmental variables because of insufficient data.
In conclusion, this meta-analysis demonstrated that the ADAM33 S2 and V4 polymorphism confers susceptibility to AR in Asians, while no association was found between the ADAM33 T1, T2, and T+1 polymorphisms and AR susceptibility. However, there was insufficient data to fully confirm the association of AR and the ADAM33 S2 and V4 polymorphism, and these results should be interpreted with caution. Well-designed studies with larger sample sizes and more ethnic groups are required to validate the risk identified in the current meta-analysis.
Ethical disclosuresProtection of human subjects and animals in researchThe authors declare that no experiments were performed on humans or animals for this investigation.
Confidentiality of dataThe authors declare that no patient data appears in this article.
Right to privacy and informed consentThe authors declare that no patient data appears in this article.
Conflict of interestWe state explicitly that we have no potential conflict of interest.




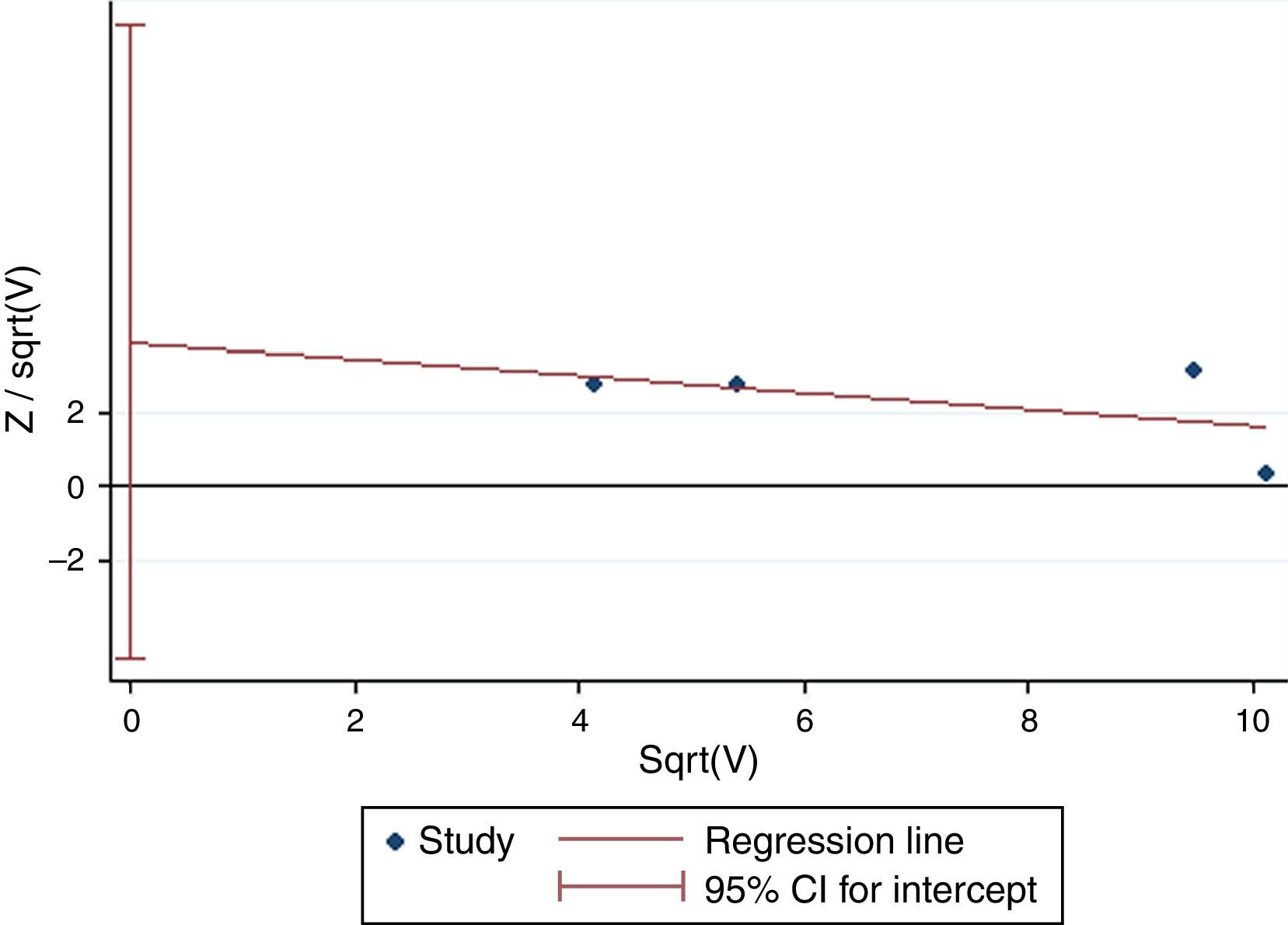
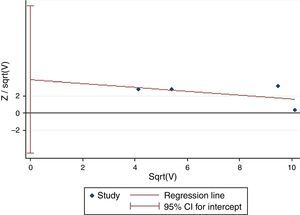 0.05).' title='Harbord's test of allele comparison of ADAM33 S2 (G vs. C). The plot was approximately symmetrical and the result of quantitative analysis was P=0.189 (P>0.05).'/>
0.05).' title='Harbord's test of allele comparison of ADAM33 S2 (G vs. C). The plot was approximately symmetrical and the result of quantitative analysis was P=0.189 (P>0.05).'/>