It is possible that imbalances in the composition of the gut microbiota or the relationship of the microbiota with the host may be implicated in the origin of allergy. Therefore, we studied the intestinal microbiota of children with atopic dermatitis (AD).
MethodsCross-sectional study with 81 children aged 5–11; 23 with AD and 58 controls. Surveys were conducted to obtain demographic, socioeconomic and neonatal data. Diagnosis of AD was made based on the International Study of Asthma and Allergies in Childhood (ISAAC) questionnaire. Eubacteria, Bacteroidetes, Firmicutes, B. fragilis, E. coli, Lactobacillus spp., S. aureus, E. faecalis, Salmonella spp., M. smithii, Bifidobacterium spp., C. difficile and C. perfringens were quantified using real-time PCR.
ResultsThe analysis showed an association between presence of C. difficile (OR: 5.88; 95 % CI: 1.24; 27.98), greater abundance of bifidobacteria (OR: 11.09; 95 % CI: 2.14; 57.39) and a lower abundance of lactobacilli (OR: 0.07; 95 % CI: 0.01; 0.51) in the gut microbiota of children with AD. Counts of Eubacteria (0,05×103 and 8.49×103), B. fragilis (0.72×109 and 4.5×109), Lactobacillus spp. (0.02×108 and 0.38×108), E. coli (0.13×109 and 1.52×109) and M. smithii (0.02×108 and 0.31×108) were lower in children with AD (P<0.05).
ConclusionsThis study confirmed that children living in the metropolitan area of São Paulo (Brazil) with AD have a different microbiota pattern with higher prevalence of C. difficile, lower abundance of Lactobacillus and greater abundance of bifidobacteria, regardless of socioeconomic status.
- There are differences between the gut microbiota of allergic and non-allergic individuals.
- Possible imbalances in the composition of the gut microbiota or its relationship with the host may be implicated in the origin of allergic diseases
What are the new findings on this subject?- This is the first study to analyze the association of gut microbiota and atopic dermatitis (AD) in Brazilian school-age children.
- Associations between a higher prevalence of C. difficile and AD and between lower abundance of Lactobacillus and AD have been confirmed. The gut microbiota of patients with atopic dermatitis has been found to have lower abundance of B. fragilis, E. coli and M. smithii and greater abundance of bifidobacteria than the microbiota of controls.
Atopic dermatitis (AD) is one of the most common skin diseases among children in Western countries, and its prevalence is apparently rising.1 Studies carried out2 in phase three of the International Study of Asthma and Allergies in Childhood (ISAAC) protocol revealed a prevalences of 8.2 % for flexural eczema in Brazilian children aged 6–7.
AD may have a major impact on the quality of life of a child and his/her family, producing a negative influence on social and educational development. When AD starts during the first year of life, it is often associated with a family history of atopic diseases.3 An epidemiological study conducted in Poland with 18,617 individuals from eight cities and one rural area has shown AD to be present in 3.9 % of the individuals. AD has been associated with female sex, living in urban areas, paternal history of atopy, higher level of education and higher economic status.4
The etiology of AD is not fully understood, and probably involves an interaction of multiple genetic and environmental factors.5 Studies performed over the last decades have shown that possible imbalances in the composition of the gut microbiota or the relationship of the microbiota with the host may be implicated in the origin of allergic diseases.6 This has led to projects to elucidate the relationship between the development of allergic diseases and the composition and function of the gut microbiota.7 Most such studies are being performed on infants.8 The few studies on children with AD older than five have shown a low prevalence of bifidobacteria and a high proportion of clostridia in the gut microbiota of allergic children.9 These studies have also revealed a higher prevalence of Bifidobacterium adolescentis and lower prevalence of Bifidobacterium catenulatum, as well as lower bacterial diversity in patients with AD.10
Considering the small number of studies on the composition of the gut microbiota in school-age children with atopic dermatitis, and the complete absence of studies on this topic performed in Latin America, the present study was carried out to compare the gut microbiota of Brazilian school-age children with and without atopic dermatitis.
Materials and methodsStudy designThis is a cross-sectional study carried out with school-age children living in the city of Osasco, in the Metropolitan Region of São Paulo, Brazil. The children admitted into the study were either residents of two slums or enrolled in two private schools. Community leaders from the slums and the schools’ pedagogical coordinators invited families with children aged between five and eleven to take part in the research. This resulted in 184 children assessed for the study. We obtained demographic information, applied the ISAAC11 questionnaire and collected stool samples in order to study the gut microbiota of the participating children. The children admitted into the study were divided into two groups: 1. Children with atopic dermatitis and 2. Control group without allergic diseases.
The Research Ethics Committee of Federal University of São Paulo approved this project. The families were invited to participate voluntarily in the research study. The families who agreed to participate signed a Free and Informed Consent Form, while children signed an Assent Form.
Case seriesAfter recruitment, the following criteria were considered for inclusion in the study: absence of diarrhea for at least 30 days, stool sample suitable for performing a molecular analysis of the gut microbiota, and complete filling of the required forms and the three modules of the ISAAC questionnaire. Exclusion criteria were the use of antibiotics in the 30 days prior to stool collection, and the presence of clinical evidence of serious diseases such as heart disease, nephropathy or neuropathies.
The three ISAAC modules were used to characterize asthma, allergic rhinitis and AD. The groups of Children with AD and Children without allergic diseases (control) were formed according to the information provided in the ISAAC questionnaire.
The AD group comprised children whose parents answered yes to question three of the eczema module ("Has this itchy rash at any time affected any of the following places: the folds of the elbows, behind the knees, in front of the ankles, under the buttocks, or around the neck, ears or eyes?"). All children with AD were included in the study, regardless of whether or not they had associated asthma and/or allergic rhinitis. The AD group thus consisted of 23 school-age children.
The control group without allergic diseases consisted of 58 children who had negative answers for all questions in the ISAAC questionnaire (i.e. showed no evidence of allergic disease).
Fifty-six children whose questionnaires raised doubts about the possibility of allergic disease and 47 children with allergic respiratory disease without skin manifestations compatible with AD were excluded from the study.
Assessment of socioeconomic and housing conditionsData regarding family composition, housing conditions, basic sanitation and access to health services has been obtained through interviews with the parents. Socioeconomic characterization used the Brazilian Economic Classification Criteria (CCEB), which takes into account the possession of certain items and the educational level of the head of household as an estimate for the monthly family income. The CCEB distinguishes five main social classes (A, B, C, D and E). Class A corresponds to the highest socioeconomic status, and the status decreases towards class E.
Neonatal dataData on childbirth, gestational age at birth, weight and length at birth, total duration of breastfeeding and maternal history of allergy were obtained for each participant. Total time of breastfeeding was used to characterize breastfeeding behavior due to mothers’ difficulty to define accurately the age when other foods were introduced into their children's diet.
Assessment of nutritional statusWeight and height measurements were obtained according to recommendations for community studies (WHO, Geneva). Children were weighed with a mechanical scale (Filizola, São Paulo, Brazil) with a capacity of 150kg and sensitivity of 100g. Height was measured with a portable vertical anthropometer (Seca Gmbh & Co. Kg., Hamburg, Germany), which allows measurements up to 190cm, with a sensitivity of 0.1cm. The children were weighed wearing light clothing. Height was measured with the children standing barefoot with spine and legs held straight. After obtaining anthropometric data, the Z scores of height-for-age (H / A) and body mass index (BMI) adjusted for age and sex were calculated using the WHO AnthroPlus software (WHO, Geneva).
Stool collection and storageStool samples were collected by the children’s parents and placed in universal collectors, following guidelines provided previously in order to ensure good quality and conservation of bacterial DNA. The samples were processed at most six hours after defecation. Approximately one gram (g) of feces from the collected samples was transferred to a sterile microcentrifuge tube containing ASL buffer from the QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) and then frozen at −20°C until the time of DNA extraction.
Extraction of genomic DNABacterial genomic DNA was subsequently extracted according to the protocol suggested by the manufacturer of the QIAmp DNA Stool Mini Kit (Qiagen, Hilden, Germany). Purified DNA was diluted to a final volume of 200μL. The quantification of DNA concentration was performed in a Nanodroop 1000 spectrophotometer (ThermoScientific, Waltham, MA, USA). All DNA samples were diluted to the final concentration of 20ng/μL and stored at −20°C.
Bacteria assessment using real-time PCRFor real-time PCR, primers were designed based on 16S rDNA gene sequences of a variety of intestinal microbiota, obtained from GenBank (http://www.ncbi.nlm.nih.gov/genbank/). The sequences were aligned using the Lasergene Software Package (DNASTAR, Madison, WI, USA), allowing regions conserved in all species to be chosen for annealing. All designed primers were tested for their specificity in the software BLAST - Basic Local Alignment Search Tool (http://www.ncbi.nlm.nih.gov/blast/), and have proven to be specific for their purposes.
The primers used were designed to identify the following constituents of the gut microbiota, described in a previously published article12: total Eubacteria, Bacteroidetes and Firmicutes phyla, Bifidobacterium spp., Bacteroides fragilis, Clostridium difficile, Clostridium perfringens, Enterococcus faecalis, Lactobacillus spp., Staphylococcus aureus, Escherichia coli, Salmonella spp. and Methanobrevibacter smithii.
The DNA in all fecal samples was submitted to a real-time PCR trial to investigate the constituents of the gut microbiota of each participant. All reactions were made in duplicate, each with a final volume of 10μL containing 5μL of Rotor-gene SYBR Green PCR Master Mix (Qiagen, Hilden, Germany), 0.2μL (10pmol/μL) of forward and reverse primers for each constituent of the gut microbiota, 0.5μl of the DNA sample (20ng/μL) and 4.1μL of DEPC water (Qiagen, Hilden, Germany). Thermocycling was performed on a Rotor-gene Q PCR cycler (Qiagen, Hilden, Germany) under the following conditions: 5min at 95°C, followed by 40 cycles of 95°C for 10s and 60°C for 15s. The cycle of dissociation for the melting curve was 95°C for 1min, and a step for producing the melting curve, which progressed from 70°C to 95°C with gradual temperature increases of 1°C/s.
The standard curve for all analyses was produced by amplifying a TOPO-TA plasmid (Invitrogen) carrying the gene fragment for each bacterium previously amplified by conventional PCR; its specificity was confirmed by sequencing and alignment in the BLAST software. Knowing the molecular mass of both plasmid and inserted gene, the number of copies of the inserted gene was calculated according to the formula: mass in Daltons (g/mol)=(plasmid size plus size of the gene inserted in base pairs [bp]). (330Da×2 nucleotides/bp). Knowing the number of copies/g and the concentration of plasmid DNA, it is possible to calculate the number of copies present in the real-time PCR reaction and to establish the values, for the standard curve. The results were expressed in colony forming units/g feces (CFU/g), using the number of 16S rRNA genes (or copies of the quantified gene when different from the 16S rRNA gene) as the CFU count.13 A reaction containing all reagents except for the DNA sample was used as a negative control.
Statistical analysisNumerical variables were expressed as medians and 25th and 75th percentiles. The results were analyzed by comparing the two independent groups using Mann-Whitney tests for continuous numerical variables and Chi-square tests for categorical variables. The extent and strength of the association of components of the gut microbiota with AD (based on the Chi-square and Mann-Whitney tests) and the interactive effects of two or more elements of the microbiota on the outcome (AD) were assessed using a multiple logistic regression. The best model was selected to describe the relationship between gut microbiota and AD. The calculations were performed using the software EpiInfo™ 7 version 3.03a (Centers for Disease Control and Prevention, Atlanta, GA, USA) and Sigma Plot 11.2 (Systat, San Jose, CA, USA), setting the level for rejection of the null hypothesis at 0.05, or 5 %.
ResultsSix of the 23 children with AD (26.1 %) also had active asthma, three (13.1 %) had rhinoconjunctivitis and one (4.3 %) had both active asthma and rhinoconjunctivitis. Age was similar (p=0.210) between the AD and control groups, with medians of 7.5 years (25th and 75th percentiles: 6.7 and 9.4) and 7.9 years (25th and 75th percentiles: 7.0 and 9.3), respectively.
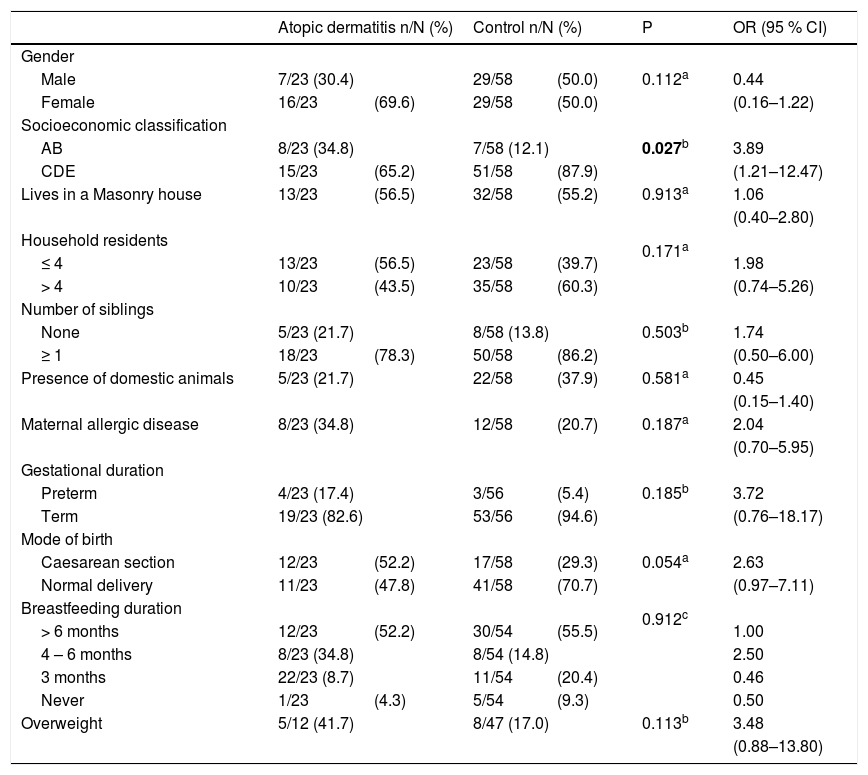
Demographic, socioeconomic, family and neonatal data for the two groups are shown is Table 1. AD has been found to be more frequent in children of socioeconomic classes A and B when compared to classes C, D and E. No differences were found regarding variables related to gender, housing conditions and neonatal data.
Demographic, socioeconomic and environmental characteristics and overweight in children aged 5–11 with atopic dermatitis or without any allergic diseases (control group).
| Atopic dermatitis n/N (%) | Control n/N (%) | P | OR (95 % CI) | |||
|---|---|---|---|---|---|---|
| Gender | ||||||
| Male | 7/23 (30.4) | 29/58 | (50.0) | 0.112a | 0.44 | |
| Female | 16/23 | (69.6) | 29/58 | (50.0) | (0.16–1.22) | |
| Socioeconomic classification | ||||||
| AB | 8/23 (34.8) | 7/58 (12.1) | 0.027b | 3.89 | ||
| CDE | 15/23 | (65.2) | 51/58 | (87.9) | (1.21–12.47) | |
| Lives in a Masonry house | 13/23 | (56.5) | 32/58 | (55.2) | 0.913a | 1.06 |
| (0.40–2.80) | ||||||
| Household residents | 0.171a | |||||
| ≤ 4 | 13/23 | (56.5) | 23/58 | (39.7) | 1.98 | |
| > 4 | 10/23 | (43.5) | 35/58 | (60.3) | (0.74–5.26) | |
| Number of siblings | ||||||
| None | 5/23 (21.7) | 8/58 (13.8) | 0.503b | 1.74 | ||
| ≥ 1 | 18/23 | (78.3) | 50/58 | (86.2) | (0.50–6.00) | |
| Presence of domestic animals | 5/23 (21.7) | 22/58 | (37.9) | 0.581a | 0.45 | |
| (0.15–1.40) | ||||||
| Maternal allergic disease | 8/23 (34.8) | 12/58 | (20.7) | 0.187a | 2.04 | |
| (0.70–5.95) | ||||||
| Gestational duration | ||||||
| Preterm | 4/23 (17.4) | 3/56 | (5.4) | 0.185b | 3.72 | |
| Term | 19/23 (82.6) | 53/56 | (94.6) | (0.76–18.17) | ||
| Mode of birth | ||||||
| Caesarean section | 12/23 | (52.2) | 17/58 | (29.3) | 0.054a | 2.63 |
| Normal delivery | 11/23 | (47.8) | 41/58 | (70.7) | (0.97–7.11) | |
| Breastfeeding duration | 0.912c | |||||
| > 6 months | 12/23 | (52.2) | 30/54 | (55.5) | 1.00 | |
| 4 – 6 months | 8/23 (34.8) | 8/54 (14.8) | 2.50 | |||
| 3 months | 22/23 (8.7) | 11/54 | (20.4) | 0.46 | ||
| Never | 1/23 | (4.3) | 5/54 | (9.3) | 0.50 | |
| Overweight | 5/12 (41.7) | 8/47 (17.0) | 0.113b | 3.48 | ||
| (0.88–13.80) | ||||||
OR: Odds ratio; 95 % CI: 95 % confidence interval.
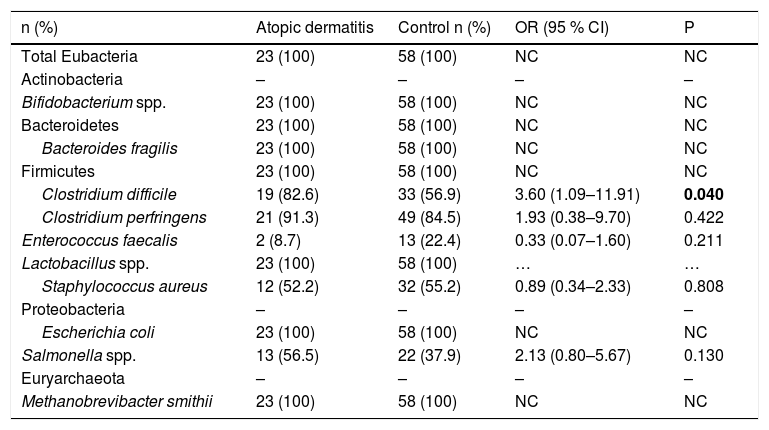
The prevalence of gut microbiota constituents is presented in Table 2. The presence of Clostridium difficile has been found to be associated with AD. Clostridium perfringens, Enterococcus faecalis, Staphylococcus aureus and Salmonella spp. presented varying prevalences; the differences between groups for these components were not statistically significant. The phyla Bacteriodetes and Firmicutes, the genera Bifidobacterium spp. and Lactobacillus spp. and the species Bacteroides fragilis, Escherichia coli and Metanobrevibacter smithii were present in all samples analyzed.
Prevalence of gut microbiota constituents in children aged 5–11 with atopic dermatitis or without any allergic diseases (control group).
| n (%) | Atopic dermatitis | Control n (%) | OR (95 % CI) | P |
|---|---|---|---|---|
| Total Eubacteria | 23 (100) | 58 (100) | NC | NC |
| Actinobacteria | – | – | – | – |
| Bifidobacterium spp. | 23 (100) | 58 (100) | NC | NC |
| Bacteroidetes | 23 (100) | 58 (100) | NC | NC |
| Bacteroides fragilis | 23 (100) | 58 (100) | NC | NC |
| Firmicutes | 23 (100) | 58 (100) | NC | NC |
| Clostridium difficile | 19 (82.6) | 33 (56.9) | 3.60 (1.09–11.91) | 0.040 |
| Clostridium perfringens | 21 (91.3) | 49 (84.5) | 1.93 (0.38–9.70) | 0.422 |
| Enterococcus faecalis | 2 (8.7) | 13 (22.4) | 0.33 (0.07–1.60) | 0.211 |
| Lactobacillus spp. | 23 (100) | 58 (100) | … | … |
| Staphylococcus aureus | 12 (52.2) | 32 (55.2) | 0.89 (0.34–2.33) | 0.808 |
| Proteobacteria | – | – | – | – |
| Escherichia coli | 23 (100) | 58 (100) | NC | NC |
| Salmonella spp. | 13 (56.5) | 22 (37.9) | 2.13 (0.80–5.67) | 0.130 |
| Euryarchaeota | – | – | – | – |
| Methanobrevibacter smithii | 23 (100) | 58 (100) | NC | NC |
Chi-square tests. NC=not calculated.
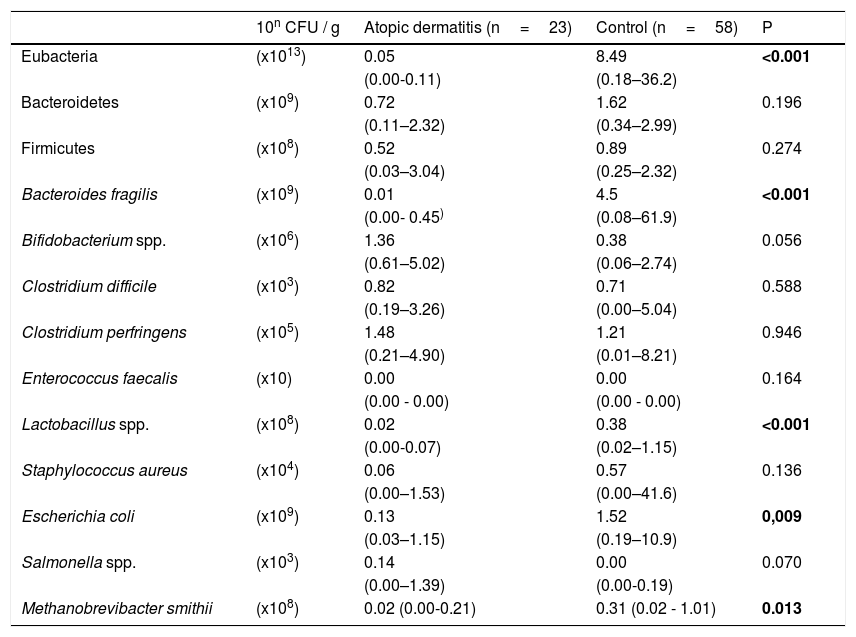
Table 3 shows the counts of Eubacteria and the phyla, genera and species analyzed. Children with AD presented lower total Eubacteria count. A statistically significant difference was also observed between groups for the counts of Lactobacillus spp., Escherichia coli, Bacteroides fragilis and Methanobrevibacter smithii. No statistically significant difference was observed between the AD and control groups for Clostridium difficile, Clostridium perfringens, Enterococcus faecalis and Staphylococcus aureus. The median counts of Bifidobacterium spp (p=0.056) and Salmonella spp. (p=0.07) were higher in children with AD, but the difference was not found to be statistically significant.
Count (colony forming units/gram feces, CFU/g) of gut microbiota constituents in children aged 5–11 with atopic dermatitis or without any allergic diseases (control group).
| 10n CFU / g | Atopic dermatitis (n=23) | Control (n=58) | P | |
|---|---|---|---|---|
| Eubacteria | (x1013) | 0.05 | 8.49 | <0.001 |
| (0.00-0.11) | (0.18–36.2) | |||
| Bacteroidetes | (x109) | 0.72 | 1.62 | 0.196 |
| (0.11–2.32) | (0.34–2.99) | |||
| Firmicutes | (x108) | 0.52 | 0.89 | 0.274 |
| (0.03–3.04) | (0.25–2.32) | |||
| Bacteroides fragilis | (x109) | 0.01 | 4.5 | <0.001 |
| (0.00- 0.45) | (0.08–61.9) | |||
| Bifidobacterium spp. | (x106) | 1.36 | 0.38 | 0.056 |
| (0.61–5.02) | (0.06–2.74) | |||
| Clostridium difficile | (x103) | 0.82 | 0.71 | 0.588 |
| (0.19–3.26) | (0.00–5.04) | |||
| Clostridium perfringens | (x105) | 1.48 | 1.21 | 0.946 |
| (0.21–4.90) | (0.01–8.21) | |||
| Enterococcus faecalis | (x10) | 0.00 | 0.00 | 0.164 |
| (0.00 - 0.00) | (0.00 - 0.00) | |||
| Lactobacillus spp. | (x108) | 0.02 | 0.38 | <0.001 |
| (0.00-0.07) | (0.02–1.15) | |||
| Staphylococcus aureus | (x104) | 0.06 | 0.57 | 0.136 |
| (0.00–1.53) | (0.00–41.6) | |||
| Escherichia coli | (x109) | 0.13 | 1.52 | 0,009 |
| (0.03–1.15) | (0.19–10.9) | |||
| Salmonella spp. | (x103) | 0.14 | 0.00 | 0.070 |
| (0.00–1.39) | (0.00-0.19) | |||
| Methanobrevibacter smithii | (x108) | 0.02 (0.00-0.21) | 0.31 (0.02 - 1.01) | 0.013 |
Wilcoxon-Mann-Whitney test: Median (25th and 75th percentiles).
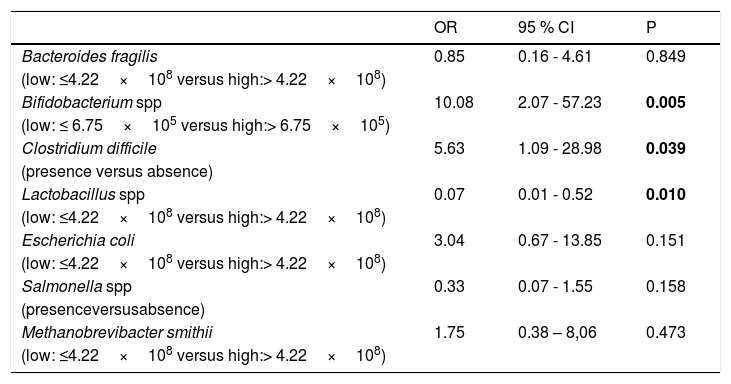
A multiple logistic regression was conducted with the occurrence of AD as the outcome variable. The dependent variables included in the model were 1. Presence or absence of Clostridium difficile and Salmonella spp., 2. Higher abundance (counts above the 50th percentile in the population studied) of Bacteroides fragilis, Bifidobacterium spp., Escherichia coli, Lactobacillus spp. and Methanobrevibacter smithii. According to the multiple logistic regression, the variables associated with AD were the presence of Clostridium difficile, greater abundance of Bifidobacterium and lower abundance of Lactobacillus (Table 4).
Multiple logistic regression analysis with atopic dermatitis as outcome variable.
| OR | 95 % CI | P | |
|---|---|---|---|
| Bacteroides fragilis | 0.85 | 0.16 - 4.61 | 0.849 |
| (low: ≤4.22×108 versus high:> 4.22×108) | |||
| Bifidobacterium spp | 10.08 | 2.07 - 57.23 | 0.005 |
| (low: ≤ 6.75×105 versus high:> 6.75×105) | |||
| Clostridium difficile | 5.63 | 1.09 - 28.98 | 0.039 |
| (presence versus absence) | |||
| Lactobacillus spp | 0.07 | 0.01 - 0.52 | 0.010 |
| (low: ≤4.22×108 versus high:> 4.22×108) | |||
| Escherichia coli | 3.04 | 0.67 - 13.85 | 0.151 |
| (low: ≤4.22×108 versus high:> 4.22×108) | |||
| Salmonella spp | 0.33 | 0.07 - 1.55 | 0.158 |
| (presenceversusabsence) | |||
| Methanobrevibacter smithii | 1.75 | 0.38 – 8,06 | 0.473 |
| (low: ≤4.22×108 versus high:> 4.22×108) |
Adjusted for socioeconomic status.
The multiple logistic regression analysis performed in our study has shown that higher prevalence of Clostridium difficile, greater abundance of Bifidobacterium spp. and lower abundance of Lactobacillus spp. in the gut microbiota are associated with AD in school-age children. The bivariate analysis has revealed a higher prevalence of Clostridium difficile and Salmonella spp. in the AD group. Regarding bacterial abundance, lower counts of Eubacteria, Bacteroides fragilis, Lactobacillus spp., Escherichia coli and Methanobrevibacter smithii have been found in the gut microbiota of children with AD. On the other hand, children in the AD group had higher counts of Bifidobacterium spp.
The relationship of gut microbiota with AD and other allergic diseases has been analyzed with both longitudinal and cross-sectional studies.8 Longitudinal studies have shown that certain characteristics are seen in the gut microbiota even before the onset of allergic diseases themselves. In this context, prospective studies with infants have shown higher clostridial counts in the first months of life in children who developed AD at both age two.14–16 In a clinical trial in which Lactobacillus rhamnosus GG was administered at the end of pregnancy and during the first months of life, increased abundance of Clostridium clusters IV and XIV have been found in children who developed eczema regardless of whether they received probiotic supplement or placebo.17 Our study, which assessed the relationship of the gut microbiota with AD in school-age children, has shown a higher prevalence of Clostridium difficile in children with AD. It can therefore be considered that bacteria of the Clostridium genus may be present both before the onset of AD as well as while the disease is active.
Certain bacteria that are part of the normal gut microbiota may have pathogenic potential, as is the case of some species of Bifidobacterium, Bacteroides, Clostridium and Eubacterium. Bacteroides fragilis and Clostridium perfringens have been associated with infectious processes, and their interaction with the immune system may participate in the pathophysiology of AD. Enterotoxigenic strains of Bacteroides fragilis and Clostridium difficile are known to cause diarrhea after the use of antibiotics.18 It is worth mentioning that interactions between the bacteria constituting the gut microbiota are known to occur. For example, several genera of bacteria, such as Bacteroides spp., Bifidobacterium spp., Lactobacillus spp., Pseudomonas spp., Staphylococcus spp. and Streptococcus spp. can inhibit the proliferation of Clostridium difficile.19 In this complex scenario of interactions involving the various components of the microbiota and the immune system, the exact mechanism by which Clostridium bacteria can contribute to the development and maintenance of AD remains unknown.
On the other hand, lactobacilli have been considered a protection factor against the development of allergic diseases in general,20,21 including AD.22 Our results confirmed a lower abundance of Lactobaccilus spp. in children with AD, in agreement with the literature.21–23 Lactobacilli are microorganisms that help maintain a healthy gut microbiota and are known for their beneficial effects on human health. The role of the lower abundance of lactobacilli in AD pathophysiology may be related to impaired T-cell regulatory response.24
Few studies have analyzed the gut microbiota of school-age children with AD. A cross-sectional study conducted in Japan that used stool culture tests showed that school-age children with AD had a higher prevalence of Staphylococcus aureus and lower counts of bifidobacteria.25 Another study in Estonia showed higher prevalence of Bifidobacterium adolescentis and lower prevalence of Bifidobacterium catenulatum, as well as lower bacterial diversity in school-age children with AD.10 Two other cross-sectional studies, carried out in Estonia26 and Italy,27 assessed the microbiota of children with allergic diseases, not necessarily AD. These studies showed a higher prevalence of Clostridium spp., a lower prevalence of bifidobacteria and lower counts of Akkermansia muciniphila, Faecalibacterium prausnitzii and Clostridium group IV in the stools of allergic children. Therefore, in general, the differences found in the gut microbiota of school-age children with AD in our study in South America are compatible with those of studies performed in countries located in other continents.
The greater presence of Clostridia in the microbiota of allergic children has been confirmed in many longitudinal studies and controlled cross-sectional studies, regardless of whether the analysis was performed before or after the onset of AD.
In our study, the abundance of bifidobacteria was higher in the AD group, but the difference found did not reach statistical significance (p=0.056). Bifidobacteria may be involved in immunoregulation mechanisms.24 This result contradicts the expectation that bifidobacteria could constitute a protective factor against AD. However, the genus Bifidobacterium was analyzed here without considering their species individually. In this context, the protective role described elsewhere could theoretically be played only by certain species of the genus. It should be noted that the literature is controversial on this issue.8
In our study, we found lower counts of Bacteroides fragilis, Escherichia coli and Methanobrevibacter smithii in children with AD, although these components have not been associated with AD in the multiple regression analysis. Lower abundance of Bacteroides fragilis and Escherichia coli was observed in the feces of Norwegian infants who later developed allergic symptoms.21 On the other hand, two other studies found a higher prevalence of Bacteroides fragilis in children with positive Asthma Predictive Index (API)28 and other allergic diseases.29
Our results have not shown differences in the prevalence and abundance of Escherichia coli in the microbiota of children with AD when compared to controls, unlike what was found for infants, for whom a greater abundance or prevalence of this bacterium has been associated with later development of AD.30 According to Avershina et al.,31Escherichia coli abundance is highest during the first year of life. However, the counts of this bacterium decrease at age two, proving that the abundance of Escherichia coli does not remain stable in the gut microbiota.
This paper is the first to document an association between AD and a lower abundance of Methanobrevibacter smithii. Methanobrevibacter smithii is associated with greater diversity in the gut microbiota, due to stimulating the growth of other fermenting bacteria in humans.32 These bacteria are also known to remain quantitatively stable over time in the human gut microbiota despite changes in diet and the use of antibiotics, which although effective against other gut bacteria do not seem to affect Methanobrevibacter smithii.33Methanobrevibacter smithii also has an impact on interactions within the intestinal ecosystem, such as promoting the production of lactobacilli and other protective bacteria in the intestinal tract, supporting greater microbial diversity and abundance. Therefore, the lower abundance of Methanobrevibacter smithii found in the microbiota of children with AD may be associated with the lower bacterial diversity found in allergic diseases.34,35
Despite differences in the prevalence of certain genera and species of bacteria in children with AD, no significant differences were observed between groups in the abundance of the Firmicutes and Bacteriodetes phyla. This result agrees with another study carried out in Italy.27
In our sample, AD was associated with higher socioeconomic status in the bivariate analysis; however, this association was not maintained in the multiple analysis. No associations were found regarding gender, age, neonatal history or living conditions, although these variables have been reported in other studies on factors associated with allergic diseases.36,37
AD is commonly associated with asthma and rhinitis.38 We observed an association of AD with asthma in 26.1 %, with rhinoconjunctivitis in 13.1 % and with asthma and rhinoconjunctivitis in 4.3 % of the school-age children included in the study. A European epidemiological study reported associations of AD with asthma and rhinoconjunctivitis in 15 % and 26 % of the study population, respectively. The literature shows a variation between 14.2 % and 52.7 % in the prevalence of asthma in children with atopic dermatitis.39 Therefore, the population analyzed in the present study lies within the expected limits for association between AD and other allergic diseases.
ConclusionWe found that the profile of the gut microbiota of children with AD is different when compared to that of children without allergic diseases. Specially, C. difficile was associated with AD. Moreover, the microbiota of AD children is characterized by a lower abundance of Lactobacillus and greater abundance of bifidobacterial, regardless of socioeconomic status. Further research must be performed do know the mechanisms by which intestinal microbes interfere with immune system.
This study was financed by the Fundação de Apoio à Pesquisa do Estado de São Paulo - Brasil (FAPESP) - Finance code 2009/18458-8.