Evidence suggests that possible imbalances in intestinal microbiota composition may be implicated in the occurrence of allergic diseases. Although several studies published until 2006 indicated a correlation between microbiota composition and allergic symptoms, it has not been possible to distinguish protective microorganisms from those associated with increased risk of allergic diseases. Therefore, the objective of this study was to review the studies published since 2007 that address the intestinal microbiota in allergic diseases. Twenty-one studies were identified after excluding those that performed a clinical intervention before stool collection. In the early microbiota of children who later developed allergies, lower bacterial diversity was observed, with a predominance of Firmicutes; a higher count of Bacteroidaceae; a higher prevalence of the anaerobic bacteria Bacteroides fragilis, Escherichia coli, Clostridium difficile, Bifidobacterium catenulatum, Bifidobacterium bifidum, and Bifidobacterium longum; and a lower prevalence of Bifidobacterium adolescentis, B. bifidum, and Lactobacillus. In the microbiota of allergic children whose intestinal microbiota was assessed at the onset of allergic symptoms, there was a higher count of Bacteroides; a lower count of Akkermansia muciniphila, Faecalibacterium prausnitzii, and Clostridium; a higher prevalence of B. adolescentis; a lower prevalence of B. catenulatum and Staphylococcus aureus; and a lower bacterial diversity.
The intestinal microbiota is a complex ecosystem of great importance for health and is composed of both aerobic and anaerobic bacteria.1 The gastrointestinal tract may be considered sterile during intrauterine life and is colonised after birth. Bacterial colonisation depends on several factors, particularly the type of childbirth, vaginal or caesarean,2 and the method by which the child is fed during the first months of life.
Evidence suggests that possible imbalances in the composition of the intestinal microbiota and its relationship with the host may be implicated in the occurrence of allergic diseases.3 This hypothesis has led to a number of investigations to elucidate the relationship between the composition and function of the intestinal microbiota and the onset of allergic diseases.4
Penders et al.5 reviewed 18 studies on the association between the intestinal microbiota and allergic diseases. This study evaluated papers published between 1999 and 2006, primarily observational studies that compared healthy and allergic individuals. The profile of the intestinal microbiota of individuals with eczema/atopic dermatitis, food allergy, wheezing, allergic rhinitis, asthma and/or sensitisation was evaluated. The composition of the faecal microbiota was analysed using different laboratory techniques, from traditional bacterial culture to novel molecular biology techniques. Most studies indicated an association between microbiota composition and atopic symptoms and/or sensitisation. However, it has not yet been possible to distinguish protective microorganisms from those associated with increased risk of allergic diseases. The differences in the results were due to differences in the types of studies and laboratory techniques used to assess the composition of the intestinal microbiota.
In this context, this systematic study aimed to review the literature published between 2007 and 2013 that addresses the intestinal microbiota in allergic diseases.
MethodIn this review, all studies published between 2007 and 2013 that compared the intestinal microbiota of allergic patients (atopic dermatitis, food allergy, wheezing, allergic rhinitis, asthma and/or atopic sensitisation) with those of healthy individuals were evaluated. Clinical trials with interventions were not included except when the composition of the intestinal microbiota on admission was evaluated before the intervention.
The studies were evaluated for sample size, place of study, age range, study design, criteria for the diagnosis of allergic diseases, and composition of the faecal microbiota.
To identify studies addressing this topic, an extensive literature search was conducted using the following databases: 1. PubMed, a database developed by the National Center for Biotechnology Information (NCBI) at the National Library of Medicine (NLM), which integrates information from NCBI databases with the PubMed database and includes MEDLINE, nucleotide sequences, protein sequences, macromolecular structures, and whole genome sequences; 2. IBECS, a database that uses the LILACS methodology and allows the selection of scientific studies on health sciences published in Spain; and 3. LILACS, a database produced by the institutions that form the Latin American and Caribbean System on Health Sciences Information. LILACS has catalogued technical–scientific data in the health sciences produced by Latin American and Caribbean authors published since 1982.6
For the search, the following keywords and limits were used: (“Intestines” [Mesh] OR intestin* OR gut OR gastrointestin* OR enteric) AND (flora OR microbiolog* OR microflora OR bacteria OR bacterial OR colonisation OR colonisation OR microbes OR microbial OR microbiota) AND (hypersensitivity OR atopic dermatitis OR allergic OR allergy OR atopic OR atopy OR eczema OR rhinitis OR asthma), as previously reported.5
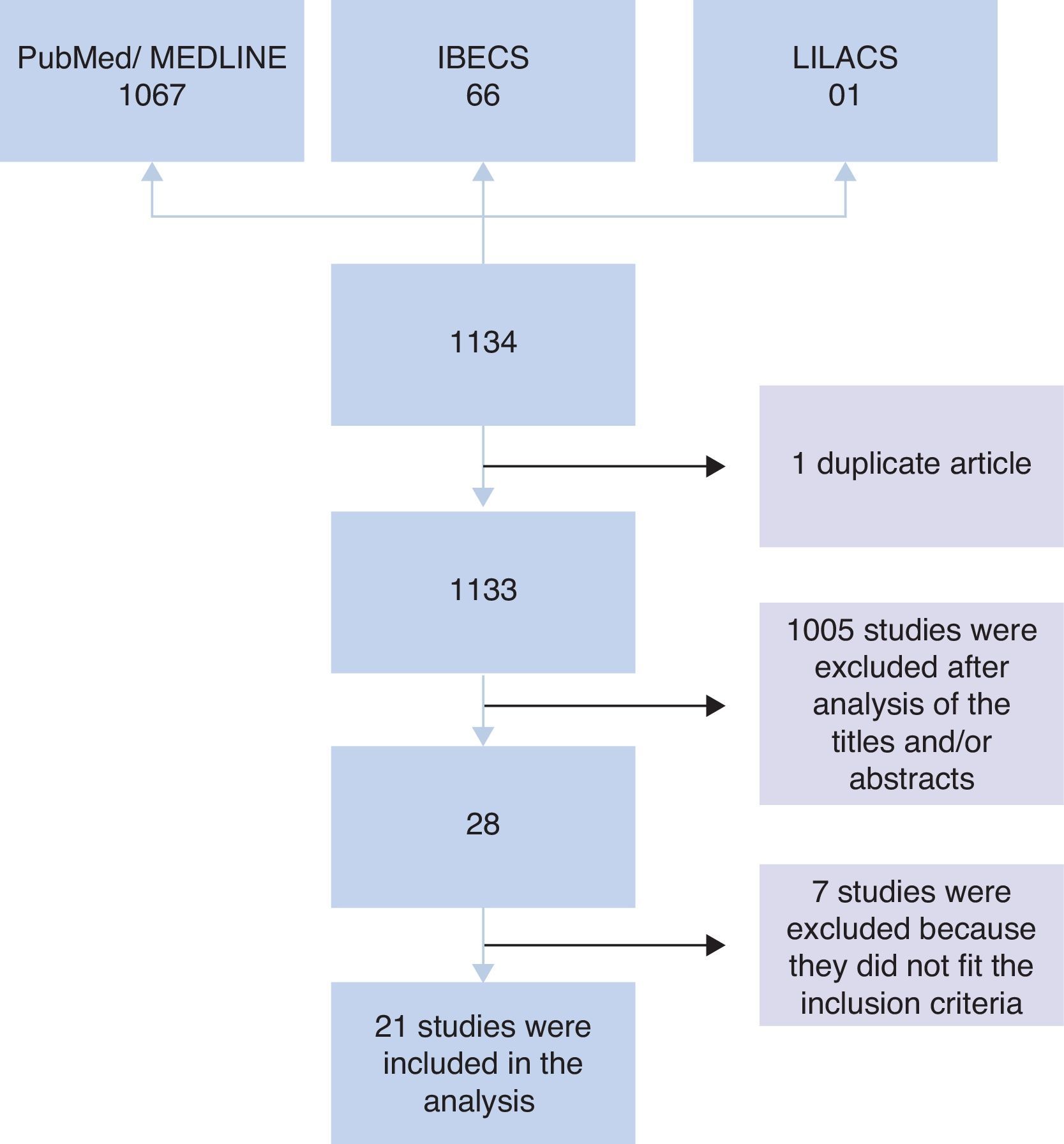
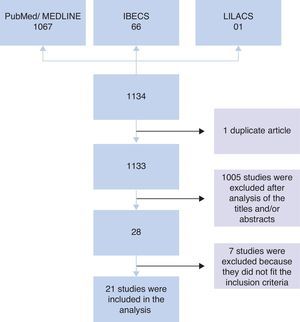
ResultsInitially, 1134 studies were found in PubMed, IBECS, and LILACS databases. Of these, one study was found in duplicate that was catalogued in both IBECS and LILACS. Therefore, an analysis of titles and/or abstracts of 1133 studies was performed considering the inclusion criteria. After this analysis, 1105 studies were excluded, and 28 studies remained. Of the 28 studies, seven were excluded after analysis of the full article because they did not meet the inclusion criteria; i.e., four were intervention studies that did not provide information on the microbiota at admission,7–10 two involved children born to mothers who received probiotic intervention during gestation,11,12 and one did not compare the microbiota of allergic and non-allergic patients.13 Therefore, 21 articles were included in the analysis (Fig. 1).
In Tables 1 and 2, the abstracts are presented according to the period in which stool collections were performed in relation to the assessment of allergic diseases. Table 1 lists 16 studies wherein stool collection was performed before assessment of the presence of allergic diseases; and Table 2 lists five studies wherein stool collection was performed during the assessment.
Studies on the association between the intestinal microbiota and the incidence of allergic diseases wherein stool collection was performed before outcome assessment.
| Author, year location | Design | Definition of allergy | Analysis of faecal microbiota | Results of allergic compared with non-allergic subjects | |
|---|---|---|---|---|---|
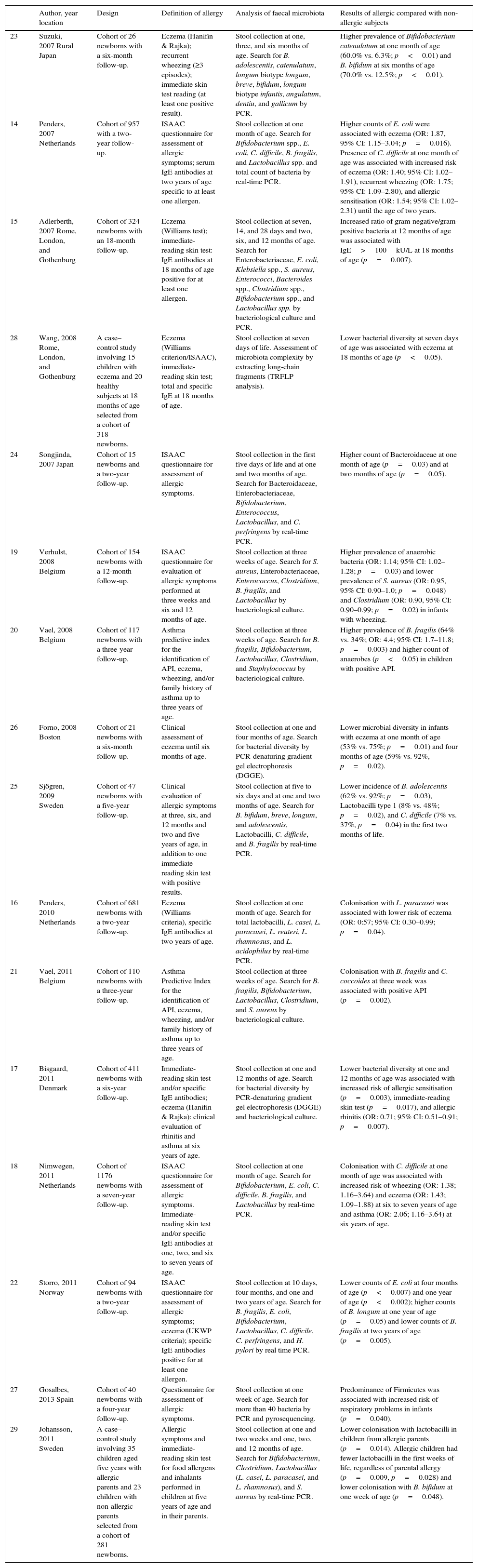
| 23 | Suzuki, 2007 Rural Japan | Cohort of 26 newborns with a six-month follow-up. | Eczema (Hanifin & Rajka); recurrent wheezing (≥3 episodes); immediate skin test reading (at least one positive result). | Stool collection at one, three, and six months of age. Search for B. adolescentis, catenulatum, longum biotype longum, breve, bifidum, longum biotype infantis, angulatum, dentiu, and gallicum by PCR. | Higher prevalence of Bifidobacterium catenulatum at one month of age (60.0% vs. 6.3%; p<0.01) and B. bifidum at six months of age (70.0% vs. 12.5%; p<0.01). |
| 14 | Penders, 2007 Netherlands | Cohort of 957 with a two-year follow-up. | ISAAC questionnaire for assessment of allergic symptoms; serum IgE antibodies at two years of age specific to at least one allergen. | Stool collection at one month of age. Search for Bifidobacterium spp., E. coli, C. difficile, B. fragilis, and Lactobacillus spp. and total count of bacteria by real-time PCR. | Higher counts of E. coli were associated with eczema (OR: 1.87, 95% CI: 1.15–3.04; p=0.016). Presence of C. difficile at one month of age was associated with increased risk of eczema (OR: 1.40; 95% CI: 1.02–1.91), recurrent wheezing (OR: 1.75; 95% CI: 1.09–2.80), and allergic sensitisation (OR: 1.54; 95% CI: 1.02–2.31) until the age of two years. |
| 15 | Adlerberth, 2007 Rome, London, and Gothenburg | Cohort of 324 newborns with an 18-month follow-up. | Eczema (Williams test); immediate-reading skin test: IgE antibodies at 18 months of age positive for at least one allergen. | Stool collection at seven, 14, and 28 days and two, six, and 12 months of age. Search for Enterobacteriaceae, E. coli, Klebsiella spp., S. aureus, Enterococci, Bacteroides spp., Clostridium spp., Bifidobacterium spp., and Lactobacillus spp. by bacteriological culture and PCR. | Increased ratio of gram-negative/gram-positive bacteria at 12 months of age was associated with IgE>100kU/L at 18 months of age (p=0.007). |
| 28 | Wang, 2008 Rome, London, and Gothenburg | A case–control study involving 15 children with eczema and 20 healthy subjects at 18 months of age selected from a cohort of 318 newborns. | Eczema (Williams criterion/ISAAC), immediate-reading skin test; total and specific IgE at 18 months of age. | Stool collection at seven days of life. Assessment of microbiota complexity by extracting long-chain fragments (TRFLP analysis). | Lower bacterial diversity at seven days of age was associated with eczema at 18 months of age (p<0.05). |
| 24 | Songjinda, 2007 Japan | Cohort of 15 newborns and a two-year follow-up. | ISAAC questionnaire for assessment of allergic symptoms. | Stool collection in the first five days of life and at one and two months of age. Search for Bacteroidaceae, Enterobacteriaceae, Bifidobacterium, Enterococcus, Lactobacillus, and C. perfringens by real-time PCR. | Higher count of Bacteroidaceae at one month of age (p=0.03) and at two months of age (p=0.05). |
| 19 | Verhulst, 2008 Belgium | Cohort of 154 newborns with a 12-month follow-up. | ISAAC questionnaire for evaluation of allergic symptoms performed at three weeks and six and 12 months of age. | Stool collection at three weeks of age. Search for S. aureus, Enterobacteriaceae, Enterococcus, Clostridium, B. fragilis, and Lactobacillus by bacteriological culture. | Higher prevalence of anaerobic bacteria (OR: 1.14; 95% CI: 1.02–1.28; p=0.03) and lower prevalence of S. aureus (OR: 0.95, 95% CI: 0.90–1.0; p=0.048) and Clostridium (OR: 0.90, 95% CI: 0.90–0.99; p=0.02) in infants with wheezing. |
| 20 | Vael, 2008 Belgium | Cohort of 117 newborns with a three-year follow-up. | Asthma predictive index for the identification of API, eczema, wheezing, and/or family history of asthma up to three years of age. | Stool collection at three weeks of age. Search for B. fragilis, Bifidobacterium, Lactobacillus, Clostridium, and Staphylococcus by bacteriological culture. | Higher prevalence of B. fragilis (64% vs. 34%; OR: 4.4; 95% CI: 1.7–11.8; p=0.003) and higher count of anaerobes (p<0.05) in children with positive API. |
| 26 | Forno, 2008 Boston | Cohort of 21 newborns with a six-month follow-up. | Clinical assessment of eczema until six months of age. | Stool collection at one and four months of age. Search for bacterial diversity by PCR-denaturing gradient gel electrophoresis (DGGE). | Lower microbial diversity in infants with eczema at one month of age (53% vs. 75%; p=0.01) and four months of age (59% vs. 92%, p=0.02). |
| 25 | Sjögren, 2009 Sweden | Cohort of 47 newborns with a five-year follow-up. | Clinical evaluation of allergic symptoms at three, six, and 12 months and two and five years of age, in addition to one immediate-reading skin test with positive results. | Stool collection at five to six days and at one and two months of age. Search for B. bifidum, breve, longum, and adolescentis, Lactobacilli, C. difficile, and B. fragilis by real-time PCR. | Lower incidence of B. adolescentis (62% vs. 92%; p=0.03), Lactobacilli type 1 (8% vs. 48%; p=0.02), and C. difficile (7% vs. 37%, p=0.04) in the first two months of life. |
| 16 | Penders, 2010 Netherlands | Cohort of 681 newborns with a two-year follow-up. | Eczema (Williams criteria), specific IgE antibodies at two years of age. | Stool collection at one month of age. Search for total lactobacilli, L. casei, L. paracasei, L. reuteri, L. rhamnosus, and L. acidophilus by real-time PCR. | Colonisation with L. paracasei was associated with lower risk of eczema (OR: 0:57; 95% CI: 0.30–0.99; p=0.04). |
| 21 | Vael, 2011 Belgium | Cohort of 110 newborns with a three-year follow-up. | Asthma Predictive Index for the identification of API, eczema, wheezing, and/or family history of asthma up to three years of age. | Stool collection at three weeks of age. Search for B. fragilis, Bifidobacterium, Lactobacillus, Clostridium, and S. aureus by bacteriological culture. | Colonisation with B. fragilis and C. coccoides at three week was associated with positive API (p=0.002). |
| 17 | Bisgaard, 2011 Denmark | Cohort of 411 newborns with a six-year follow-up. | Immediate-reading skin test and/or specific IgE antibodies; eczema (Hanifin & Rajka): clinical evaluation of rhinitis and asthma at six years of age. | Stool collection at one and 12 months of age. Search for bacterial diversity by PCR-denaturing gradient gel electrophoresis (DGGE) and bacteriological culture. | Lower bacterial diversity at one and 12 months of age was associated with increased risk of allergic sensitisation (p=0.003), immediate-reading skin test (p=0.017), and allergic rhinitis (OR: 0.71; 95% CI: 0.51–0.91; p=0.007). |
| 18 | Nimwegen, 2011 Netherlands | Cohort of 1176 newborns with a seven-year follow-up. | ISAAC questionnaire for assessment of allergic symptoms. Immediate-reading skin test and/or specific IgE antibodies at one, two, and six to seven years of age. | Stool collection at one month of age. Search for Bifidobacterium, E. coli, C. difficile, B. fragilis, and Lactobacillus by real-time PCR. | Colonisation with C. difficile at one month of age was associated with increased risk of wheezing (OR: 1.38; 1.16–3.64) and eczema (OR: 1.43; 1.09–1.88) at six to seven years of age and asthma (OR: 2.06; 1.16–3.64) at six years of age. |
| 22 | Storro, 2011 Norway | Cohort of 94 newborns with a two-year follow-up. | ISAAC questionnaire for assessment of allergic symptoms; eczema (UKWP criteria); specific IgE antibodies positive for at least one allergen. | Stool collection at 10 days, four months, and one and two years of age. Search for B. fragilis, E. coli, Bifidobacterium, Lactobacillus, C. difficile, C. perfringens, and H. pylori by real time PCR. | Lower counts of E. coli at four months of age (p<0.007) and one year of age (p<0.002); higher counts of B. longum at one year of age (p=0.05) and lower counts of B. fragilis at two years of age (p=0.005). |
| 27 | Gosalbes, 2013 Spain | Cohort of 40 newborns with a four-year follow-up. | Questionnaire for assessment of allergic symptoms. | Stool collection at one week of age. Search for more than 40 bacteria by PCR and pyrosequencing. | Predominance of Firmicutes was associated with increased risk of respiratory problems in infants (p=0.040). |
| 29 | Johansson, 2011 Sweden | A case–control study involving 35 children aged five years with allergic parents and 23 children with non-allergic parents selected from a cohort of 281 newborns. | Allergic symptoms and immediate-reading skin test for food allergens and inhalants performed in children at five years of age and in their parents. | Stool collection at one and two weeks and one, two, and 12 months of age. Search for Bifidobacterium, Clostridium, Lactobacillus (L. casei, L. paracasei, and L. rhamnosus), and S. aureus by real-time PCR. | Lower colonisation with lactobacilli in children from allergic parents (p=0.014). Allergic children had fewer lactobacilli in the first weeks of life, regardless of parental allergy (p=0.009, p=0.028) and lower colonisation with B. bifidum at one week of age (p=0.048). |
Studies on the association between the intestinal microbiota and allergic diseases wherein stool collection was performed during outcome assessment.
| Author, year location | Design | Definition of allergy | Analysis of faecal microbiota | Results of allergic compared with non-allergic subjects | |
|---|---|---|---|---|---|
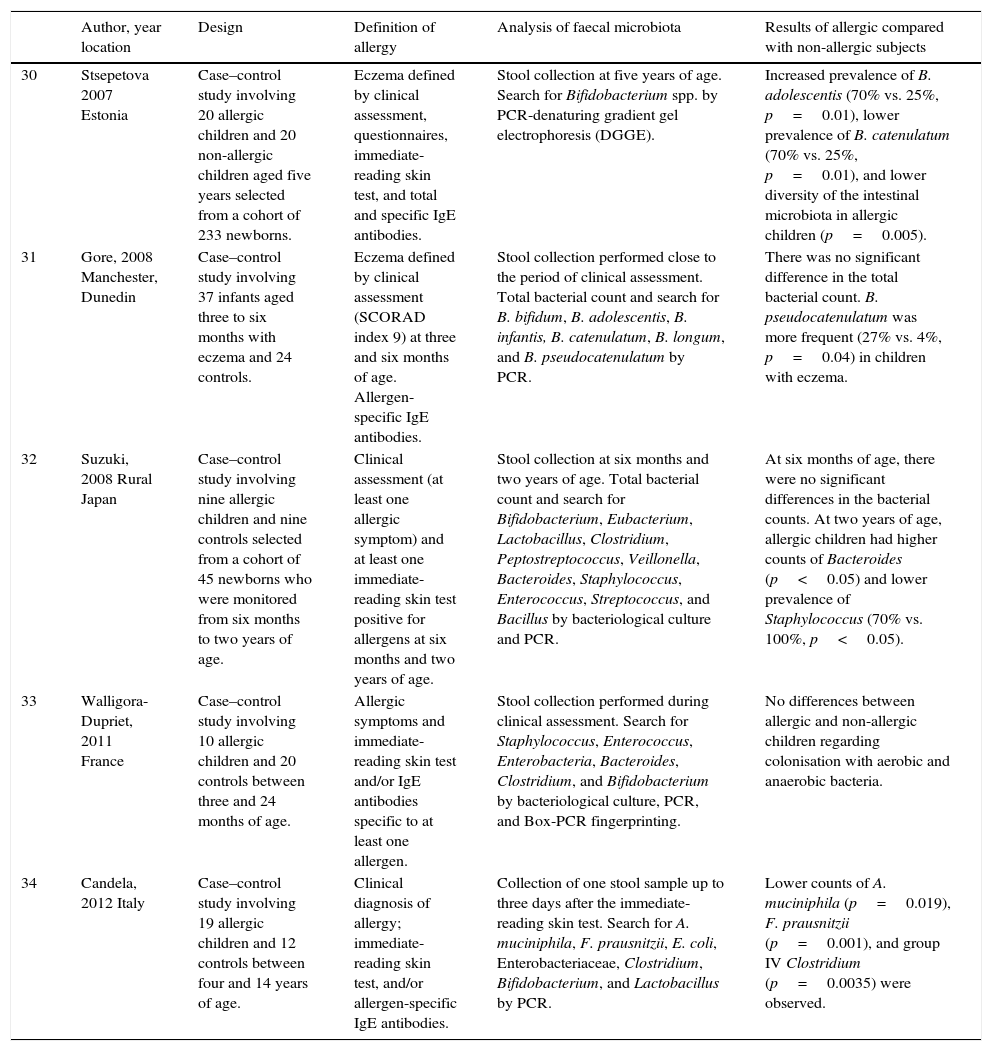
| 30 | Stsepetova 2007 Estonia | Case–control study involving 20 allergic children and 20 non-allergic children aged five years selected from a cohort of 233 newborns. | Eczema defined by clinical assessment, questionnaires, immediate-reading skin test, and total and specific IgE antibodies. | Stool collection at five years of age. Search for Bifidobacterium spp. by PCR-denaturing gradient gel electrophoresis (DGGE). | Increased prevalence of B. adolescentis (70% vs. 25%, p=0.01), lower prevalence of B. catenulatum (70% vs. 25%, p=0.01), and lower diversity of the intestinal microbiota in allergic children (p=0.005). |
| 31 | Gore, 2008 Manchester, Dunedin | Case–control study involving 37 infants aged three to six months with eczema and 24 controls. | Eczema defined by clinical assessment (SCORAD index 9) at three and six months of age. Allergen-specific IgE antibodies. | Stool collection performed close to the period of clinical assessment. Total bacterial count and search for B. bifidum, B. adolescentis, B. infantis, B. catenulatum, B. longum, and B. pseudocatenulatum by PCR. | There was no significant difference in the total bacterial count. B. pseudocatenulatum was more frequent (27% vs. 4%, p=0.04) in children with eczema. |
| 32 | Suzuki, 2008 Rural Japan | Case–control study involving nine allergic children and nine controls selected from a cohort of 45 newborns who were monitored from six months to two years of age. | Clinical assessment (at least one allergic symptom) and at least one immediate-reading skin test positive for allergens at six months and two years of age. | Stool collection at six months and two years of age. Total bacterial count and search for Bifidobacterium, Eubacterium, Lactobacillus, Clostridium, Peptostreptococcus, Veillonella, Bacteroides, Staphylococcus, Enterococcus, Streptococcus, and Bacillus by bacteriological culture and PCR. | At six months of age, there were no significant differences in the bacterial counts. At two years of age, allergic children had higher counts of Bacteroides (p<0.05) and lower prevalence of Staphylococcus (70% vs. 100%, p<0.05). |
| 33 | Walligora-Dupriet, 2011 France | Case–control study involving 10 allergic children and 20 controls between three and 24 months of age. | Allergic symptoms and immediate-reading skin test and/or IgE antibodies specific to at least one allergen. | Stool collection performed during clinical assessment. Search for Staphylococcus, Enterococcus, Enterobacteria, Bacteroides, Clostridium, and Bifidobacterium by bacteriological culture, PCR, and Box-PCR fingerprinting. | No differences between allergic and non-allergic children regarding colonisation with aerobic and anaerobic bacteria. |
| 34 | Candela, 2012 Italy | Case–control study involving 19 allergic children and 12 controls between four and 14 years of age. | Clinical diagnosis of allergy; immediate-reading skin test, and/or allergen-specific IgE antibodies. | Collection of one stool sample up to three days after the immediate-reading skin test. Search for A. muciniphila, F. prausnitzii, E. coli, Enterobacteriaceae, Clostridium, Bifidobacterium, and Lactobacillus by PCR. | Lower counts of A. muciniphila (p=0.019), F. prausnitzii (p=0.001), and group IV Clostridium (p=0.0035) were observed. |
Among the 16 studies that performed stool collection before the assessment, 14 were prospective, and two were non-prospective studies, totalling 4246 stool samples included for analysis. In five prospective studies, the experimental groups contained more than 300 children14–18 who were monitored for 18 months to seven years. In four studies, the groups comprised between 90 and 160 children19–22 who were monitored for 12 months to three years, and in five studies, the groups comprised fewer than 60 children23–27 who were monitored for six months to five years. Among the non-prospective studies, 35 children aged 18 months28 and 58 children aged five years29 were evaluated (Table 1).
The other five studies in which stool collection was performed close to the period of allergy assessment comprised case–control studies involving 18–61 children aged between three months and 14 years, totalling 170 analyses.30–34 In the study of Suzuki et al.,32 children (both cases and controls) were monitored from six months to two years of age (Table 2).
The criteria used to define allergic diseases involved mostly clinical assessment and the performance of skin tests and/or quantity of IgE antibodies in 11 studies,15–17,23,25,28,29,31–34 in addition to the use of questionnaires in two other studies.22,30 Three studies20,21,26 performed only clinical assessments, two studies14,18 used tests and questionnaires, and three studies19,24,27 only used questionnaires. The studies that used tests include those by Penders et al.,10,14 Gore et al.,31 and Storro et al.,22 wherein IgE antibodies were quantified in foods and/or inhalants, and those by Suzuki et al.,23,32 Sjögren et al.,25 and Johansson et al.,29 wherein skin tests were used. Other studies measured IgE antibodies and performed skin tests. The only criterion for the classification of allergies included the use of the International Study of Asthma and Allergies in Childhood (ISAAC)36 questionnaires in the studies by Songjinda et al.24 and Verhulst et al.19 and the Asthma Predictive Index35 in the studies by Vael et al.20,21
For the analysis of faecal microbiota, culture-independent techniques, such as real-time PCR, which was used in seven studies,14,16,18,22,24,25,29 or conventional PCR,23,27,31,34 predominated. Techniques that allowed the assessment of microbial diversity were used in four studies.17,26,28,30 Four other studies19–21,32 used only culture-dependent techniques, and both culture-dependent and culture-independent techniques were used in two studies15,33 (Tables 1 and 2).
In stool samples analysed during the first year of life, a lower bacterial diversity was observed in three studies,17,26,28 with a predominance of Firmicutes,27 higher counts of Bacteroidaceae,24 and a higher prevalence of anaerobic bacteria,19,20 such as Bacteroides fragilis,20,21Escherichia coli,14 and Clostridium difficile,14,18 in the microbiota of children who developed allergies later in life (Table 1). In three studies, the prevalence of C. difficile,25E. coli,22 and Staphylococcus aureus19 in the microbiota of allergic children was lower than that of the controls (Table 1), and three other studies found no significant differences in bacterial counts31–33 (Table 2).
Bifidobacteria were investigated in ten studies where the stool was collected before the assessment of the clinical outcome (allergy) and in five studies where the stool was collected during the assessment. The most prevalent species in the early microbiota among infants (up to 12 months of age) who developed allergies were Bifidobacterium catenulatum,23,31Bifidobacterium bifidum,23 and Bifidobacterium longum,22 and the least prevalent were Bifidobacterium adolescentis25 and B. bifidum29 (Table 1). In the faecal microbiota of five-year-old children, a higher prevalence of B. adolescentis and a lower prevalence of B. catenulatum30 were found (Table 2).
Allergic children exhibited a lower prevalence of Lactobacillus25,29 and Lactobacillus paracasei16 in the first weeks of life. An increased gram-negative/gram-positive ratio at 12 months of age was associated with IgE>100kU/L at 18 months of age15 (Table 1).
Allergic children at two years of age had higher counts of Bacteroides and a lower prevalence of S. aureus32 in their faecal microbiota, and children between four and 14 years of age had lower counts of Akkermansia muciniphila, Faecalibacterium prausnitzii, and Clostridium.34 Stsepetova et al.30 also found a lower microbial diversity in the faecal microbiota of children younger than five years old (Table 2).
DiscussionOur study indicated differences in the microbiota of allergic children compared with non-allergic children, as previously reported by Penders et al.5 Twenty studies out of the 21 published between 2007 and 2013 indicated associations between some abnormalities in the intestinal microbiota and atopic symptoms or sensitisation, whereas in the review conducted previously, 14 studies out of the 17 published between 1999 and 2006 indicated such an association. Furthermore, an increased number of studies in the period from 2007 to 2013 used advanced laboratory techniques and a larger sample size.
As reported in three pioneering studies conducted in Estonia and Sweden and reviewed by Penders et al.,5 investigations into the role of the faecal microbiota in the aetiology of allergic diseases began in the 1990s and involved healthy one-year-old children in Estonia, where the prevalence of childhood allergy was low, and in Sweden, where allergy prevalence was high.37 Lactobacilli and eubacteria were the most prevalent bacterial groups, particularly among Estonian children, and Swedish children harboured higher counts of C. difficile. Based on these observations, the first study comparing the composition of the faecal microbiota of allergic and healthy children was conducted.38 The results of this study indicated that allergic children in both countries were colonised less frequently with lactobacilli compared with non-allergic children and harboured higher counts of facultative aerobic microorganisms, particularly coliforms in Estonia and S. aureus in Sweden. A prospective study39 by the same research group found a lower prevalence of colonisation with bifidobacteria and enterococci in the first year of life, higher counts of clostridia at three months of age, and a higher prevalence of S. aureus at six months of age among children who developed allergic diseases.
Of the 21 studies reviewed herein, 16 studies performed stool collection before the assessment of allergies and evaluated 4246 individuals, most of whom were children monitored for up to seven years. In the other studies where stool collection was performed concomitantly with the assessment of allergies, the small number of subjects evaluated (N=170) should be noted, with small experimental groups (≤37 individuals) that included infants to 14-year-olds. We observed differences in both the study design and the techniques used to analyse the faecal microbiota, with a predominance of molecular techniques.
Molecular biology techniques, which were performed in 85.7% (18/21) of the studies analysed, are able to overcome the limitations of traditional bacterial culture techniques, such as low sensitivity and low reproducibility, due to the large number of species able to be identified.40 These techniques, including fluorescence in situ hybridisation (FISH), PCR combined with denaturing- or temperature-gradient gel electrophoresis (PCR-DGGE/PCR-TGGE), and real-time PCR, are molecular techniques with wide applicability for the study of intestinal microbiota.41 The greatest advantages of these methods are speed, specificity, sensitivity, and the possibility of quantification. In addition, samples can be frozen for future analysis.42 These tools have provided detailed molecular characterisation of the composition and concentration of faecal microorganisms and led to the discovery of a large number of bacterial genera and species, thereby increasing by more than 50% the number of organisms known to compose the faecal microbiota.4
Many epidemiological methods have been used to assess the distribution, determinants of disease prevalence and to analyse cause-and-effect associations. The selection of the methods used to evaluate the association between disease and environment depends on several factors because each of these methods has advantages and limitations.5 Observational studies serve a variety of purposes, including providing an initial assessment of the possible causes of a disease and the magnitude of previously observed associations. Therefore, cohort, case–control, and cross-sectional studies represent different approaches that can be employed to investigate the incidence of related events in a population in a given period of time.43
In contrast to studies published before 2007, most of which were cross-sectional or transversal, the studies evaluated in this review were predominantly cohort studies. In 16 birth cohort studies, stool collection was performed in the first weeks of life; other studies involved stool collection from subjects by the age of two years.
Assuming that the chronology of events can be easily determined in cohort studies, i.e., exposure is followed by the clinical outcome,44 it can be hypothesised that in the first weeks of life, the process of microbiota development would determine the level of exposure, which would then culminate in the outcome studied, i.e., allergic disease. In the early years of life, the intestinal microbiota of children is simple but becomes more complex over time, and bacterial diversity increases. Bacteria that can be identified in this initial phase include the genera Bifidobacterium, Ruminococcus, Enterococcus, Clostridium, and Enterobacter.41 In a recent study using molecular techniques, changes were observed in the intestinal microbial population at different ages; e.g., E. coli was more frequent at 10 days of life among children whose mothers were also positive for this bacterium, and this frequency remained stable during the first year of life in 80–85% of the study group. However, at two years of age, this frequency decreased to 45% in children who were previously positive. In contrast, B. longum is considered to be one of the most persistent colonisers; at 10 days of life, B. longum was detected in almost all newborns who harboured the bacteria at three days of life, and even at two years of age, this species persisted in most children who were previously positive. At the age of four months, bifidobacteria represented 57.6% of the total intestinal microbiota, whereas the prevalence of Lactobacillales and Streptococcus organisms had decreased.45
Studies that involved stool collection before clinical outcome assessmentStudies that used real-time PCR to investigate microbiota indicated a lower colonisation with lactobacilli16,25,29 and a higher prevalence of C. difficile14,18 in children who developed allergic diseases by age five. Real-time PCR is a valuable tool for the quantification of intestinal populations because it combines the specificity of fluorescent oligonucleotide probes with the sensitivity of PCR.46 Of the two studies that identified E. coli, the study by Penders et al.,14 which was conducted in the Netherlands, indicated a correlation between the presence of E. coli and eczema, whereas the study by Storro et al.,22 which was conducted in Norway, found a lower concentration of E. coli among allergic individuals. Other differences were observed for bifidobacteria, depending on the species studied. B. adolescentis and B. bifidum exhibited decreased prevalence or colonisation and B. longum exhibited increased prevalence in allergic individuals.22,25,29 With regard to Bacteroides, B. fragilis was less prevalent22 and the family Bacteroidaceae was more prevalent in allergic individuals.24
Three of these studies found that lactobacilli species were less prevalent in allergic individuals regardless of the species studied. Lactobacilli and bifidobacteria are known for their beneficial effects to human health, and they ensure a healthy intestinal flora by inhibiting the proliferation of pathogenic bacteria and promoting immunoregulation, which triggers regulatory T-cell responses.47 However, differences were observed in the prevalence and species composition of bifidobacteria. Differences in the species composition of bifidobacteria were investigated for the first time in 2001 in stool samples from seven allergic children and six healthy children who were breastfed; the allergic children were more frequently colonised with B. adolescentis, whereas the healthy children were more frequently colonised with B. bifidum.48
According to Penders et al.,5 the differences in the prevalence and cell count of bifidobacteria between allergic and healthy children in studies involving traditional bacterial culture should be interpreted with caution because of possible biases in the techniques used. The effects of bifidobacteria may be species dependent, similar to the effects reported for lactobacilli. In this review, the bifidobacteria species were identified using molecular techniques alone, but doubts persist about their role in protecting against the development of allergic diseases.
Three other studies used PCR alone or associated23 with other techniques, such as cell culture15 and pyrosequencing.27 There was a predominance of Firmicutes at one week of age, B. catenulatum at one month of age, and B. bifidum at six months of age, in addition to an increased ratio of gram-negative bacteria at 12 months of age, in children who developed allergic complications later in life. In three studies that used culture techniques,19–21 higher counts of anaerobic bacteria were detected, and in two other studies,20,21 a higher prevalence of B. fragilis was observed among children who later developed allergies. In the study by Verhulst et al.,19 children who developed wheezing had a lower prevalence of C. difficile and S. aureus.
Staphylococcus, Streptococcus, Enterococcus, and Enterobacter spp. are facultative anaerobic bacteria that first develop in newborns and create an anaerobic environment that promotes the growth of obligate anaerobes such as Bifidobacterium spp., Bacteroides spp., Clostridium spp., and Eubacterium spp.49 These anaerobic bacteria are normal components of the oral, intestinal, and vaginal microbiota and may have important pathogenic potential. Some species of B. fragilis and C. perfringens have been associated with infections. Enterotoxigenic B. fragilis strains are highly prevalent in diarrheal diseases,50 and the presence of C. difficile is associated with changes in the endogenous microbiota, which usually occurs with antibiotic use. Moreover, various strains of Bacteroides spp., Bifidobacterium spp., Lactobacillus spp., Pseudomonas spp., Staphylococcus spp., and Streptococcus spp. can inhibit the proliferation of C. difficile.51
The DGGE and terminal restriction fragment length polymorphism (T-RFLP) techniques, which are employed to assess the diversity of microbial communities, were used in three studies17,26,28 that found that a lower microbial diversity was associated with increased allergic sensitisation, allergic rhinitis, and eczema.
It is difficult to elucidate the differences and conflicting results of these studies. Many factors may be involved in the pathogenesis of intestinal microbiota from the geographic area of residence, to type of delivery, duration of breastfeeding, diet, use of antibiotics, allergy definition and laboratory techniques, among others. A study conducted in two different cohorts52 showed greater abundance of Bifidobacteria and less of Clostridium leptum in Singapore infants compared to Indonesian infants. An association of greater abundance of Bifidobacteria with greater number of siblings was observed, regardless of region. Bacterial diversity was similar in both regions, however, when the type of delivery was considered a higher diversity in intestinal microbiota was shown in those born vaginally. Thus, many factors can interfere with the evaluation of the association of the faecal microbiota composition and predisposition to diseases.
For example, in two studies14,19 there was contradiction in prevalence in relation to C. difficile as a risk factor or protective factor for wheezing. Verhulst et al.19 reported that this difference could be due to the criteria used for allergy definition. In these studies the allergy classification occurred in the first two years of life, during which the allergic manifestations are mainly limited to the skin and intestine.25 In most studies in which the collection occurred before the clinical outcome, the infants were followed until the age of two. Vael et al.20,21 state that the differences in the microbiota of allergic compared to non-allergic, could not be explained by differences in gender, diet, use of antibiotics, maternal smoking during pregnancy or socioeconomic status.
Studies that involved stool collection during clinical outcome assessmentThe five studies where stool collection was performed during the assessment of the clinical outcome used molecular techniques,30–34 and the study by Suzuki et al.32 used molecular and culture techniques. Three studies performed stool collection between three months and two years of age and indicated a higher prevalence of B. catenulatum in children with eczema31 and higher counts of Bacteroides and a lower prevalence of Staphylococcus32 in allergic children. The study by Walligora–Dupriet et al.33 found no difference in the colonisation rate with aerobic and anaerobic bacteria.
Studies that performed stool collection for microbiota analysis after the age of four, when the microbiota is presumably stabilised, reported a higher prevalence of B. adolescentis and a lower prevalence of B. catenulatum, as well as a lower bacterial diversity, among allergic children at five years of age.30 Other bacteria of the phylum Firmicutes, A. muciniphila and F. prausnitzii, in addition to Clostridium group IV, had lower counts in the colonic microbiota of allergic children aged four to 14 years.34
With regard to F. prausnitzii, its anti-inflammatory effects are used in models of cell colitis, and its use as a probiotic has been suggested to balance the intestinal dysbiosis found in inflammatory intestinal disease.53,54 In contrast, A. muciniphila has been found to a lesser extent in the microbiota of individuals with type-2 diabetes, and it exerted anti-diabetic effects in a murine model.55
These recent findings support the hypothesis that decreased intestinal microbial diversity is associated with increased risk of atopic sensitisation and allergic disease, which are characterised by the decreased colonisation of lactobacilli and bifidobacteria and increased colonisation of C. difficile, which have been associated with sensitisation and allergic diseases in childhood.56
Under certain circumstances, when the intestinal microbiota is less diversified, certain species appear to have a detrimental role in allergic disease.25 Differences in the initial communities can lead to differences in microbial succession of patterns in the intestine and other habitats of the body which may persist over time. Likewise, the composition of the initial microflora may have implications for nutritional and immune functions associated with microbiota in development.57
ConclusionCertain bacteria are associated with a reduced risk of allergic diseases, whereas others were reported to be associated with an increased risk of allergic diseases. The comparison of these results is difficult because of differences in several variables, including the methodology used in these studies, sample size, differences in the protocols, and the selection of the microorganisms present in the colonic microbiota.
Despite these limitations, we observed lower biodiversity in the colonic microbiota of allergic children compared with non-allergic children, with a predominance of Firmicutes, in addition to an increased count of Bacteroidaceae; a higher prevalence of the anaerobic bacteria B. fragilis, E. coli, C. difficile, B. catenulatum, B. bifidum, and B. longum; and a lower prevalence of B. adolescentis, B. bifidum, and Lactobacillus.
Conflict of interestThe authors have no conflict of interest to declare.
Ethical disclosuresProtection of human subjects and animals in researchThe authors declare that no experiments were performed on humans or animals for this investigation.
Patients’ data protectionConfidentiality of data. The authors declare that no patient data appears in this article.
Right to privacy and informed consentThe authors declare that no patient data appears in this article.