Cytokines, including interleukin-1 (IL-1), seem to contribute towards the pathogenesis of juvenile idiopathic arthritis (JIA), so this study was designed to evaluate the associations of IL-1 gene cluster and IL-1 receptor (IL-1R) gene single nucleotide polymorphisms (SNPs) with JIA proneness in Iranian population.
Materials and methodsGenomic DNA of 55 Iranian patients with JIA and 140 controls were extracted and typed for IL-1α gene at position −889, IL-1β gene at positions −511 and +3962, IL-1R gene at position Pst-I 1970, and interleikin-1 receptor antagonist (IL-1Ra) gene at position Mspa-I 11100, using polymerase chain reaction with sequence-specific primers method, and compared between patients and controls.
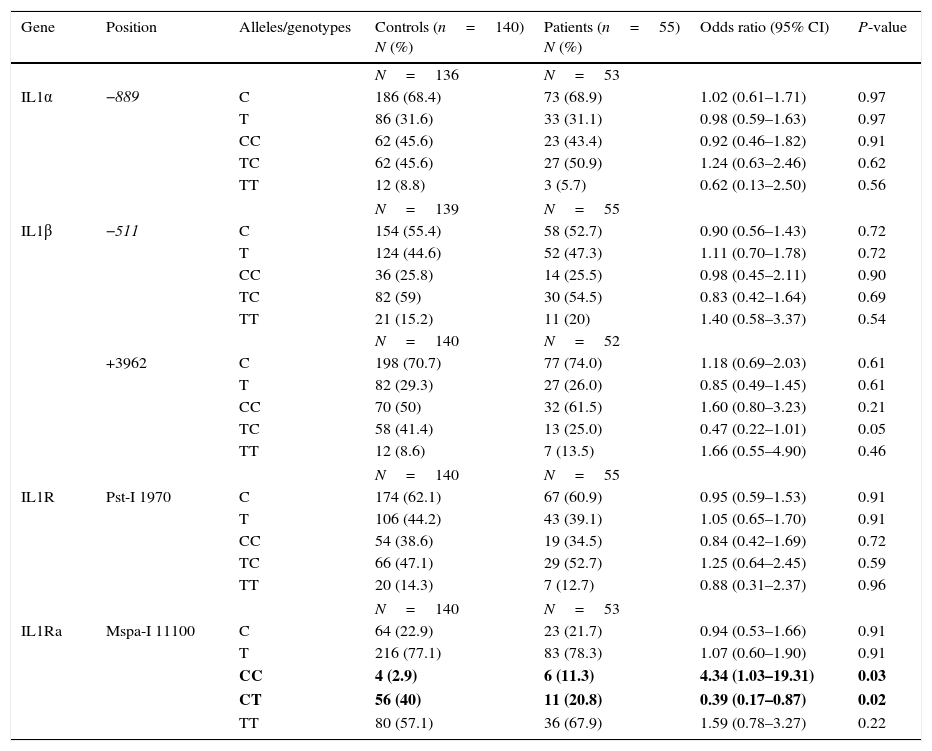
ResultsThe CC genotype of IL-1Ra at Mspa-I 11100 position was found to be more frequent in patients with JIA compared to healthy individuals (P=0.03), although the CT genotype at the same position was significantly higher in the control group in comparison with patients with JIA (P=0.02). No significant differences were observed between the two groups of case and control for IL-1α (−889 C/T), IL-1β (−511 C/T and +3962 C/T) and IL-1R (Pst-1 1970 C/T).
ConclusionThe results of the present investigation suggest that certain IL-1Ra gene variants are associated with individuals’ susceptibility to JIA. Nevertheless, further studies are required to establish the results of the current study.
Juvenile idiopathic arthritis (JIA) is a common autoimmune disease, characterised by chronic arthritis of unknown aetiology with onset prior to the age of 16 years. It has been previously propounded that numerous risk alleles from various susceptibility genes, including a multitude of major histocompatibility complex and non-major histocompatibility complex regions, predispose individuals to development of JIA following exposure to as yet unravelled environmental factors.10 Given the increased efficacy of the early prescription of disease-modifying anti-rheumatic drugs, it stands to reason that identification of other genetic markers associated with individual susceptibility to JIA would be valuable to initiate therapy at an early stage of the disease.
Cytokines such as interleukin-1 (IL-1) are known to be the central mediators of joint inflammation, found in both the synovial fluid and sera of patients with JIA. The aforementioned immune mediators with polymorphic gene sequences have been suggested as potential markers of individuals’ susceptibility to JIA.5,9,18
To date, various single nucleotide polymorphisms (SNPs) in different cytokines genes, influencing their level of synthesis, have been studied in the context of rheumatologic disorders, such as juvenile-onset systemic lupus erythematosus.14,20,22,24 However, to the best of our knowledge, no association study has been conducted on interleukin-1 gene cluster and interleukin-1 receptor polymorphisms in an Iranian population with JIA.
The aim of the current investigation was to study possible genetic contributions of selected cytokine polymorphisms (IL-1α at position −889, IL-1β at positions −511 and +3962, interleukin-1 receptor (IL-1R) at position Pst-1 1970 and interleukin-1 receptor antagonist (IL-1Ra) at position Mspa-I 11100 C/T) on JIA vulnerability in a population of Iranian patients with JIA.
Patients and methodsSubjectsA total of 55 consecutive JIA patients, recruited from the Rheumatology Clinic of the Children's Medical Centre Hospital, the Paediatrics Centre of Excellence in Iran, were enrolled in the present study as the case group and compared to 140 healthy unrelated controls, randomly selected from blood donors at Iranian blood transfusion organisations.3 The ILAR classification criteria for JIA were used to establish the diagnosis of JIA.19 Our patients’ group consisted of 25 individuals with oligoarticular JIA, 19 with polyarticular JIA, and 11 with systemic disease subtype.
Written informed consents were obtained from all participants according to the guidelines of the Ethical Committee of Tehran University of Medical Sciences prior to enrolment.
Sampling and genotypingFor all of the entrants to this study, 5ml of peripheral blood samples were obtained and kept with EDTA at −20°C until the extraction of genomic DNA using the “salting out” technique.16 Polymerase chain reaction with sequence-specific primers (PCR-SSP) assay (PCR-SSP kit, Heidelberg University, Heidelberg, Germany) was employed for cytokine gene typing as discussed previously.3 This assay uses the self-same amplification and detection conditions, resulting in speedy and cost-efficient analysis of polymorphisms. Amplification of the extracted DNA was carried out by a thermal cycler Techne Flexigene apparatus (Rosche, Cambridge, UK) under the following conditions: prime denaturation at 94°C for 2min; denaturation at 94°C for 10s; annealing+extension at 65°C for 1min (10 cycles); denaturation at 94°C for 10s; annealing at 61°C for 50s; extension at 72°C for 30s (20 cycles). The availability of polymerase chain reaction (PCR) products was visualised by 2% agarose gel electrophoresis, followed by ultraviolet transillumination. Additionally, photography for interpretation and documentation was performed. Each of the primer mixes contained a control primer pair that amplified either a part of the C-reactive protein (CRP) gene or a part of the β-globin gene. The β-globin control primers produce an 89bp fragment, while the primer pairs amplifying the CRP gene produced a 440bp amplicon.13 The five single nucleotide polymorphisms (SNPs) investigated were IL-1α (−889 C/T; rs1800587), IL-1β (−511 C/T; rs16944 and +3962 C/T; rs1143634), IL-1R (Pst-1 1970 C/T; rs2234650) and IL-1Ra (Mspa-I 11100 C/T; rs315952).
Statistical analysisWe assessed the allele and genotype frequencies by direct counting and compared with the controls using the chi-square test. The odds ratio (OR) and 95% confidence interval (CI) were calculated for each allele and genotype in both case and control groups. Adherence to the Hardy–Weinberg equilibrium constant was evaluated using chi-square test. The P value of less than 0.05 was considered to be statistically significant.
ResultsAllelic and genotype frequencies in Iranian patients with JIA and healthy individuals are depicted in Table 1.
Allele and genotype frequencies in Iranian patients with juvenile idiopathic arthritis and healthy individuals.
| Gene | Position | Alleles/genotypes | Controls (n=140) N (%) | Patients (n=55) N (%) | Odds ratio (95% CI) | P-value |
|---|---|---|---|---|---|---|
| N=136 | N=53 | |||||
| IL1α | −889 | C | 186 (68.4) | 73 (68.9) | 1.02 (0.61–1.71) | 0.97 |
| T | 86 (31.6) | 33 (31.1) | 0.98 (0.59–1.63) | 0.97 | ||
| CC | 62 (45.6) | 23 (43.4) | 0.92 (0.46–1.82) | 0.91 | ||
| TC | 62 (45.6) | 27 (50.9) | 1.24 (0.63–2.46) | 0.62 | ||
| TT | 12 (8.8) | 3 (5.7) | 0.62 (0.13–2.50) | 0.56 | ||
| N=139 | N=55 | |||||
| IL1β | −511 | C | 154 (55.4) | 58 (52.7) | 0.90 (0.56–1.43) | 0.72 |
| T | 124 (44.6) | 52 (47.3) | 1.11 (0.70–1.78) | 0.72 | ||
| CC | 36 (25.8) | 14 (25.5) | 0.98 (0.45–2.11) | 0.90 | ||
| TC | 82 (59) | 30 (54.5) | 0.83 (0.42–1.64) | 0.69 | ||
| TT | 21 (15.2) | 11 (20) | 1.40 (0.58–3.37) | 0.54 | ||
| N=140 | N=52 | |||||
| +3962 | C | 198 (70.7) | 77 (74.0) | 1.18 (0.69–2.03) | 0.61 | |
| T | 82 (29.3) | 27 (26.0) | 0.85 (0.49–1.45) | 0.61 | ||
| CC | 70 (50) | 32 (61.5) | 1.60 (0.80–3.23) | 0.21 | ||
| TC | 58 (41.4) | 13 (25.0) | 0.47 (0.22–1.01) | 0.05 | ||
| TT | 12 (8.6) | 7 (13.5) | 1.66 (0.55–4.90) | 0.46 | ||
| N=140 | N=55 | |||||
| IL1R | Pst-I 1970 | C | 174 (62.1) | 67 (60.9) | 0.95 (0.59–1.53) | 0.91 |
| T | 106 (44.2) | 43 (39.1) | 1.05 (0.65–1.70) | 0.91 | ||
| CC | 54 (38.6) | 19 (34.5) | 0.84 (0.42–1.69) | 0.72 | ||
| TC | 66 (47.1) | 29 (52.7) | 1.25 (0.64–2.45) | 0.59 | ||
| TT | 20 (14.3) | 7 (12.7) | 0.88 (0.31–2.37) | 0.96 | ||
| N=140 | N=53 | |||||
| IL1Ra | Mspa-I 11100 | C | 64 (22.9) | 23 (21.7) | 0.94 (0.53–1.66) | 0.91 |
| T | 216 (77.1) | 83 (78.3) | 1.07 (0.60–1.90) | 0.91 | ||
| CC | 4 (2.9) | 6 (11.3) | 4.34 (1.03–19.31) | 0.03 | ||
| CT | 56 (40) | 11 (20.8) | 0.39 (0.17–0.87) | 0.02 | ||
| TT | 80 (57.1) | 36 (67.9) | 1.59 (0.78–3.27) | 0.22 | ||
Values in bold indicate significant P-value.
The CC genotype of IL-1Ra at Mspa-I 11100 position was shown to be more frequent in patients with JIA, compared to healthy individuals (11.3% vs. 2.9%; P value, 0.03), although the CT genotype at the same position was significantly higher in the control group in comparison with patients with JIA (40% vs. 20.8%; P value, 0.02). The allelic frequencies of IL-1Ra at Mspa-I 11100 position were similar in the two groups of patients and controls. Additionally, no significant difference was observed between the two groups of case and control for IL-1α (−889 C/T), IL-1β (−511 C/T and +3962 C/T) and IL-1R (Pst-1 1970 C/T).
Furthermore, we observed no significant difference between the aforementioned gene variants and individuals’ susceptibility to different JIA subtypes, including systemic, polyarticular, and oligoarticular JIA.
DiscussionAccording to our knowledge, this is the first investigation assessing the association between IL-1α, IL-1β, IL-1R and IL-1Ra gene polymorphisms and JIA susceptibility in Iranian population. We found the CC genotype of IL-1Ra at Mspa-I 11100 position to be more frequent in patients with JIA in comparison with healthy controls, while the CT genotype at the same position was significantly lower in the patients with JIA than the healthy controls. On the other hand, we observed no significant differences between the two groups of case and control for IL-1α (−889 C/T), IL-1β (−511 C/T and +3962 C/T) and IL-1R (Pst-1 1970 C/T).
The superfamily of IL-1 comprises pro-inflammatory cytokines, including IL-1α as well as IL-1β, receptors (IL-1R) and antagonist molecules (IL-1Ra), playing the role as the regulator of the innate immunity altogether.6 This gene cluster maps to 2q12.11 IL-1α mostly exerts autocrine and paracrine functions, stimulating local inflammation, while IL-1β mainly triggers systemic inflammation. Binding of IL-1 to its receptor, expressed as both a membrane receptor on the B lymphocytes and macrophages surfaces together with a soluble form, initiates the pro-inflammatory cascade, eventually culminating in the activation of immune cells, the stimulation of the differentiation of T helper (Th) lymphocytes in Th17, the activation of endothelial cells, synovial fibroblasts, and osteoclasts, the promotion of the synthesis of colony stimulating factors, and the stimulation of the chondrocytes to secrete matrix degrading enzyme.6,17 IL-1 signalling is inhibited by IL-1Ra, a glycosylated protein of 22kDa with high affinity to IL-1R, which is expressed in many cells, such as keratinocytes and intestinal epithelial cells.7 It has been previously demonstrated that deficiency in IL-1Ra could result in inflammatory arthritis suggestive of rheumatoid arthritis (RA) in mice.4 The present study has evaluated the possible correlations of the IL-1α (−889 C/T), IL-1β (−511 C/T and +3962 C/T), IL-1R (Pst-1 1970 C/T) and IL-1Ra (Mspa-I 11100 C/T) to JIA vulnerability. Our results are in line with the findings of the study conducted by Allam et al.,2 which found no significant association between IL-1β −511 SNP and RA susceptibility in an Algerian population. Similarly, You et al. observed no significant differences in the distribution of IL-1β −511 SNP between the two groups of RA patients and controls in a Chinese Han population.23 In another study performed by Aggarwal et al., no difference in genotype frequency among patients with enthesitis related arthritis (ERA) category of JIA and controls for IL-1Ra, IL-1α −889 and IL-1β −511 was detected.1 Consistently, Havemose-Poulsen et al. observed no significant correlations between IL-1α −889 and IL-1β −511 SNPs and individuals proneness to both JIA and RA in a Danish population.12 Moreover, Donn et al. detected no association between IL-1α -889 polymorphism and UK oligoarticular JIA patients.8 On the other hand, Shafia et al. suggested that the IL1β −511 C allele could have a protective role from RA development in the ethnic Kashmiri population.21 Additionally, McDowell et al. have reported an increased carriage of IL-1α −889 T allele in patients with early-onset, polyarticular JIA compared with controls.15
The present study has a number of limitations to be acknowledged. Firstly, the relatively small number of the entrants to this study could both diminish the statistical power of our investigation and inhibit us from comparing different JIA subgroups so as to evaluate the probable variations in the distribution of IL-1 family gene polymorphisms among JIA subgroups. Furthermore, we were unable to discover the effects of different IL-1 family gene variants on the level of cytokine production due to our inability to measure both the synovial and serum levels of IL-1.
ConclusionTo conclude, the results of the current study suggest that certain IL-1Ra gene variants could render individuals more susceptible to the development of JIA, while the aforementioned IL-1α, IL-1β and IL-1R gene polymorphisms are not the significant factors influencing the pathogenesis of JIA in Iranian population. However, further investigations, using larger sample sizes in different ethnicities are required so as to fully understand the possible role of IL-1 family gene polymorphisms in the aetiopathogenesis of JIA.
It should be noted that there is no ethical problem (approved by the research ethics committee of Tehran University of Medical Sciences) or conflict of interest in our research. There was no honorarium, grant, or other form of payment to authors to produce the manuscript.
Ethical disclosuresConfidentiality of dataThe authors declare that no patient data appears in this article.
Right to privacy and informed consentThe authors have obtained the informed consent of the patients and/or subjects mentioned in the article. The author for correspondence is in possession of this document.
Protection of human subjects and animals in researchThe authors declare that the procedures followed were in accordance with the regulations of the responsible Clinical Research Ethics Committee and in accordance with those of the World Medical Association and the Helsinki Declaration.
FundingThis study was funded by Tehran University of Medical Sciences and Health Services (grant number: 92-03-30-24902).
Conflict of interestThe authors have no conflict of interest to declare.