To investigate whether the genetic variants of TGFB1, TLE4, MUC22 and IKZF3 are associated with the development of asthma in Chinese children.
Methods572 adolescent asthma patients and 590 age-matched healthy controls were included in this study. A total of four SNPs were genotyped, including rs2241715 of TGFB1, rs2378383 of TLE4, rs2523924 of MUC22, and rs907092 of IKZF3. Allele frequencies of the patients and the control group were compared by the Chi-square test. The Student t test was used to analyse the relationship between genotypes and clinical feature of the patients.
ResultsPatients were found to have significantly different frequencies of allele A of rs2241715, allele G of rs2378383 and allele A of rs2523924 as compared with the controls (40.4% vs. 45.9%, p=0.01 for rs2241715; 17.2% vs. 13.4%, p=0.01 for rs2378383; 15.3% vs. 11.9%, p=0.02 for rs2523924). For patients with severe asthma, those with genotype AA/AG of rs2241715 had remarkably higher FEV1% as compared with those with genotype GG (59.1±4.3% vs. 55.4±3.7%, p<0.001). Moreover, those with genotype GG/GA of rs2378383 had remarkably lower FEV1% as compared with those with genotype AA (54.6±2.9% vs. 58.6±4.1%, p<0.001).
ConclusionsGenes TGFB1, TLE4 and MUC22 are associated with the risk of childhood asthma in Chinese population. Our results associating TGFB1 and TLE4 with clinical features of asthma suggest potential application of these parameters in the management of asthma children.
Asthma is a common chronic disease among children, with a high prevalence rate.1,2 It is characterised by airway inflammation and bronchoconstriction followed by airflow obstruction.3,4 To date, the mechanisms leading to asthma development have not been fully understood. Previous studies have identified certain clinical risk factors of asthma,5–7 which however can only explain a small part of the disease. As indicated by the familial aggregation of asthma,8–10 genetic determinants are promising targets to elucidate the inheritable causes of asthma. Twin studies support a strong genetic component to asthma with genetic risk factors heritability estimated to contribute to 48–79% of asthma risk.11,12 From this perspective, identification of genetic variants associated with asthma could be helpful for a better understanding of its aetiopathogenesis.
Two traditional approaches have been applied to the identification of asthma susceptibility genes, including candidate gene association studies and linkage studies. Through these two methods, previous genetic association studies have revealed over 200 asthma susceptible genes in the past decade. However, few genetic loci show consistent associations across populations.13,14 In recent years, it became feasible to investigate the genetic architecture of asthma by genome-wide association studies (GWASs). In the first GWAS of asthma, Moffatt et al.15 found significant associations of SNPs adjacent to ORMDL3 and GSDMB with risk of childhood asthma in German and British populations. More recently, Himes et al.16 found SNPs in PDE4D are associated with risk of asthma in whites from the United States and replicated this finding in two other white populations. Choudhry et al.17 implicated chromosome 5q23 SNPs for association with asthma in patients from Puerto Ricans. Despite these intriguing findings, few genetic variants have been observed to confer large risks of asthma. It is apparent that numerous genetic risk factors are involved in the incidence of asthma with each of them conferring a small relative risk.
To date, most reported susceptibility loci were discovered in European or American populations. The different modes of linkage disequilibrium (LD) among Europeans and Asians highlight the importance of studying diverse populations in genetic association analyses. Replication of these novel loci in different populations may provide insights into racial disparities in asthma prevalence and severity. Recently, GWASs conducted in Latino population have revealed several susceptible genes of childhood asthma with genome-wide significance,18–20 which greatly reinforced their role in the aetiology of the disease. Therefore, they are of great interest to be replicated in a different population. To the best of our knowledge, the association between asthma and the genetic variants of these genes remains obscure in the Chinese population. In this study, we aimed to investigate whether the genetic variants of TGFB1, TLE4, MUC22 and IKZF3 are associated with the development of asthma in Chinese children.
MethodsSubjectsUnder the approval of the local institutional review board, the current case–control study included 572 adolescent asthma patients and 590 age-matched healthy controls. The asthma was diagnosed according to the criteria published by the American Thoracic Society,21 and the severity was determined according to the classification of the Global Initiative for Asthma guidelines.22 The following criteria were used for the inclusion of cases: (1) aged between 8 and 18 years; (2) being atopic. Atopic status was defined by one or more positive skin scratch test responses to common aeroallergens, or by specific IgE to any allergen >0.35kU/l. Patients with other chronic inflammatory diseases were excluded from the study. The control group had no history of respiratory disease and no first-degree relative with asthma patients. Moreover, all the controls had a negative skin-prick result.
The baseline characteristics of the subjects were recorded, including gender, age, asthma severity, level of IgE and results of pulmonary function test (PFT). Asthma severity was rated as mild, moderate, or severe, according to symptoms of the patients.22 PFT was performed with a Vitalograph 2150 spirometer (Compact, Buckingham, UK) and the baseline was determined by the best of three recordings. Values of forced expiratory volume in the first second of expiration (FEV1) were recorded, and we used the spirometric reference values for healthy Chinese children aged between 5 and 15 years to calculate the FEV1%.23 All the subjects gave their informed consent for the collection of the blood samples. Blood sample was used for total IgE analysis and specific IgE analysis. Age-adjusted serum total IgE levels and specific IgE levels for common aeroallergens were determined as previously reported. The value of IgE was transformed with nature logarithm (Log10).
Genotyping of target variantsGenomic DNA was extracted from the blood samples with a commercial kit (Qiagen K.K., Tokyo, Japan) following standard protocol. TaqMan Genotyping Assay was carried out for the genotyping of target variants. The assay was performed with each reaction unit containing 1.5μl DNA, 5μl TaqMan Universal PCR master mix and 3.5μl sterile water. Thermal cycling conditions began at 95°C for 10min and then proceeded with 45 cycles of 92°C for 15s and 60°C for 5min. The four asthma-related SNPs reported in the Latino populations were selected for genotyping, including rs2241715 (A/C) of TGFB1, rs2378383 (G/A) of TLE4, rs2523924 (A/G) of MUC22, and rs907092 (A/G) of IKZF3. The results of genotyping assay were analysed by ABI 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA). Twenty percent of the samples were randomly selected for a duplicate for the high quality of the genotyping assay.
Functional annotationTo explore the regulatory properties of the susceptible variant, we used chromatin state segmentation in LCL data generated by the ENCODE project to determine whether the variants or its proxies can annotate enhancer elements or putative transcription factor binding.
Statistical analysisSPSS version 19.0 (SPSS Inc., Chicago, IL, USA) was used for data analysis. Baseline characteristics of the cases and the controls were compared by the Student t test. The Hardy–Weinberg equilibrium (HWE) test was performed for the controls. Genotype and allele frequencies of the patients and the control group were compared by the Chi-square test. Odds ratio (OR) was calculated using the minor allele as a reference. The Student t test was used to analyse the relationship between genotypes and clinical feature of the patients in the three subgroups classified according to the asthma severity. Logistic regression analysis was used to examine the effect of each SNP with three genetic models, including the codominant model, dominant model, and recessive model. A p value of <0.05 was considered to be statistically significant.
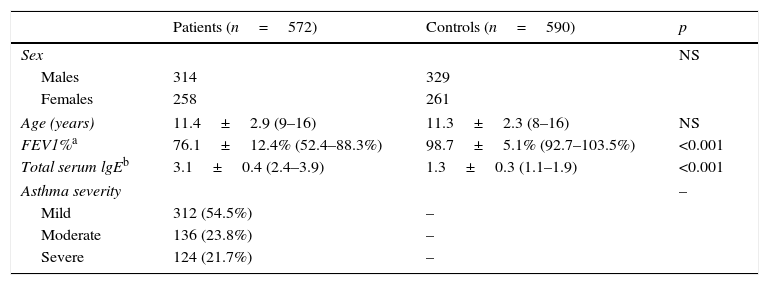
ResultsDemographic data of the participantsTable 1 summarises the baseline characteristics of the subjects. The cases and the controls were matched in terms of age (11.4±2.9 years vs. 11.3±2.3 years, p=0.51) and the ratio of male to female (314/258 vs. 328/261, p=0.78). Patients were found to have remarkably decreased FEV1% as compared with the controls (76.1±12.4% vs. 98.7±5.1%, p<0.001). Besides, there was a remarkably higher level of total serum lgE in the patients than in the controls (3.1±0.4 vs. 1.3±0.3, p<0.001). More patients were found to have mild asthma (54.5%) as compared with those having moderate (23.8%) or severe asthma (21.7%).
Baseline characteristics of the subjects.
| Patients (n=572) | Controls (n=590) | p | |
|---|---|---|---|
| Sex | NS | ||
| Males | 314 | 329 | |
| Females | 258 | 261 | |
| Age (years) | 11.4±2.9 (9–16) | 11.3±2.3 (8–16) | NS |
| FEV1%a | 76.1±12.4% (52.4–88.3%) | 98.7±5.1% (92.7–103.5%) | <0.001 |
| Total serum lgEb | 3.1±0.4 (2.4–3.9) | 1.3±0.3 (1.1–1.9) | <0.001 |
| Asthma severity | – | ||
| Mild | 312 (54.5%) | – | |
| Moderate | 136 (23.8%) | – | |
| Severe | 124 (21.7%) | – | |
NS: not significant; FEV1: forced expiratory volume in the first second of expiration.
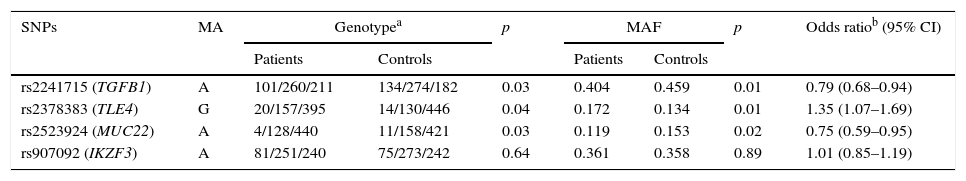
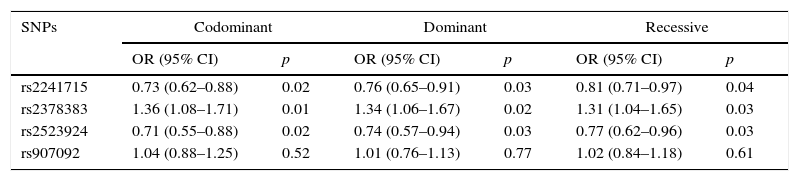
The genotyping results of the four variants are summarised in Table 2. HWE test showed no remarkable difference in the genotype frequency of the controls. Compared with the controls, patients were found to have significantly higher frequency of genotype GG of rs2378383 and significantly lower frequencies of genotype AA of rs2241715 and rs2523924 (3.5% vs. 2.4%, p=0.04 for rs2378383; 17.7% vs. 22.7%, p=0.03 for rs2241715; 0.7% vs. 1.9%, p=0.03 for rs2523924). Additionally, patients were found to have significantly higher frequencies of allele G of rs2378383 and significantly lower frequencies of allele A of rs2241715 and rs2523924 than the controls (17.2% vs. 13.4%, p=0.01 for rs2378383; 40.4% vs. 45.9%, p=0.01 for rs2241715; 15.3% vs. 11.9%, p=0.02 for rs2523924). The OR values were 0.79 for rs2241715, 1.35 for rs2378383 and 0.75 for rs2523924, respectively. As for the allele or genotype frequencies of rs907092, no significant difference was found between the patients and the controls. Table 3 summarises the results of three logistic regression models. Statistically significant associations with the susceptibility to asthma were observed with rs2378383, rs2241715 and rs2523924 under the codominant, dominant, and recessive models, respectively.
Association of the four variants with development of asthma.
| SNPs | MA | Genotypea | p | MAF | p | Odds ratiob (95% CI) | ||
|---|---|---|---|---|---|---|---|---|
| Patients | Controls | Patients | Controls | |||||
| rs2241715 (TGFB1) | A | 101/260/211 | 134/274/182 | 0.03 | 0.404 | 0.459 | 0.01 | 0.79 (0.68–0.94) |
| rs2378383 (TLE4) | G | 20/157/395 | 14/130/446 | 0.04 | 0.172 | 0.134 | 0.01 | 1.35 (1.07–1.69) |
| rs2523924 (MUC22) | A | 4/128/440 | 11/158/421 | 0.03 | 0.119 | 0.153 | 0.02 | 0.75 (0.59–0.95) |
| rs907092 (IKZF3) | A | 81/251/240 | 75/273/242 | 0.64 | 0.361 | 0.358 | 0.89 | 1.01 (0.85–1.19) |
M: minor allele; MAF: minor allele frequency; CI: confidential interval.
Logistic regression analysis of the four variants with the codominant model, dominant model, and recessive model.
| SNPs | Codominant | Dominant | Recessive | |||
|---|---|---|---|---|---|---|
| OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | |
| rs2241715 | 0.73 (0.62–0.88) | 0.02 | 0.76 (0.65–0.91) | 0.03 | 0.81 (0.71–0.97) | 0.04 |
| rs2378383 | 1.36 (1.08–1.71) | 0.01 | 1.34 (1.06–1.67) | 0.02 | 1.31 (1.04–1.65) | 0.03 |
| rs2523924 | 0.71 (0.55–0.88) | 0.02 | 0.74 (0.57–0.94) | 0.03 | 0.77 (0.62–0.96) | 0.03 |
| rs907092 | 1.04 (0.88–1.25) | 0.52 | 1.01 (0.76–1.13) | 0.77 | 1.02 (0.84–1.18) | 0.61 |
OR: odds ratio; CI: confidential interval.
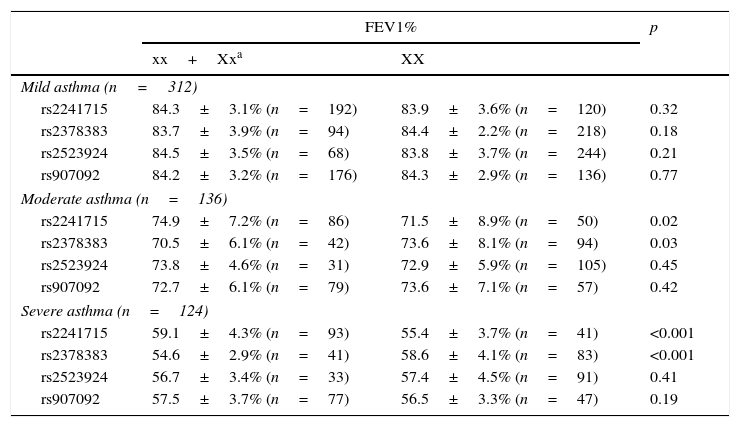
Results of the associations between four SNPs and asthma-related phenotype are summarised in Table 4. In the subgroups with moderate or severe asthma, patients with genotype AA/AG of rs2241715 could have remarkably higher FEV1% as compared with those with genotype GG. Moreover, patients with genotype GG/GA of rs2378383 could have remarkably lower FEV1% as compared with those with genotype AA. As for the other two SNPs, no significant associations with the asthma-related phenotype were found.
Association between the genotypes of four variants and clinical feature of patients with different severity of asthma.
| FEV1% | p | ||
|---|---|---|---|
| xx+Xxa | XX | ||
| Mild asthma (n=312) | |||
| rs2241715 | 84.3±3.1% (n=192) | 83.9±3.6% (n=120) | 0.32 |
| rs2378383 | 83.7±3.9% (n=94) | 84.4±2.2% (n=218) | 0.18 |
| rs2523924 | 84.5±3.5% (n=68) | 83.8±3.7% (n=244) | 0.21 |
| rs907092 | 84.2±3.2% (n=176) | 84.3±2.9% (n=136) | 0.77 |
| Moderate asthma (n=136) | |||
| rs2241715 | 74.9±7.2% (n=86) | 71.5±8.9% (n=50) | 0.02 |
| rs2378383 | 70.5±6.1% (n=42) | 73.6±8.1% (n=94) | 0.03 |
| rs2523924 | 73.8±4.6% (n=31) | 72.9±5.9% (n=105) | 0.45 |
| rs907092 | 72.7±6.1% (n=79) | 73.6±7.1% (n=57) | 0.42 |
| Severe asthma (n=124) | |||
| rs2241715 | 59.1±4.3% (n=93) | 55.4±3.7% (n=41) | <0.001 |
| rs2378383 | 54.6±2.9% (n=41) | 58.6±4.1% (n=83) | <0.001 |
| rs2523924 | 56.7±3.4% (n=33) | 57.4±4.5% (n=91) | 0.41 |
| rs907092 | 57.5±3.7% (n=77) | 56.5±3.3% (n=47) | 0.19 |
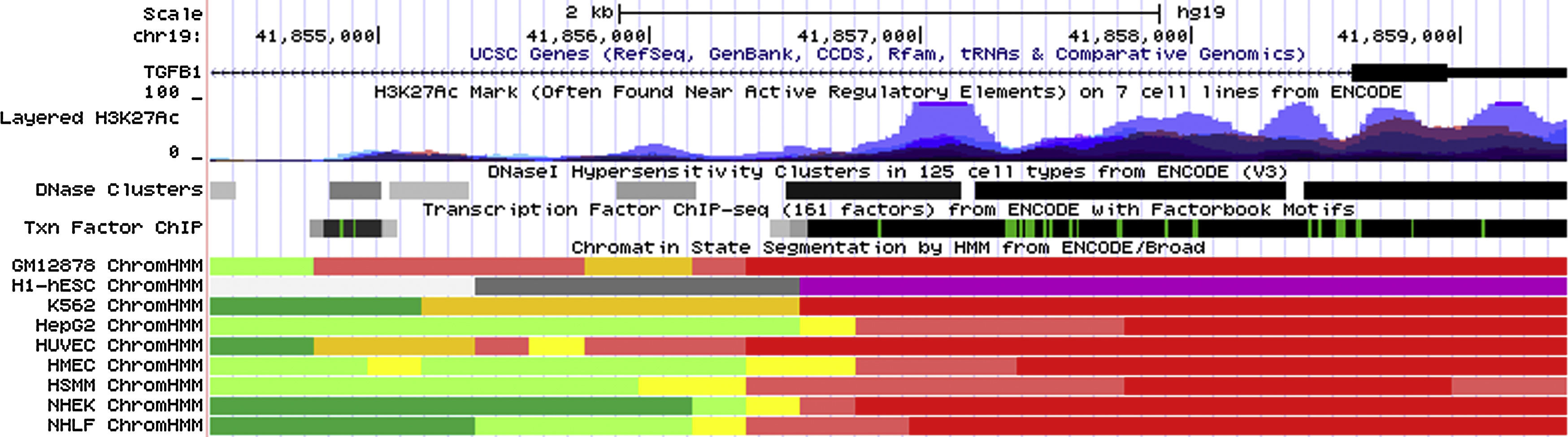
Data from ENCODE suggested that rs2241715 is located in the intronic region of TGFB1 gene marked by promoter and enhancer histone marks in multiple cell lines. As shown in Fig. 1, rs2241715 resides within DNase I hypersensitive regions across multiple tissues marked by a number of histone marks and motifs. As for rs2378383 and rs2523924, there was no strong regulatory signal around these two loci as indicated by the ENCODE.
Functional annotation of rs2241715 using UCSC genome browser. rs2241715 is located in the intronic region of TGFB1 gene marked by promoter and enhancer histone marks as well as DNase I hypersensitive regions across multiple tissues. Regulatory elements of these regions were annotated by ENCODE as follows: bright red, active promoter; light red, weak promoter; orange, strong enhancer; yellow, weak/poised enhancer; blue, insulator; dark green, transcriptional transition and transcriptional elongation; light green, weak transcribed; grey, Polycomb repressed; light grey, heterochromatin, low signal.
New genetic variants associated with the risk of asthma have been recently revealed through GWASs. Hancock et al.19 conducted a GWAS in 492 Mexican children with asthma and identified TLE4 as a novel genetic variation for childhood asthma. Based on the same dataset, Wu et al.20 performed a separate candidate gene analyses for 2933 SNPs in the 237 selected genes, and identified TGFB1, IL1RL1, IL18R1, and DPP10 as the genes most significantly associated with asthma. In a meta-analysis of several Latino populations, Galanter et al.18 found a novel locus on 6p21 (MUC22) and confirmed a previously identified locus on 17q21 (IKZF3) as well. Among these genes, IL1RL1, IL18R1, and DPP10 have been replicated in the Chinese children,24,25 except for TGFB1, TLE4, MUC22 and IKZF3. In the current case–control study, we herein investigated the effects of those four variants in the Chinese adolescents, and demonstrated for the first time that rs2241715 of TGFB1,rs2378383 of TLE4 and rs2523924 of MUC22 are associated with the risk of childhood asthma in a Chinese population.
TGFB1 is a multi-functional cytokine expressed in many cell types including inflammatory cells and airway epithelial cells. It may modulate the development of allergic inflammation and airway remodelling in asthma. As a widely investigated asthma candidate gene, variants of TGFB1 have been associated with asthma in different populations, including Mexicans, Costa Ricans, Japanese and Caucasian French Canadians.26–28 The SNP rs2241715 that was found significantly associated with asthma in the present analysis was in moderate to high LD with the asthma-associated SNPs rs1800469 and rs1982073 reported in previous paper.26 As a functional variant, rs1800469 is located in the promoter region and can influence the circulating levels of TGFB1.26 Interestingly, in the current study we found that rs2241715 was associated with significantly decreased PFT in patients with asthma. We speculate that over-expression of TGFB1 can be indicative of more severe symptoms of asthma. The precise functional effect of rs2241715 on expression levels of TGFB1 remains to be determined by more in vivo cellular experiments.
SNP rs2378383 is located 147kb upstream of TLE4 in the chromosome 9q21.31. The TLE family of proteins participates in cell fate determination for neurogenesis and segmentation.29 The TLE4 gene shows ubiquitous expression across many tissues and functions as a transcriptional co-repressor in several key developmental pathways.30 It is speculated that TLE4 could be implicated in asthma through interactions with immune-related factors such as PAX5 and RUNX3.31,32 In the GWAS of Hancock et al.,19 a cluster of SNPs spanning TLE4 and its upstream region rank among the top significant SNPs. To date, there is still a paucity of evidence supporting the impact of rs2378383 on TLE4 expression. But it is possible that these SNPs reside in a regulatory region of TLE4. Further fine-mapping of the region surrounding rs2378383 may be helpful to reveal the role of TLE4 in the development of asthma.
SNP rs2523924 is located intron of MUC22in the chromosome 6p21 near the HLA region. MUC are highly glycosylated macromolecules and function as the major components of mucus secretions.33 Inflammatory lung diseases such as bronchial asthma and chronic obstructive pulmonary disease are often characterised by high production of mucus.34 MUC22 was also identified as a susceptible gene of diffuse panbronchiolitis, another obstructive lung disease.35 Hijikata et al.35 observed that MUC22 was expressed in the lung and bronchial epithelial cells differentiated at an air-liquid interface. Future study conducted in the context of expression analysis of MUC22 can add to the understanding on its relationship with asthma.
Although rs907092 of IKZF3 gene was reported as susceptible variants of asthma, we failed to validate the association of this SNP with asthma in our study. It is noteworthy that rs907092 is located 50kb upstream from GSDMB, and there is a high degree of LD in this region. It was validated that there was also a lack of association between GSDMB and childhood asthma in the Chinese population.25 The inconsistency between our result and that of the previous study might be resulted from the ethnic heterogeneity. However, we cannot entirely exclude the association between IKZF3 and the risk of asthma in the Chinese population. Further investigation is warranted to determine whether other polymorphisms of IKZF3 gene might be responsible for the reported association with asthma.
The primary limitation of this study lies in that some other covariates such as parental history of asthma and socioeconomic status should be adjusted when performing the statistical analysis. Due to the inherent limitation of the retrospective design of our study, however, we failed to collect the data of these variables for most of the patients. In a future study we shall control these variables to exclude potential bias.
ConclusionsThe findings of our study provide insight into the role of the genetic polymorphism in asthma. We confirmed that the genes TGFB1, TLE4 and MUC22 are associated with the risk of childhood asthma in the Chinese population. Besides, TGFB1 and TLE4 can be involved in clinical phenotypes of asthmatic children. However, variants of these three genes can only explain limited variance of asthma as indicated by the low OR. Therefore, more susceptible variants of asthma await to be investigated. And the functional role of these susceptible genes needs to be explored for a better understanding of the aetiology of asthma.
Ethical disclosuresConfidentiality of dataThe authors declare that they have followed the protocols of their work centre on the publication of patient data and that all the patients included in the study have received sufficient information and have given their informed consent in writing to participate in that study.
Right to privacy and informed consentThe authors have obtained the informed consent of the patients and/or subjects mentioned in the article. The author for correspondence is in possession of this document.
Protection of human subjects and animals in researchThe authors declare that no experiments were performed on humans or animals for this investigation.
Conflict of interestThe authors involved in this study do not have anything to disclose regarding funding or any conflicts of interest with respect to this manuscript.