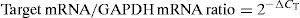ErbB family receptors and tight junction proteins participate in the pathologic process including tissue remodelling of inflammatory diseases in the upper and lower respiratory tracts. This study aimed at investigating the expressions of erbB1, 2, 3, 4, and a tight junction protein, claudin-1, in the nasal mucosa of patients with chronic hypertrophic rhinitis.
MethodsInferior turbinates were collected from 10 turbinectomised patients with allergic and non-allergic chronic hypertrophic rhinitis. The expressions of erbB1, 2, 3, 4, and claudin-1 were examined by fluorescence immunohistochemistry and by quantitative real-time transcription-polymerase chain reaction (qRT-PCR).
ResultsAll erbB1-4 and claudin-1 were detected, and mainly localised in the epithelial cells and nasal gland cells. The immunoreactivity for claudin-1 was positively correlated with the expressions of erbB1, 2 and 4, but negatively correlated with that of erbB3. The mRNA expressions of erbB1, 2 and 4 were positively correlated with one another, whereas the expression of erbB3 showed negative correlation with the immunoreactivity for erbB2 and 4.
ConclusionsThese results suggest a possible participation of erbBs and claudin-1 in tissue remodelling in chronic hypertrophic rhinitis.
Chronic hypertrophic rhinitis is one of the common sequelae of long-term allergic/non-allergic inflammation, habitual use of topical nasal vasoconstrictors, and vasomotor reaction. Patients with chronic hypertrophic rhinitis generally complain of intractable nasal obstruction, which often causes headache, fatigue, thirst, lack of concentration, daytime cognitive deficits, daytime sleepiness, and sleep disturbance, eventually leading to a decline in quality of life. Other nasal symptoms such as nasal discharge, sneezing, and postnasal drip can usually be managed by drug therapy. However, nasal obstruction caused by irreversibly hypertrophied nasal mucosa is resistant to conservative treatment, and often forces patients to sustain surgical treatment, including inferior turbinectomy.1,2 Despite a number of clinical studies on chronic hypertrophic rhinitis,3,4 the pathogenesis of hypertrophied nasal mucosa is not fully understood.
ErbB family receptors, also referred to as type I receptor tyrosine kinases, consist of four subtypes, erbB1, erbB2, erbB3, and erbB4. They play important roles in the proliferation, activation, survival, differentiation, migration, and neoplastic transformation of epithelial cells, mesenchymal cells, endothelial cells, and nerve cells.5–10 In recent years, erbBs have been reported to participate in the pathological process of inflammatory diseases in the upper and lower respiratory tracts.11–14 Recent research into cell biology has also revealed that regulation and activation of erbBs are associated with tight junction integrity,15,16 which is closely related to epithelial development, injury and tumorigenesis.17 This study aimed at investigating the expressions of erbB1, 2, 3, 4, and a tight junction protein, claudin-1, in the nasal mucosa of patients with chronic hypertrophic rhinitis.
Materials and methodsPatients and sample collectionSamples were obtained from 10 patients with chronic hypertrophic rhinitis, consisting of six allergic and four non-allergic patients (seven men and three women ranging in age from 17 to 63 years, with an average age of 48 years). Chronic hypertrophic rhinitis was diagnosed by clinical history, rhinoscopic examination, and computed tomography. Patients with sinonasal tumours, acute rhinosinusitis, acute upper/lower airway infections, chronic bronchitis, and/or bronchial asthma were excluded from the study. Total serum IgE levels were measured by a radioimmunosorbent test (RIST), and those over 170U/ml were considered to be positive. Specific serum IgE levels were determined by radioallergosorbent tests (RAST) for allergic antigens, including house dust mites, Japanese cedar pollen, orchard grass pollen, short ragweed pollen, cypress pollen, mugwort pollen, timothy grass pollen, Candida, Aspergillus, and Alternaria. RAST was considered to be positive when at least one item was positive. Patients were judged allergic when RIST and/or RAST were positive.
Inferior turbinectomy was performed via a transnasal endoscopic approach under general anaesthesia. For immunohistochemistry, the specimens were fixed with 4% paraformaldehyde in 0.1M phosphate buffer at pH 7.4 (PB) at 4°C overnight. For quantitative reverse transcription-polymerase chain reaction (qRT-PCR), the specimens were soaked in RNA stabilisation reagent (Qiagen Inc., Valencia, CA, USA) at 4°C overnight and then stored at −80°C until use.
The state of disease was considered severe in all the patients from clinical symptoms and macro/microscopic findings; they had complained of persistent nasal obstruction for years, and exhibited marked and irreversible hypertrophy of the inferior turbinates on rhinoscopy. The turbinates histologically showed noticeable changes such as submucosal fibrosis, goblet cell hyperplasia, and squamous metaplasia with loss of cilia. Informed consent was obtained from the patients, and the study was approved by the Institutional Review Board of the University of Occupational and Environmental Health.
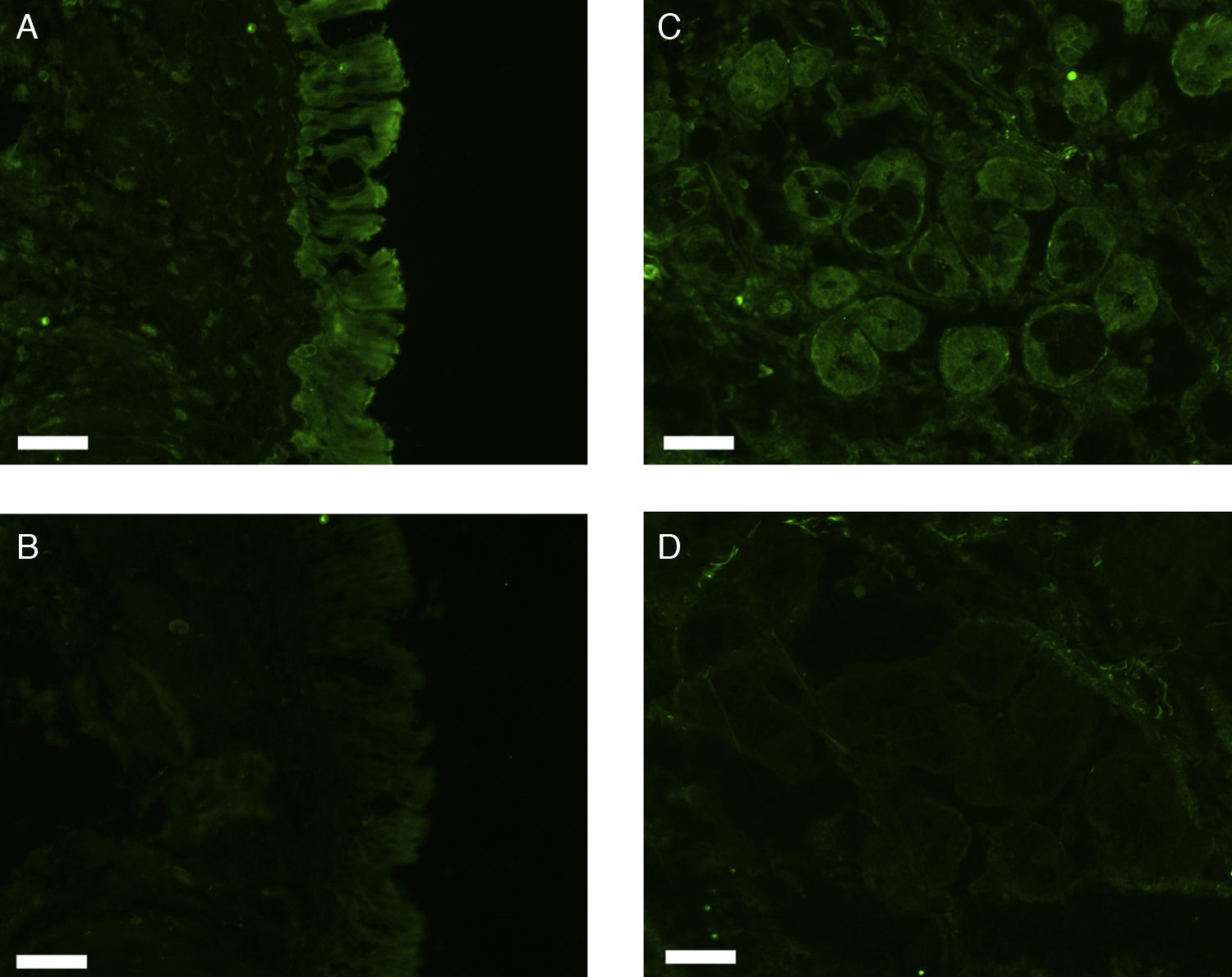
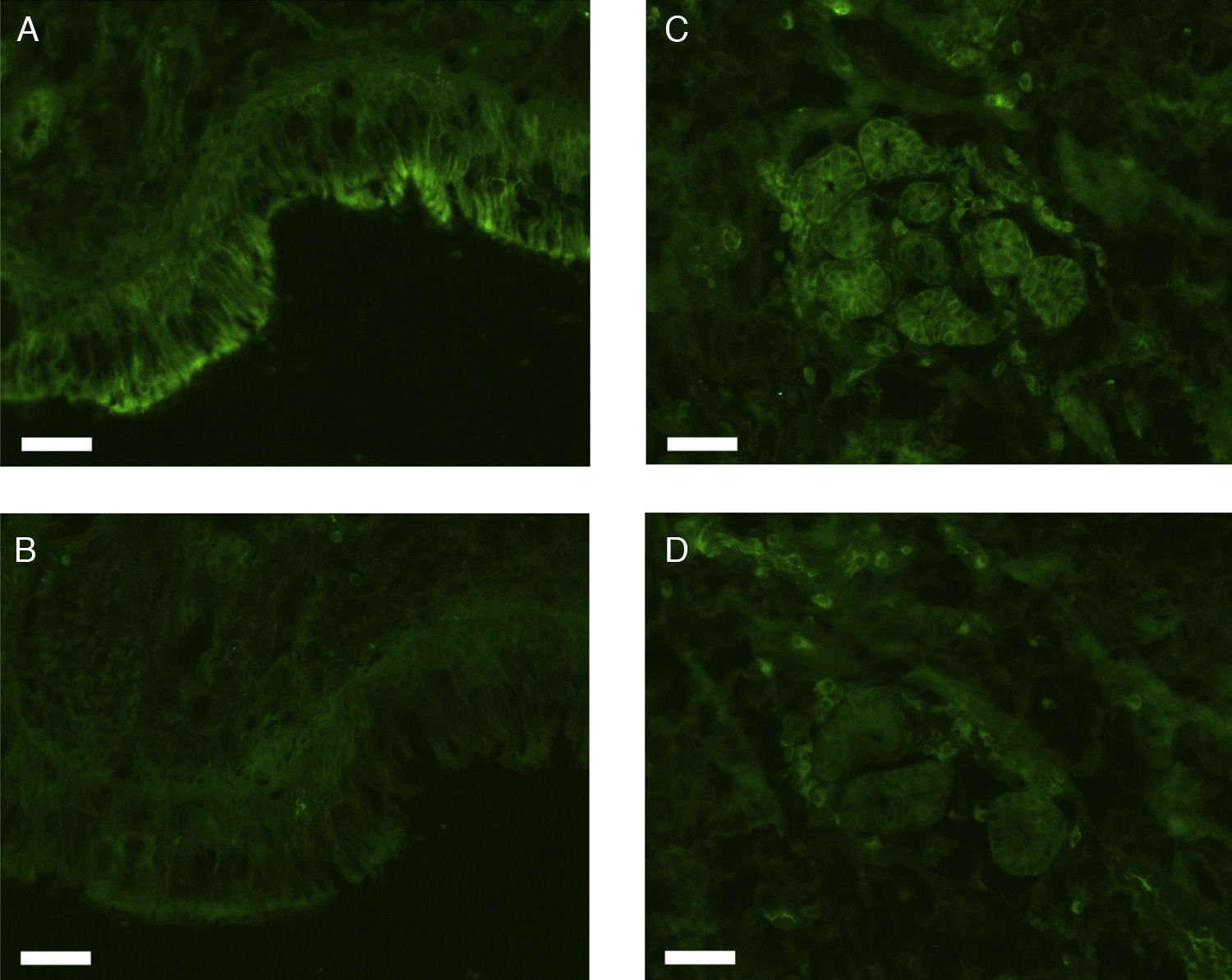
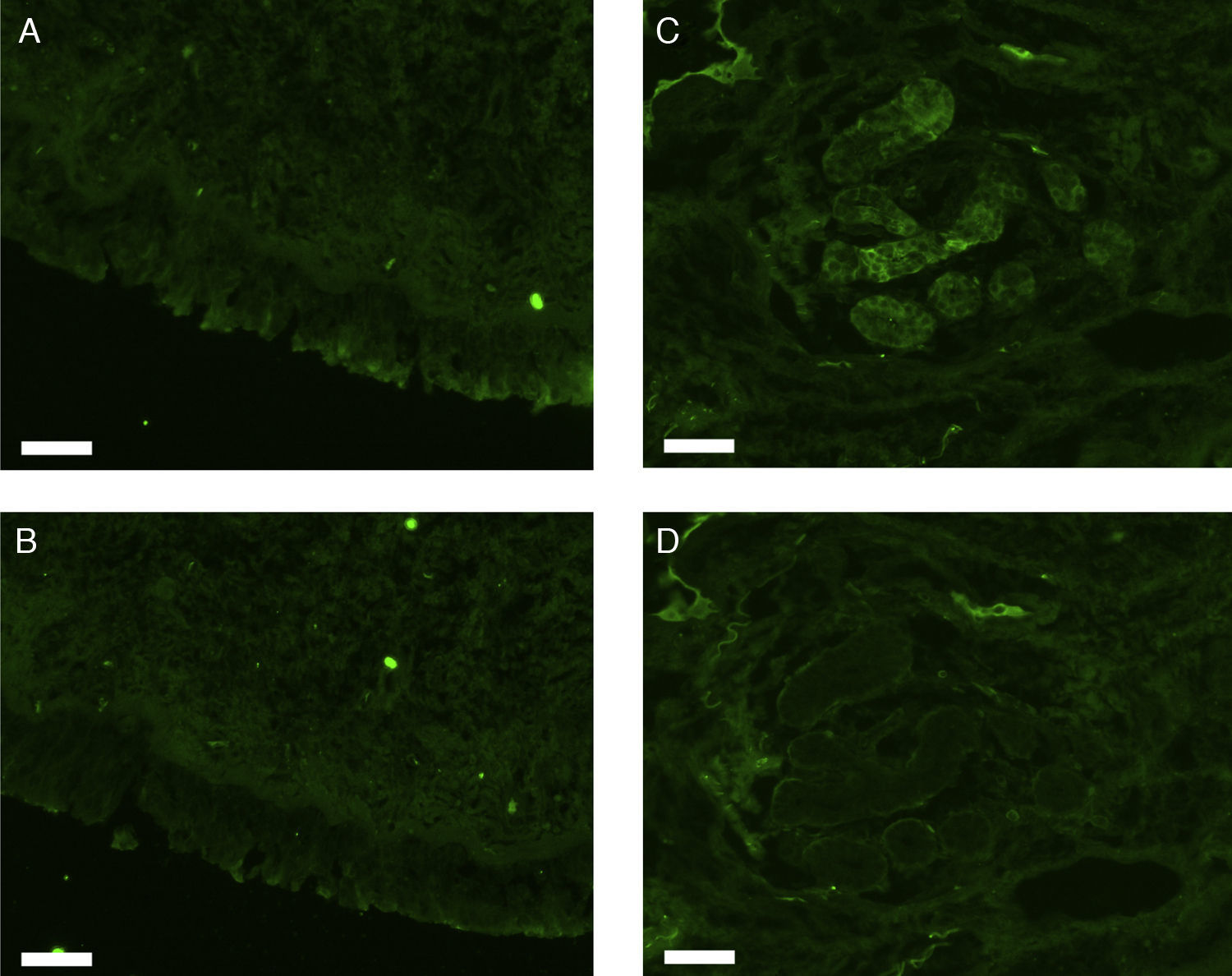
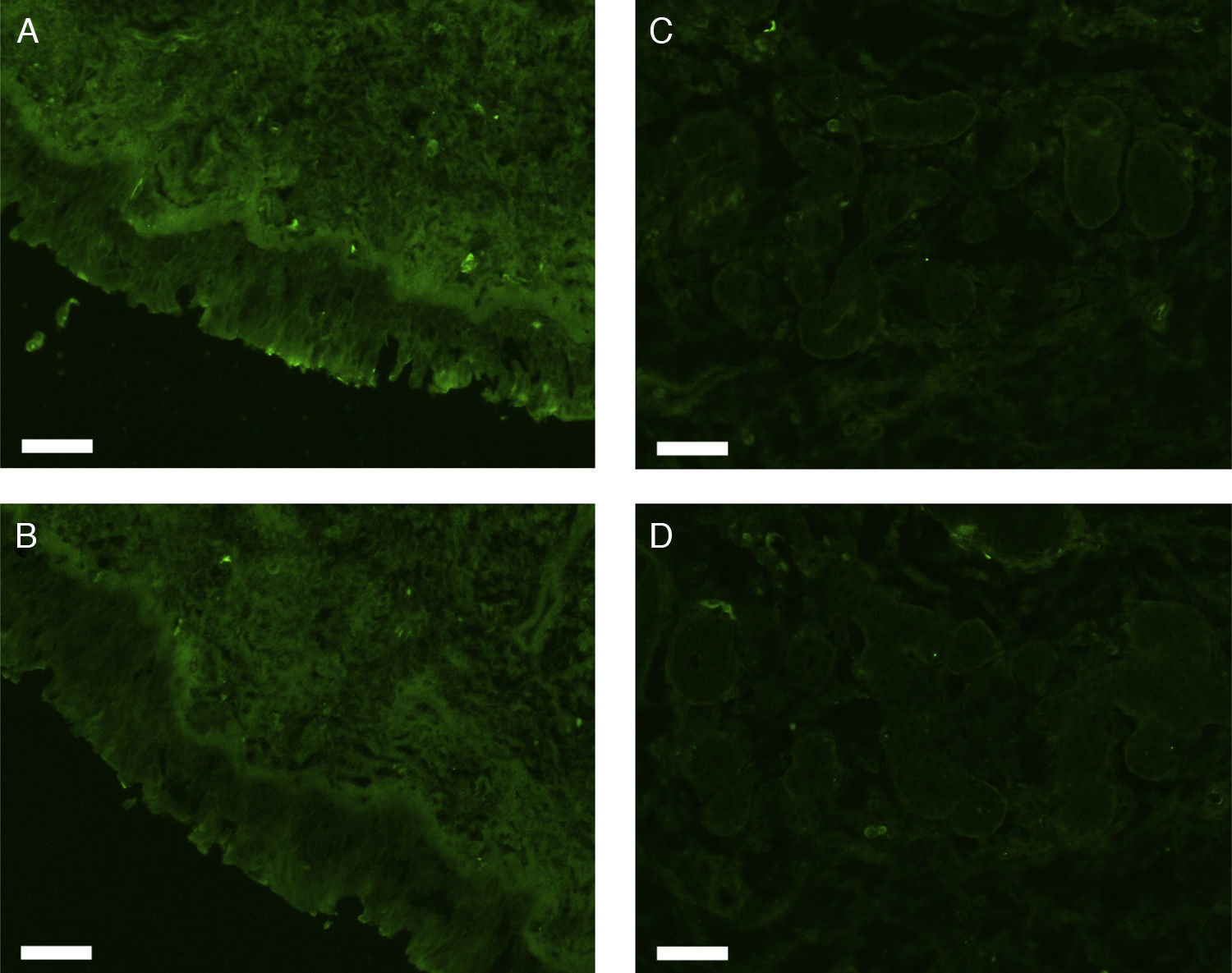
Fluorescence immunohistochemistryThe fixed samples were transferred into 20% sucrose in 0.1M phosphate buffered saline at pH 7.4 (PBS), and incubated at 4°C for two nights with 3–4 changes of the solution. The samples were then embedded while frozen in optimum cutting temperature (OCT) compound and stored at −80°C before sectioning. Seven-μm-thick sections were prepared using a cryostat, mounted on silane-coated glass slides (Superfrost; Matsunami Glass Industries, Osaka, Japan), and air-dried. The sections were hydrated in 0.1M phosphate buffered saline with 0.3% Triton X-100 (PBST) for 20min, and treated with 1.5% normal donkey serum (for erbB1) or normal goat serum (for erbB2, 3, 4, and claudin-1) in 0.1M PBST for 1h. They were then incubated with rabbit anti-human erbB1 antibody (Abgent, Flanders Court, San Diego, CA, USA), mouse anti-human erbB2 antibody (NeoMarkers Inc., Fremont, CA, USA), mouse anti-human erbB3 antibody (NeoMarkers Inc.), mouse anti-human erbB4 antibody (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA), or mouse anti-human claudin-1 antibody (Invitrogen Co., Camarillo, CA, USA) at 4°C overnight. The primary antibodies were used at a dilution of 1:50 (for anti-erbB1 and anti-claudin-1) or 1:100 (for anti-erbB2, 3 and 4) in 0.1M PBST containing 0.5% bovine serum albumin (BSA). As a control, primary antibodies were omitted from the process.
After a brief rinse with PBST, the sections were reacted with Alexa Fluor 488-conjugated donkey anti-rabbit IgG for erbB1 or Alexa Fluor 488-conjugated goat anti-mouse IgG for erbB2, 3, 4, and claudin-1 (Invitrogen; Molecular Probes, Eugene, OR) diluted 1:1000 in PBST containing 0.5% BSA at room temperature for two hours. The sections were coverslipped with Prolong Gold antifade reagent and examined under a Carl Zeiss Axioskop 2 plus fluorescence microscope. The light source was an HBO 103W/2 mercury vapour lamp. The light was let through a 475–495nm bandpass filter for the excitation of Alexa Fluor 488. The emitted fluorescence was allowed to pass through a 515–565nm bandpass filter. Images were captured using a Carl Zeiss AxioCam digital camera attached to the microscope.
Measurement of fluorescence intensity was performed in quadruplicate (four sections for each sample), using Axio Vision software (version 4.7.2.0; copyright 2006–2008; Carl Zeiss Imaging Solutions GmbH). The fluorescence intensity was displayed in a 256-step arbitrary scale of 0 (no fluorescence) to 255 (most intense fluorescence) in each pixel of the image. A region of interest in the image was manually defined, and its average pixel value (fluorescence intensity value) was calculated in order to quantitatively estimate the fluorescence intensity of the region. Then the fluorescence intensity value of the corresponding region in the control slide was subtracted to obtain the net fluorescence intensity value.
Preparation of total RNATotal RNA was extracted with an RNeasy Midi Kit (Qiagen Inc., Valencia, CA, USA) according to the manufacturer's instructions. The purity of RNA was assessed by the ratio of light absorption at 260 to that at 280nm (an A260/A280 ratio between 1.9 and 2.1 was considered acceptable). The RNA concentration was determined from A260.
qRT-PCRTwo μg of the total RNA was reverse-transcribed to cDNA with a High Capacity RNA-to-cDNA Kit (Applied Biosystems Inc., Foster City, CA, USA), which uses random primers. qRT-PCR analysis was performed with an Applied Biosystems StepOnePlus real-time PCR system using the TaqMan Fast Universal PCR Master Mix (Applied Biosystems) for five target genes (erbB1, erbB2, erbB3, erbB4, and claudin-1) and for glyceraldehyde-3-phosphate dehydrogenase mRNA (GAPDH) as a housekeeping gene according to the manufacturer's specification. The TaqMan Gene Expression Assays for erbB1 (assay identification number: Hs00193306_m1), erbB2 (assay identification number: Hs01001580_m1), erbB3 (assay identification number: Hs00951455_m1), erbB4 (assay identification number: Hs00171783_m1), claudin-1 (assay identification number: Hs00221623_m1), and GAPDH (assay identification number: Hs00951455_m1) were purchased from Applied Biosystems. One hundred ng/μl of cDNA was mixed with TaqMan Universal PCR Master Mix with AmpErase (uracil N-glycosylase) and a target primer/probe set of the TaqMan Gene Expression Assays, and subjected to PCR amplification with real-time detection. The thermal cycler conditions were as follows: holding at 95°C for 2min, followed by two-step PCR of 40 cycles at 95°C for 1s followed by 60°C for 20s. Each sample was assayed in duplicate.
The measured threshold cycle (CT) was normalised by subtracting CT for the housekeeping gene (GAPDH) of each sample from that for the target genes. From the obtained ΔCT, the ratio of the target gene to the housekeeping gene was calculated as follows:
Statistical analysisThe statistical significance of Pearson's correlation coefficients was tested using the t-test. P values less than 0.05 were considered significant.
ResultsThe total serum IgE level ranged from 2 to 438U/ml with an average of 163.4U/ml. RIST and RAST were positive in five and six patients, respectively. All five patients with positive RIST showed positive RAST.
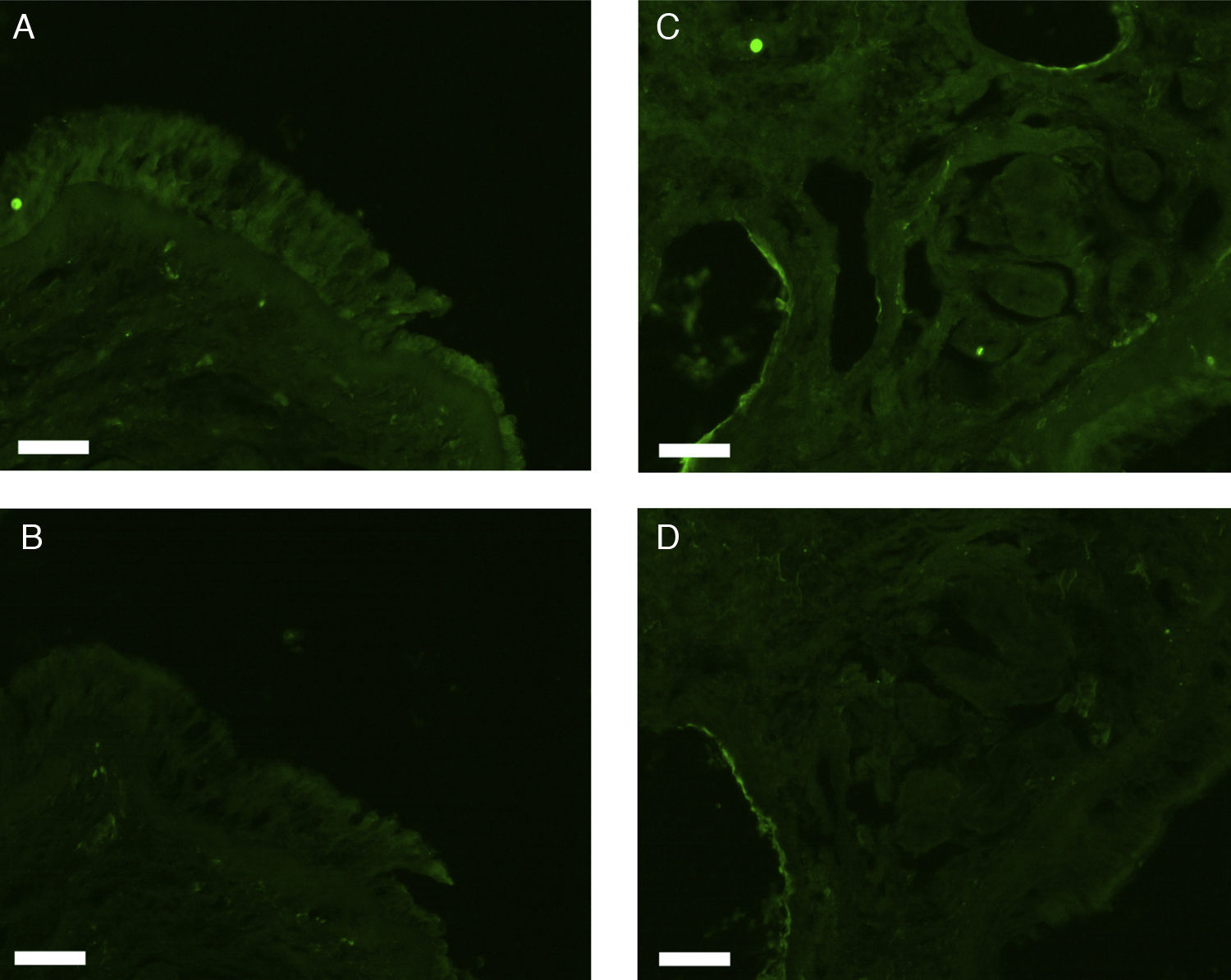
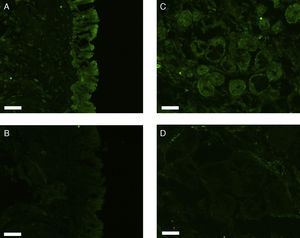
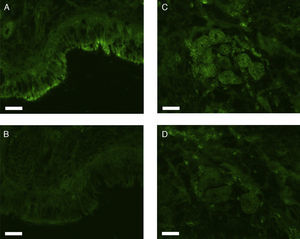
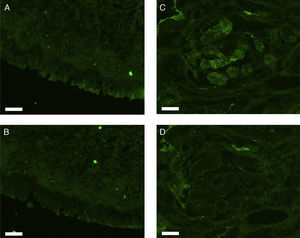
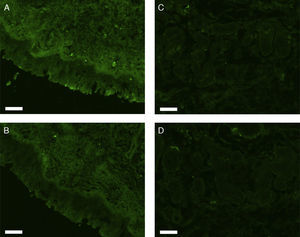
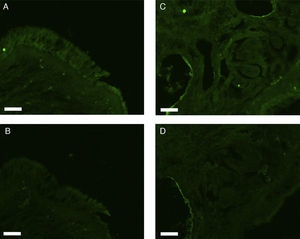
All erbB1-4 and claudin-1 were detected immunohistochemically and at the mRNA level. The immunoreactivity was mainly localised in the epithelial layer and in the nasal glands. Representative photomicrographs of immunohistochemical staining for erbB1, 2, 3, 4 and claudin-1 are presented in Figs. 1–5, respectively.
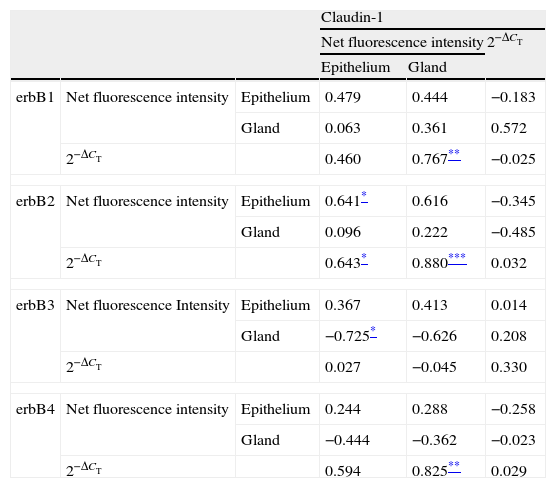
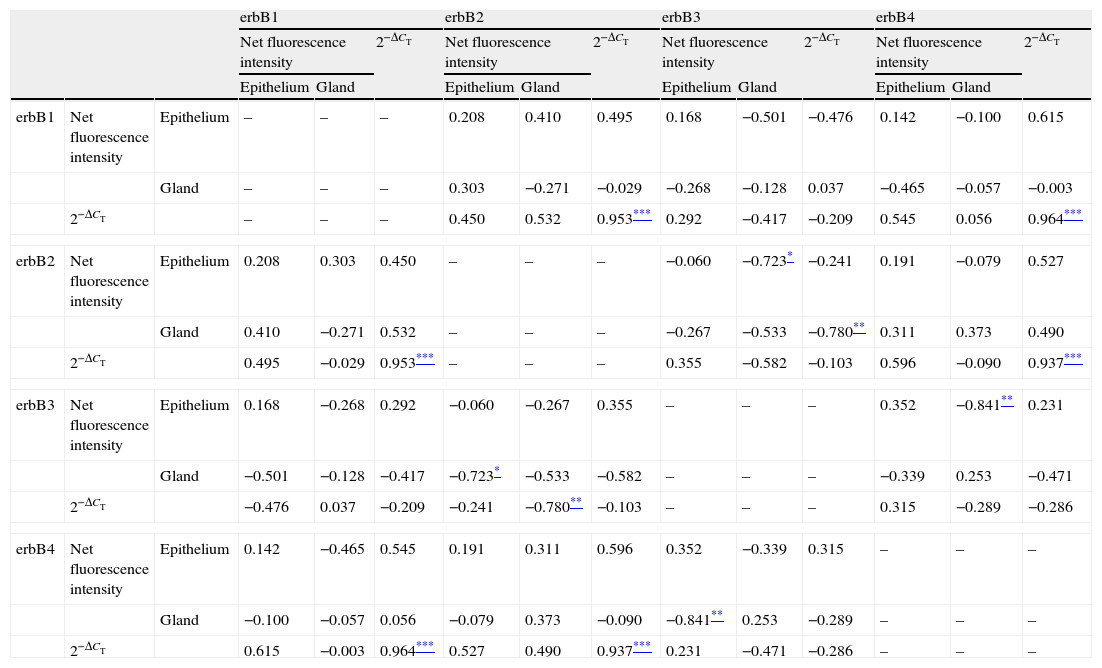
Correlations of net fluorescence intensities and 2−ΔCT values between erbBs and claudin-1 are shown in Table 1. Immunoreactivity for claudin-1 was positively correlated with the expressions of erbB1, 2 and 4, but negatively correlated with immunoreactivity for erbB3. Table 2 represents correlations among the expressions of erbB1-4. The mRNA expressions of erbB1, 2 and 4 were positively correlated with one another, whereas the expression of erbB3 showed negative correlation with the immunoreactivity for erbB2 and 4. These results imply that erbB1, 2, 4, and claudin-1 are similarly regulated in parallel, whereas erbB3 is regulated inversely to the other molecules in the hypertrophied nasal turbinate mucosa.
Correlation between the expressions of erbBs and claudin-1.
| Claudin-1 | |||||
| Net fluorescence intensity | 2−ΔCT | ||||
| Epithelium | Gland | ||||
| erbB1 | Net fluorescence intensity | Epithelium | 0.479 | 0.444 | −0.183 |
| Gland | 0.063 | 0.361 | 0.572 | ||
| 2−ΔCT | 0.460 | 0.767** | −0.025 | ||
| erbB2 | Net fluorescence intensity | Epithelium | 0.641* | 0.616 | −0.345 |
| Gland | 0.096 | 0.222 | −0.485 | ||
| 2−ΔCT | 0.643* | 0.880*** | 0.032 | ||
| erbB3 | Net fluorescence Intensity | Epithelium | 0.367 | 0.413 | 0.014 |
| Gland | −0.725* | −0.626 | 0.208 | ||
| 2−ΔCT | 0.027 | −0.045 | 0.330 | ||
| erbB4 | Net fluorescence intensity | Epithelium | 0.244 | 0.288 | −0.258 |
| Gland | −0.444 | −0.362 | −0.023 | ||
| 2−ΔCT | 0.594 | 0.825** | 0.029 | ||
The values shown are Pearson's correlation coefficients. 2−ΔCT indicates the ratio of target mRNA/GAPDH mRNA.
Correlation between the expressions of erbBs.
| erbB1 | erbB2 | erbB3 | erbB4 | |||||||||||
| Net fluorescence intensity | 2−ΔCT | Net fluorescence intensity | 2−ΔCT | Net fluorescence intensity | 2−ΔCT | Net fluorescence intensity | 2−ΔCT | |||||||
| Epithelium | Gland | Epithelium | Gland | Epithelium | Gland | Epithelium | Gland | |||||||
| erbB1 | Net fluorescence intensity | Epithelium | – | – | – | 0.208 | 0.410 | 0.495 | 0.168 | −0.501 | −0.476 | 0.142 | −0.100 | 0.615 |
| Gland | – | – | – | 0.303 | −0.271 | −0.029 | −0.268 | −0.128 | 0.037 | −0.465 | −0.057 | −0.003 | ||
| 2−ΔCT | – | – | – | 0.450 | 0.532 | 0.953*** | 0.292 | −0.417 | −0.209 | 0.545 | 0.056 | 0.964*** | ||
| erbB2 | Net fluorescence intensity | Epithelium | 0.208 | 0.303 | 0.450 | – | – | – | −0.060 | −0.723* | −0.241 | 0.191 | −0.079 | 0.527 |
| Gland | 0.410 | −0.271 | 0.532 | – | – | – | −0.267 | −0.533 | −0.780** | 0.311 | 0.373 | 0.490 | ||
| 2−ΔCT | 0.495 | −0.029 | 0.953*** | – | – | – | 0.355 | −0.582 | −0.103 | 0.596 | −0.090 | 0.937*** | ||
| erbB3 | Net fluorescence intensity | Epithelium | 0.168 | −0.268 | 0.292 | −0.060 | −0.267 | 0.355 | – | – | – | 0.352 | −0.841** | 0.231 |
| Gland | −0.501 | −0.128 | −0.417 | −0.723* | −0.533 | −0.582 | – | – | – | −0.339 | 0.253 | −0.471 | ||
| 2−ΔCT | −0.476 | 0.037 | −0.209 | −0.241 | −0.780** | −0.103 | – | – | – | 0.315 | −0.289 | −0.286 | ||
| erbB4 | Net fluorescence intensity | Epithelium | 0.142 | −0.465 | 0.545 | 0.191 | 0.311 | 0.596 | 0.352 | −0.339 | 0.315 | – | – | – |
| Gland | −0.100 | −0.057 | 0.056 | −0.079 | 0.373 | −0.090 | −0.841** | 0.253 | −0.289 | – | – | – | ||
| 2−ΔCT | 0.615 | −0.003 | 0.964*** | 0.527 | 0.490 | 0.937*** | 0.231 | −0.471 | −0.286 | – | – | – | ||
The values shown are Pearson's correlation coefficients. 2−ΔCT indicates the ratio of target mRNA/GAPDH mRNA.
Chronic hypertrophic rhinitis is clinically characterised by an irreversibly enlarged inferior turbinate, and its pathogenesis has been controversial. The enlarged turbinates histologically exhibit irreversible changes such as submucosal fibrosis, goblet cell hyperplasia, and squamous metaplasia with the loss of cilia,18 suggesting the occurrence of tissue damage and repair – that is, remodelling. Tissue remodelling in varying degrees is thought to be involved in the pathological process of rhinitis and sinusitis, although to a lesser extent than in that of asthma.19 The sinus mucosa of chronic sinusitis/rhinosinusitis exhibits histological findings characteristic of tissue remodelling, such as shedding of the epithelium, thickening of the basement membrane, and submucosal collagen deposition.20,21 Nasal epithelial damage in allergic rhinitis is milder, but has been shown at the ultrastructural level; i.e., vacuolation of the epithelial cells and widening of the intercellular spaces.22,23
Several studies have demonstrated the expression of erbBs in the lower respiratory tract of normal and asthmatic humans,12,24 implying their potential roles in the repair and regeneration of the airway epithelium.11 The presence of erbBs in the upper respiratory tract has also been documented since 2000.25–29 ErbB1, 2 and 3 are constitutively expressed in the normal human nasal epithelium.25 The observation that the expression of erbB1 increases in the inflammatory nasal mucosa and polyps27–29 strongly suggests the occurrence of tissue repair in these pathological states.
The expression and activation of erbBs in the airway epithelium are connected with tissue damage, which is accompanied by the breakdown of the tight junction. Vermeer et al.15 proposed a hypothesis that erbBs and their ligands are confined in the basolateral and apical surfaces of the epithelial cells, respectively, but mixed with each other when epithelial integrity is disrupted, leading to the activation of erbBs. Takeyama et al.30 and Petecchia et al.31 have found that airway epithelia damaged by cigarette smoke exposure exhibit activation of erbB1 and disassembly of tight junction components.The claudin family, consisting of 20 subtypes, is one of the major groups of tight junction proteins, and constitutes a physiological epithelial barrier.32 For example, overexpression of claudin-1 by transfection induces several-fold reinforcement of the electrical barrier function of cultured epithelial cells.33 In the human nasal mucosa, claudin-1, 4 and 7 are expressed throughout the epithelium.34 In vitro studies using cultured human nasal epithelial cells have revealed that the expression of these claudins is altered after exposure to inflammatory stimuli such as cytokine and virus.35–37
The present study demonstrated that the expressions of erbBs correlated with that of claudin-1, suggesting the participation of these molecules in the tissue remodelling of chronic hypertrophic rhinitis. In particular, erbBs 1, 2 and 4 showed positive correlation with one another and with claudin-1.
Generally, erbBs are activated by ligand-induced dimerisation.38 In addition to homodimerisation, erbBs can combine with one another and form heterodimers. Of the four types of erbBs, erbB2 has a unique property; it has no known ligand but can combine with the other 3 erbBs, and its heterodimerisation has been extensively studied.38 Activated erbBs then share common intracellular signalling pathways involving mitogen-activated protein kinase, STAT3, STAT5, Src family kinases, phospholipase C-γ, and phosphatidylinositol 3-kinase,17 and may work together for the repair and regeneration of impaired tissue. Interestingly, in the present study, erbB3 was inversely correlated with claudin-1 and other erbBs, as shown in Tables 1 and 2. The reason for this finding is unclear. In in vitro experiments, the biological half-life of erbB3 has been shown to be much shorter than that of the other erbBs.39 Moreover, unlike the other erbBs, erbB3 can be degraded via a unique pathway that involves neuregulin receptor degradation protein-1 (Nrdp1).40 It has been reported that Nrdp1 is stabilised by neuroregulin, resulting in a decrease of erbB3 expression.40 Such distinct characteristics of erbB3 relative to the other members may, at least partially, account for the aberrant expression of erbB3 in the present study.
In conclusion, we investigated the expressions of erbB1-4 and claudin-1 in the nasal mucosa of patients with chronic hypertrophic rhinitis by means of fluorescence immunohistochemistry and qRT-PCR. All erbB1-4 and claudin-1 were detected, and were mainly localised in the epithelial cells and nasal gland cells. Quantitative analyses revealed positive correlations among erbB1, 2, 4 and claudin-1, but a negative correlation of erbB3 with other erbBs and claudin-1. These results suggest a possible participation of these molecules in tissue remodelling in chronic hypertrophic rhinitis. Studies including in vitro experiments remain to be performed to further elucidate the pathogenesis of this disease.
Conflict of interestThe authors have no conflict of interest.
Ethical disclosuresPatients’ data protectionConfidentiality of data. The authors declare that no patient data appears in this article.
Right to privacy and informed consentRight to privacy and informed consent. The authors declare that no patient data appears in this article.
Protection of human subjects and animals in researchProtection of human and animal subjects. The authors declare that the procedures followed were in accordance with the regulations of the responsible Clinical Research Ethics Committee and in accordance with those of the World Medical Association and the Helsinki Declaration.
This study was partially supported by a Grant-in-Aid for Scientific Research (C) (no. 18591899; 2006–2008) from the Japan Society for the Promotion of Science.