It has been suggested that polymorphisms of histamine metabolising enzymes can be a risk factor for developing histamine-involving diseases. The aim of the present study is to research the possible association between two functional single nucleotide polymorphisms (SNPs): C314T in the Histamine-N-Methyl Transferase gene and C2029G in the Diamine Oxidase gene, with the severity of allergic rhinitis and the number of allergic diseases, in a group of allergic Mexican children.
MethodsWe studied 154 unrelated allergic children. SNPs were analysed by RT-PCR. The total serum IgE was measured by chemiluminescence and the serum histamine by ELISA. We used logistic regression analysis to determine OR.
ResultsPatients carrying the mutant allele for any SNP had more risk to develop higher rhinitis severity or a bigger number of allergic diseases. Haplotype analysis revealed that this effect is synergistic. In patients carrying one or two mutant alleles, serum histamine levels were higher than those of patients carrying only wild alleles. Serum IgE levels were not associated with the presence of mutant alleles.
ConclusionThe presence of these SNPs in patients with allergic rhinitis can lead to higher serum histamine, therefore to a higher risk of developing more severe symptoms or more associated allergic diseases, even if the serum IgE remains low.
Allergic rhinitis (AR) is the most common allergic disease. With a prevalence of 20–23%, it affects more than 500 million people worldwide.1,2 It is characterised by nasal symptoms including sneezing, rhinorrhoea and nasal blockage. AR, as other allergic diseases (AD) (asthma and eczema), is associated with an IgE-mediated immune response against allergens.3 It is believed that the disease may be the result of the interaction between different genetic alterations, each of which would contribute to a small defect. The heritability of AR has been estimated to be as high as 70–90%.3,4 Recently, special attention has been given to genes that may be implicated in AR and other AD. A series of genomic researches have been made, yielding different chromosomal associations, the most common being those involving chromosomes 2, 3, 4, 9 and 21. Single-nucleotide polymorphism (SNP) studies related to genes encoding for molecules implicated in the pathogenesis of AR have also been carried out. Such molecules comprise chemokines and their receptors, interleukins and their receptors, eosinophil peroxidase and leukotrienes, among others.5,6
As in other AD, histamine plays a principal role in the pathophysiology of AR. Some patients with AR are symptomatic only during the pollen season, while many others are allergic to multiple allergens including indoor allergens, which lead to perennial symptoms. Circulating IgE antibodies bind to the high affinity IgE receptor on mast cells and basophils, then they are crosslinked by allergen, initiating the secretion of inflammatory mediators including histamine.7 In this way, synthesis, activity and degradation of histamine could impact on the severity of AR or other AD symptoms.
It has been suggested that an over-expression of histidine decarboxylase (HD) gen is associated with rhinitis8 and that a polymorphism of this enzyme is more common among patients with rhinitis.9 Another study demonstrated that three SNPs of the H4 Histamine receptor are associated with infection-induced asthma.10
Histamine is degraded through two enzymes: histamine N-methyltransferase (HNMT, EC. 2.1.1.8) and diamine oxidase (DAO) or amiloride binding protein 1 (ABP1, EC 1.4.3.6). HNMT is involved in the inactivation of intracellular histamine11 while DAO is secreted and plays a role in the inactivation of extracellular histamine.12
The genes coding for HNMT and DAO are polymorphic. The HNMT gene, located in chromosome 2q22.1, shows eight SNPs, but only one is non-synonymous. This SNP is located in exon 4 and causes the amino acid substitution Thr105Ile (rs11558538).13 This SNP is unambiguously related to decreased enzyme activity.14
Three non-synonymous SNPs have been mapped to the DAO gene, located in chromosome 7q34–36. Functional impairment has been shown on serum DAO activity only for the SNP rs1049793, which codes for an altered protein with the amino acid substitution His645Asp.15 Because of this, we focused on both SNPs: Thr105Ile for HNMT, and His645Asp for DAO, which will be named C314T and C2029G respectively (for the allelic nucleotide substitution) in this study.
HNMT and DAO SNPs involvement has been investigated in several pathologies implying histamine participation, mainly in AD. The association of the HNMT C314T polymorphism with asthma was initially reported,16 although further independent studies failed to identify such an association.17,18 Recently, two studies have supported a positive association between these polymorphisms with asthma.19,20 On the other hand, two studies suggest that C314T polymorphism is a risk factor for eczema.21,22 DAO C2029G polymorphism has not been associated with the presence of any histamine related pathology. However, two studies were able to associate the severity of symptoms in Ulcerative Colitis23 and the intensity of symptoms of asthma and rhinitis24 with this polymorphism.
The aim of the present study is to investigate the possible association between the functional SNPs C314T in the HNMT gene and C2029G in the DAO gene, with the severity of rhinitis and the number of AD in a group of allergic Mexican children.
Materials and methodsWe studied 154 unrelated children from ages 3 to 18 years, with a mean of 7.8±3.84 (±SD). Patients were recruited from the Paediatric Allergy Department of the Health Secretary in Torreon, Coahuila, Mexico, between March 2013 and May 2014.
The diagnosis of AR was made according to the ARIA workshop.3 The Hanifin and Rajka Diagnostic Criteria were used for atopic dermatitis, while the GINA criteria were used for asthma. Patients who had a history of antihistaminic drugs within one month prior to the study were excluded from the study. The total serum IgE level was measured by chemiluminescence (kit Cat.LKIE1 IMMULITE IgE Total). The serum histamine was measured by ELISA (kit Cat. IB89128). An immediate hypersensitivity skin prick test was used to detect allergic reactions for a panel of 55 allergens: 4 epithelia, 2 mites, 9 fungi, 13 trees, 5 grasses, 12 weeds and 10 foods. Histamine was used as a positive control and saline solution as a negative control. A flare or induration higher than 3mm was considered positive.
All patients were invited to participate and signed an informed consent. This study was done according to the principles of the Declaration of Helsinki. The protocol was approved by the Ethics Committee of the Medicine School, University of Coahuila, Mexico.
Polymorphism analysisDNA was obtained from peripheral blood (5ml) using the Salting Out method.25 The polymorphisms were analysed by real-time polymerase chain reaction (RT-PCR). Genotyping was performed using TaqMan assays (supplied by Applied Biosystem®) designed to detect the following SNPs: for DAO, rs1049793 (C_7599774_10) a non-synonymous variant causing the amino acid substitution His664Asp, and for HNMT rs11558538 (C_11650812_20) a non-synonymous variant causing the amino acid substitution Thr105 Ile.
Polymorphisms were carried out by RT-PCR using the RT-PCR equipment (7300 real time PCR system by Applied Biosystem®, with software 7300 system). We used a concentration of 8ng of DNA. The amplification conditions were: after a denaturation time of 10min at 95°C, 40 cycles of 95°C for 15s and at 60°C for 90s were carried out. 308 samples were analysed (154 for HNMT and 154 for DAO polymorphisms).
Statistical analysisWe counted the frequencies for the HNMT and DAO SNPs. The statistical analysis was performed with the χ2 test, ANOVA or Kruskal–Wallis test, each when appropriate. The magnitude of association between HNMT and DAO mutations and phenotypic severity of AR were estimated by the odds ratio (OR) with a 95% confidence interval. To assess whether HNMT and DAO variants influence synergically upon the course of AR or upon the number of AD, subjects were classified as carriers and non-carriers of both SNPs, resulting in four different haplotypes. Logistic regression analysis was performed to assess if haplotypes were correlated with the severity of AR. A statistical analysis was performed using the software STATA 11.1®.
The Hardy–Weinberg equilibrium was confirmed using the software Arlequinver.2.000 (CMPG Zoological Institute, University of Berne, Berne, Switzerland).
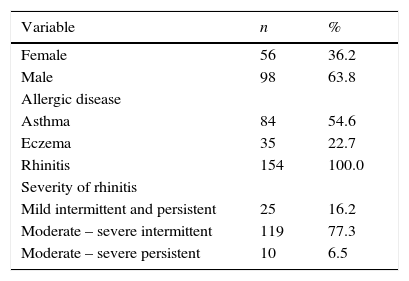
ResultsA total of 154 consecutive allergic children, of 7.8±3.84 (mean±SD.) years old were studied. All of them were diagnosed with AR and out of these, 84 also presented asthma, and 35 had eczema. The main clinical characteristics are shown in Table 1. Forty-five children (35.7%) only had one AD, 80 (51.9%) had two AD, and 19 (12.3%) had three AD (asthma, eczema and rhinitis). The range of serum IgE was 4.9–2224.0IU/L, and according to the cut-off point of 100IU/L, there were 41 children (26.6%) with low IgE levels. Five children (3.2%) were negative for the 56 allergens tested; one child was positive for 23 allergens.
Characteristics of the patients.
| Variable | n | % |
|---|---|---|
| Female | 56 | 36.2 |
| Male | 98 | 63.8 |
| Allergic disease | ||
| Asthma | 84 | 54.6 |
| Eczema | 35 | 22.7 |
| Rhinitis | 154 | 100.0 |
| Severity of rhinitis | ||
| Mild intermittent and persistent | 25 | 16.2 |
| Moderate – severe intermittent | 119 | 77.3 |
| Moderate – severe persistent | 10 | 6.5 |
| Mean±SD | Range | |
| Age (years) | 7.8±3.8 | 3–18 |
| No. of allergic diseases | 1.76±0.654 | 1–3 |
| STP (+) | 7.01±3.7 | 0–23 |
| Serum histamine (μg/L) | 0.81±0.555 | 0.13–2.90 |
| Median | IQ range | |
| Serum IgE (IU/L) | 215 | 215–488 |
All children had allergic symptoms and all had allergic rhinitis. Some children (19, 12.3%) had the three AD (asthma, eczema and rhinitis). 56 allergens were proven in each child and the range of positivity to these Skin Prick Test (SPT +) was of 0 to 23 allergens (median=7), 42 children (27.3%) had serum IgE below 100IU/L.
Genotype distribution of HNMT polymorphism was in concordance with Hardy–Weinberg law, but genotype distribution of DAO polymorphism was in discordance with it, that is, there was a significant difference (p=0.02) between genotype distribution of this SNP in our allergic patients and the healthy Hispanic population.26
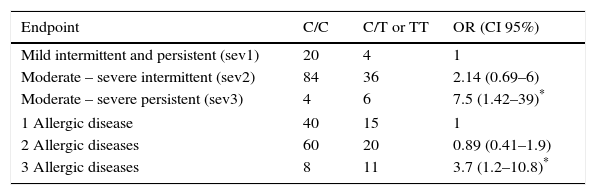
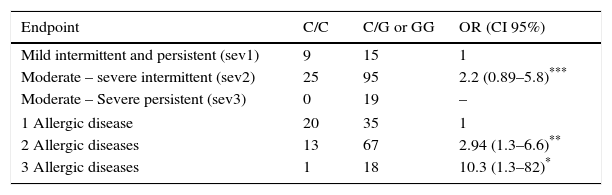
Association was found between the mutant allele of C314T HNMT polymorphism and the highest severity (moderate-severe persistent) of AR (OR=7.5, p=0.017), and with the highest number (3) of AD (OR=3.7, p=0.019) (Table 2). On the other hand, an association between the mutant allele of C2029G DAO polymorphism and the severity of AR was not found. However it was found with the number of AD: two AD (OR=2.9, p=0.009) and three AD (OR=10.3, p=0.029) (Table 3).
Association between genotypes for HNMT polymorphism, the intensity of symptoms of rhinitis and the number of allergic diseases in each child (asthma, eczema and rhinitis).
| Endpoint | C/C | C/T or TT | OR (CI 95%) |
|---|---|---|---|
| Mild intermittent and persistent (sev1) | 20 | 4 | 1 |
| Moderate – severe intermittent (sev2) | 84 | 36 | 2.14 (0.69–6) |
| Moderate – severe persistent (sev3) | 4 | 6 | 7.5 (1.42–39)* |
| 1 Allergic disease | 40 | 15 | 1 |
| 2 Allergic diseases | 60 | 20 | 0.89 (0.41–1.9) |
| 3 Allergic diseases | 8 | 11 | 3.7 (1.2–10.8)* |
OR was obtained comparing sev2 or sev3 with sev1; as well as comparing 2 or 3 AD with 1 AD. OR were calculated by logistic regression analysis and adjusted by age, sex and BMI.
Association between genotypes for DAO polymorphism with the intensity of symptoms of rhinitis and with the number of allergic diseases in each child (asthma, eczema and rhinitis).
| Endpoint | C/C | C/G or GG | OR (CI 95%) |
|---|---|---|---|
| Mild intermittent and persistent (sev1) | 9 | 15 | 1 |
| Moderate – severe intermittent (sev2) | 25 | 95 | 2.2 (0.89–5.8)*** |
| Moderate – Severe persistent (sev3) | 0 | 19 | – |
| 1 Allergic disease | 20 | 35 | 1 |
| 2 Allergic diseases | 13 | 67 | 2.94 (1.3–6.6)** |
| 3 Allergic diseases | 1 | 18 | 10.3 (1.3–82)* |
OR were calculated by logistic regression and adjusted by age, sex and BMI. OR were obtained comparing sev2 or sev3 with sev1; as well as comparing 2 or 3 AD with 1 AD. OR for sev3 could not be calculated because of the absence of patients with this severity in the group of wild homozygous individuals.
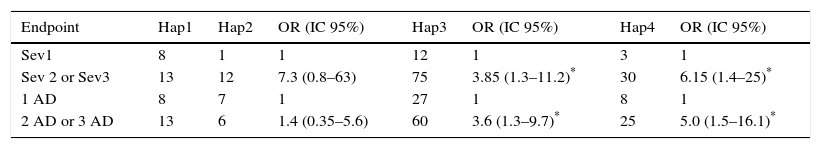
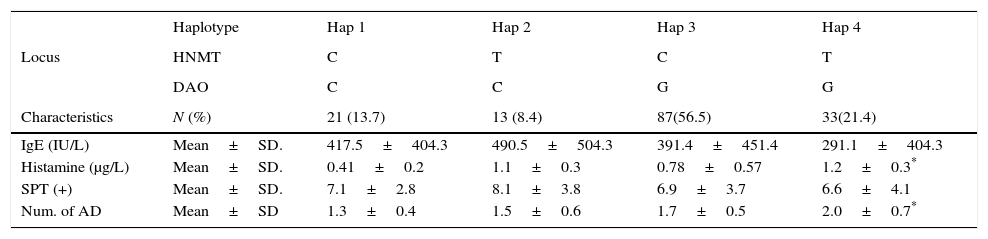
We considered four haplotypes: Haplotype 1 (Hap1) only has wild alleles for the two locus, haplotype 2 (Hap2) has the mutated allele in the HNMT locus, haplotype 3 (Hap3) has the mutated allele in the DAO locus, and haplotype 4 (Hap 4) contains mutated alleles in both locus (either as homozygous or heterozygous individuals). We found that Hap3 (OR=3.8, p=0.014) and Hap4 (OR=6.1, p=0.016) were associated with the severity of AR. Association between these haplotypes with the number of AD was also found: Hap3 (OR=3.6, p=0.011) and Hap4 (OR=5, p=0.007) (Table 4).
Association between haplotype with the intensity of symptoms of rhinitis and with the number of allergic diseases in each child (asthma, eczema and rhinitis).
| Endpoint | Hap1 | Hap2 | OR (IC 95%) | Hap3 | OR (IC 95%) | Hap4 | OR (IC 95%) |
|---|---|---|---|---|---|---|---|
| Sev1 | 8 | 1 | 1 | 12 | 1 | 3 | 1 |
| Sev 2 or Sev3 | 13 | 12 | 7.3 (0.8–63) | 75 | 3.85 (1.3–11.2)* | 30 | 6.15 (1.4–25)* |
| 1 AD | 8 | 7 | 1 | 27 | 1 | 8 | 1 |
| 2 AD or 3 AD | 13 | 6 | 1.4 (0.35–5.6) | 60 | 3.6 (1.3–9.7)* | 25 | 5.0 (1.5–16.1)* |
Haplotypes constitutions are explained in the text. Hap1 are wild homozygous for both alleles. OR were obtained comparing Hap2, Hap3 or Hap4 with Hap1. OR were calculated by logistic regression analysis and adjusted by age, sex and BMI.
Comparing some characteristics of the patients grouped by haplotypes, it was evident that the presence of the mutated alleles increases the level of serum histamine (p=0.0001) and the number of AD in the patients (p=0.0020). The mutated alleles did not have any effect on the serum levels of IgE nor on the positivity to allergens tested by Skin Prick Test (SKP+), as can be observed in Table 5.
Main characteristics of the allergic children grouped by haplotype.
| Haplotype | Hap 1 | Hap 2 | Hap 3 | Hap 4 | |
|---|---|---|---|---|---|
| Locus | HNMT | C | T | C | T |
| DAO | C | C | G | G | |
| Characteristics | N (%) | 21 (13.7) | 13 (8.4) | 87(56.5) | 33(21.4) |
| IgE (IU/L) | Mean±SD. | 417.5±404.3 | 490.5±504.3 | 391.4±451.4 | 291.1±404.3 |
| Histamine (μg/L) | Mean±SD. | 0.41±0.2 | 1.1±0.3 | 0.78±0.57 | 1.2±0.3* |
| SPT (+) | Mean±SD. | 7.1±2.8 | 8.1±3.8 | 6.9±3.7 | 6.6±4.1 |
| Num. of AD | Mean±SD | 1.3±0.4 | 1.5±0.6 | 1.7±0.5 | 2.0±0.7* |
Haplotypes constitutions are explained in the text. STP+: Skin Prick Test positive (number of positivity to tested allergens). AD (asthma, eczema and rhinitis). IgE values were compared by Kruskal–Wallis Test. Histamine, STP (+) and number of AD were compared by ANOVA.
The AR, like other atopic diseases, is the result of an allergen-triggered response mediated by a series of inflammatory cells and mediators (including histamine). Histamine related genes may be important in the pathogenesis of rhinitis. These results indicate that the polymorphism of HNMT and DAO enzymes involved in the degradation of circulating histamine may influence the clinical presentation of AR. Individuals who are carriers of HNMT C314T mutated allele developed significantly more severe clinical symptoms of AR and more AD (even with low levels of serum IgE). Individuals who are carriers of DAO C2029G mutated allele tend to develop more severe symptoms of rhinitis too, but this did not have any significance for severity 2 (moderate-severe intermittent). However, for severity 3 (moderate-severe persistent), all the patients carrying the mutant G allele had this severity. Because of this, it was not possible to calculate the OR for severity 3, but it is evident that individuals carrying the mutant allele of DAO C2029G polymorphism have a high risk of developing a severe AR. The presence of the mutant allele clearly increases the risk to develop more AD additionally to AR.
Haplotypes analysis suggests that the presence of both mutations in the same individuals not only increases the risk of higher severity of AR, but also the risk of more AD (Table 4), which is accompanied by a significant increase in the serum level of histamine, but not by an increment in IgE levels (Table 5). Thus, allergic patients carrying the mutant allele for HNMT C314T or DAO C2029G polymorphisms have a higher risk to develop a more severe AR. If both mutant alleles are present, the risk is even higher, even if the serum IgE remains low. There is more risk to develop a bigger number of AD too.
These findings are consistent with those observed in Hispanic patients with Ulcerative Colitis, in which DAO C2029G polymorphism was associated with the severity of the disease,23 and with those observed in Hispanic patients with asthma and rhinitis, in which carriers of the DAO mutate allele are more prone to developing symptoms, even with low levels of IgE.24 In these studies, HNMT C314T polymorphism does not seem to be associated with the severity of the disease. The interaction between DAO and HNMT polymorphisms was not proven.
We hypothesised that the low frequency of the HNMT SNP did not allow to see some effects, as others have demonstrated for asthma19,20 or eczema.21,22
Garcia-Martin (2007) demonstrated lower levels of IgE in patients carrying the DAO mutant allele24; for the high variability of IgE levels in our patients, this cannot be demonstrated (although a tendency to lower levels of IgE was observed in Hap 2 and Hap3 patients). However, from all the 41 patients with low serum IgE (<100IU/L), only two were in the Hap1 group, and the rest were in the haplotype containing mutate alleles groups. This differential distribution reaches statistical significance (Xi2 test, p=0.04), suggesting that in another condition (with an adequate degradation of histamine) these patients would not have developed symptoms. In this condition (low IgE) the histamine production should be low, but if its degradation was decreased, the levels of serum histamine could be high. In fact, this could be observed, and our patients had increasing levels of histamine through Hap1, Hap 2, Hap 3 and Hap 4 (Table 5).
Because histamine is metabolised by two polymorphic enzymes, it could be possible that individuals with impaired ability to metabolise histamine may be more likely to develop histamine-related symptoms. Although histamine plasma levels are highly variable due to circadian rhythm and inter-individual variability in patients with AD.27 Several independent studies indicate an increase of histamine and a decrease of the histamine metabolite, N-methylhistamine in plasma among patients with asthma, as compared with healthy subjects.28–30 Thus, it may be speculated that individuals with altered histamine degradation may display atopic symptoms even though the amount of histamine released is not high.
The development of AR entails a complex interaction between genetic predisposition and environmental exposure to different factors, of which the most important is the implicated allergen. There is a clear hereditary component in AR that has been well corroborated by segregation studies and investigations in twins.6 From the strictly genetic perspective, it is believed that the disease may be the result of the interaction of different genetic alterations, each one of which would contribute to a small defect. Among these altered genes, HNMT and DAO histamine metabolising enzymes can be relevant, as is suggested by this study.
In this study the sample size prevented us from observing some potential relevant associations. However, this study was conducted as a pilot study to explore potential associations for further exploration in larger and better defined cohorts. We recognise that stratification with a small sample may lead to erroneous results, however a Type II error would more likely be the outcome, instead of finding an association when none truly exists.
Future studies investigating the functional activity of histamine receptors and enzymes are needed to clearly elucidate potential implications of gene variants in the histamine degradation pathway on AD. Such findings may be important for better understanding AR pathophysiology and in guiding improved and more directed therapies for patients with AR.
Ethical disclosuresPatients’ data protection and confidentiality of dataThe authors declare that they have followed the protocols of the WMA Declaration of Helsinki, and that they have received the approval of an Ethics Committee, attending the rules on the publication of patient data and that all the patients included in the study have received sufficient information and have given their informed consent in writing to participate in that study.
Right to privacy and informed consentThe authors have obtained the informed consent of the patients and/or subjects mentioned in the article. The author for correspondence is in possession of this document.
Protection of human subjects and animals in researchThe authors declare that the procedures followed were in accordance with the regulations of the responsible Clinical Research Ethics Committee and in accordance with those of the World Medical Association and the Helsinki Declaration.
Conflict of interestThe authors declare no conflicts of interest.