Common variable immunodeficiency (CVID) is the most common symptomatic form of primary immunodeficiency (PID). LPS-responsive beige-like anchor protein (LRBA) deficiency is an autosomal recessive disease characterized by a CVID-like phenotype. T cell abnormality was reported in patients with CVID and LRBA deficiency. The study's aim was to evaluate IL-4, IL-5, IL-10 and GATA3 expression in patients with LRBA deficiency and CVID with no known monogenic disease, and further evaluate its relevance with immunological futures and clinical complications of patients.
MethodsThe study population comprised patients with CVID, LRBA deficiency and age–sex matched healthy controls. Mutation analysis was done by whole exome sequencing in CVID patients to rule out monogenic PIDs. After CD4+ T cell stimulation with anti-CD3 and anti-CD28 monoclonal antibodies, gene expression of IL-4, IL-5, IL-10 and transcription factor GATA3 was evaluated by real-time polymerase chain reaction. The protein of mentioned cytokines was assessed by enzyme-linked immunosorbent assay.
ResultsThe main clinical presentations of CVID patients were infections only and lymphoproliferations phenotypes, but in LRBA patients were autoimmune and enteropathy phenotype. The frequencies of CD4+ T cells were significantly reduced in LRBA and CVID patients. There were no statistically significant differences among GATA3, IL4, and IL5 gene expressions by CD4+ T cells of patients and controls, however, the IL10 expressions in CVID patients was significantly lower than in LRBA patients and HCs. As compared with HCs, CVID patients showed a prominent decrease in IL-4 and IL-10 production by CD4+ T cells.
ConclusionsOur findings demonstrated that patients with CVID and LRBA deficiency (even with severe infectious and inflammatory complications) have not imbalance in Th2 response, which is in parallel with lower frequency of allergy and asthma in these patients.
Common variable immunodeficiency (CVID) is the most frequent symptomatic primary immunodeficiency disorder characterized by a marked decrease in serum IgG, IgA or IgM of at least 2 standard deviations below the mean for age; absent isohemagglutinins and/or poor response to vaccines; onset of immunodeficiency after four years of age; and exclusion of secondary causes of hypogammaglobulinemia.1 CVID has an approximate prevalence of 1 in 25,000–50,000 in the general population. The most frequent symptoms of CVID are recurrent infections, pulmonary complications, gastrointestinal diseases, granulomatous or polyclonal lymphocytic disease, autoimmunity and malignancy.2–4 Genetic mutations can be identified as the cause of disease in approximately 10–20% of CVID patients.5 These include the inducible co-stimulator (ICOS), CD19, CD20, CD21, CD81, B cell-activating factor receptor (BAFFR), transmembrane activator and calcium modulator and cyclophilin ligand interactor (TACI), Cytotoxic T-Lymphocyte Associated Protein 4 (CTLA4), nuclear factor-kappa B1 (NFKB1), phosphoinositide-3-kinase, regulatory subunit 1 (PIK3R1), and lipopolysaccharide responsive beige-like anchor (LRBA) genes.5 LRBA deficiency is a combined immunodeficiency with childhood onset, caused by loss-of-function mutation in LRBA gene. The patients have a variety of clinical symptoms including hypogammaglobulinemia, recurrent infections, autoimmunity, and enteropathy. Further, it has been reported that most LRBA patients had previously been diagnosed with CVID.6
Several studies reported that except for B-cell defect, reduced frequency of CD4+ T cells and their cytokines as well as uncontrolled T helper (TH) cell polarization may also be involved in the pathogenesis of immune dysregulation in CVID and LRBA patients.7 Although there is a lack of studies regarding some subsets such as TH2, but increased frequency of TH1 and circulating follicular T helper (cTFH) cells as well as lower frequency Th17 and regulatory T (Treg) cells has been reported in previous studies, however, data regarding cytokines [Interferon-γ (IFN-γ), Interleukin (IL)-4, IL-9, IL-10, IL-13 and IL-17) serum levels and their production by peripheral blood mononuclear cells (PBMCs) or isolated CD4+ T cells is contradictory (maybe due to the different types of specimen).8–12
In the present study, we aimed to evaluate the Th2 cells determinant cytokines (IL-4, IL-5, and IL-10) and transcription factor (GATA3) following the stimulation of isolated CD4+ cells by anti-CD3 and anti-CD28 monoclonal antibodies (mAb) in LRBA patients and CVID patients with no known monogenic disease. Furthermore, we compared the immunological futures and clinical manifestation of these two groups of patents, and evaluated their relevance with results obtained for IL-4, IL-5, IL-10 and GATA3 levels.
Subjects and methodsSubjectsThe study population comprised 12 LRBA and 12 CVID (genetically evaluated) patients registered in the Iranian national registry of PID patients13 at Children's Medical Center affiliated to Tehran University of Medical Sciences, Tehran-Iran. The mutation analysis in these patients was evaluated by whole exome sequencing, and mutations found in LRBA gene were confirmed by Sanger sequencing as described previously.6 Twelve healthy individuals with no history of immune-related disorders were selected as the healthy control (HC) group. The project has been reviewed and approved by the ethics committees of the involved institution and written informed consent was obtained from all participants and/or their parents. The demographic and clinical data of patients were collected from the Iranian national registry of PID patients13 updating by monthly visits of patients, and reviewed thoroughly by clinical immunologists.
Cell isolation and purification of CD4+ T cellThe blood samples were collected four weeks after IVIg infusion in heparin-containing tubes. PBMCs were obtained from both patients and controls using Lymphocyte Separation Media (Lymphosep, Biosera, France) and re-suspended in the RPMI-1640 medium (Lymphosep, Biosera) supplemented with 10% fetal bovine serum (Lymphosep, Biosera), penicillin (100IU), and streptomycin (100μg/mL) (Biosera, Ringmer, East Sussex, UK) for CD4+ T cell isolation. The viability of isolated PBMCs was observed more than 97% as assessed by Trypan blue viability test. CD4+ T cells were purified from PBMCs with a human CD4+ T Cell Isolation Kit (Miltenyi Biotec, Gladbach, Germany) by depletion of non-CD4+ T cells (negative selection). The purity of CD4+ T cells was routinely more than 95%, measured by flow cytometric analysis.
Quantitative real-time polymerase chain reaction (RT-PCR)The method applied for CD4+ T cell stimulation was similar to our previous studies.14,15 Briefly, CD4+ T cells were harvested and prepared in 24-well plates that were pre-coated with 3μg/mL anti-CD3 mAb at final concentration of 2×106/mL (eBioscience, San Diego, CA, USA). Additionally, 2μg/mL anti-CD28 mAb (eBioscience, San Diego, CA, USA) was added concurrently and incubated in RPMI-1640 medium at 37°C in a 5% CO2 humidified atmosphere. Stimulated cells were collected 18h afterwards and washed twice with PBS. For RT-PCR, total RNA was extracted from cultured cells using RNeasy Mini Kit (Qiagen, Hilden, Germany) and reverse transcribed into cDNA by Takara kit (Takara, Japan) according to the manufacturer's instructions. The expression levels of IL4, IL5, IL10, and GATA-binding protein 3 (GATA3) genes were measured by quantitative RT-PCR, using SYBR Green PCR Master Mix (Takara) with specific primers (Table S1). Quantitative gene expression data were normalized relative to levels of GAPDH (glyceraldehyde-3-phosphate dehydrogenase). Fold-change in gene expression was determined applying 2−ΔΔCt method. The primers were designed using Oligo 6 software (http://www.oligo.net/), and the specificity of amplification products was confirmed by NCBI Primer-Blast Tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). Moreover, single sharp melting curve peaks were observed for all products.
Cytokine assayCD4+ T cells were harvested and brought to a final concentration of 3×105/mL in 24-well plates and were subsequently stimulated by anti-CD3 mAb and anti-CD28 mAb using the same method mentioned in the previous section. Supernatants were collected after 48h, and cytokines’ production (IL-4, IL-5 and IL-10) was evaluated using quantitative enzyme-linked immunosorbent assay (ELISA) with the commercial human ELISA Ready-SET-Go kits (eBioscience, San Diego, CA, USA) according to the manufacturer's instructions. The sensitivity of detection for IL-10, IL-4 and IL-5 was 2pg/mL, 2pg/mL and 1.5pg/mL, respectively.
Statistical analysisValues were expressed as frequency (number and percentage), and median (IQR, presented as a range with 75th–25th percentiles) as appropriate. Shapiro–Wilks test was used to check the normality assumption for the variable; so according to the establishment of assumptions, a parametric or non-parametric test was done. Statistical analyses were performed using the SPSS software package, version 22 (SPSS Inc., Chicago, IL, USA).
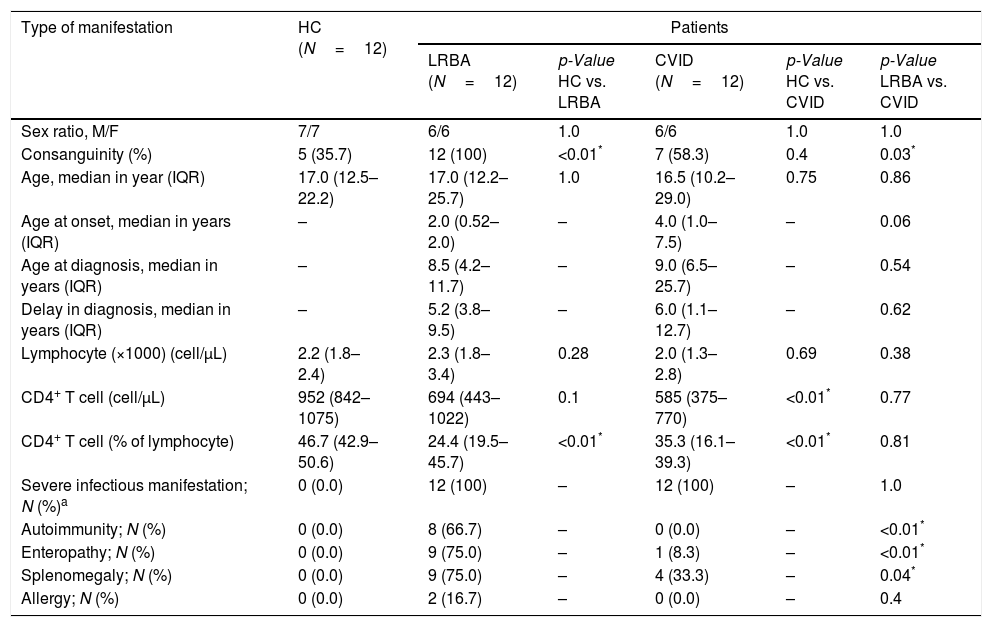
ResultsCharacteristics and clinical phenotypes of patients and controlsThe demographic characteristics of patients with CVID and LRBA deficiency and HC individuals are summarized in Table 1. As illustrated in Table 1, all patients with CVID and LRBA deficiency had a history of infectious complications, while autoimmunity (66.7 vs. 0.0, p<0.01), enteropathy (75.0 vs. 8.3, p<0.01), splenomegaly (75.0 vs. 33.3, p=0.04) and bronchiectasis (58.3 vs. 8.3, p=0.02) was more frequent in patients with LRBA deficiency than CVID. Detailed immunologic and clinical data are illustrated in Tables S2 and S3. Monthly low-dose IVIg infusion was administered in all CVID patients and 11 patients with LRBA deficiency as a maintenance therapy. A lower frequency of CD4+ T cells was observed in patients with CVID and LRBA deficiency compared to HC (Table 1 and Fig. S1). The changes in absolute count of lymphocyte and CD4+ T cell of CVID and LRBA patients compared at time of diagnosis and at the time of current study are illustrated in Table S4.
Demographic and corresponding immunologic and clinical data for patients and control.
| Type of manifestation | HC (N=12) | Patients | ||||
|---|---|---|---|---|---|---|
| LRBA (N=12) | p-Value HC vs. LRBA | CVID (N=12) | p-Value HC vs. CVID | p-Value LRBA vs. CVID | ||
| Sex ratio, M/F | 7/7 | 6/6 | 1.0 | 6/6 | 1.0 | 1.0 |
| Consanguinity (%) | 5 (35.7) | 12 (100) | <0.01* | 7 (58.3) | 0.4 | 0.03* |
| Age, median in year (IQR) | 17.0 (12.5–22.2) | 17.0 (12.2–25.7) | 1.0 | 16.5 (10.2–29.0) | 0.75 | 0.86 |
| Age at onset, median in years (IQR) | – | 2.0 (0.52–2.0) | – | 4.0 (1.0–7.5) | – | 0.06 |
| Age at diagnosis, median in years (IQR) | – | 8.5 (4.2–11.7) | – | 9.0 (6.5–25.7) | – | 0.54 |
| Delay in diagnosis, median in years (IQR) | – | 5.2 (3.8–9.5) | – | 6.0 (1.1–12.7) | – | 0.62 |
| Lymphocyte (×1000) (cell/μL) | 2.2 (1.8–2.4) | 2.3 (1.8–3.4) | 0.28 | 2.0 (1.3–2.8) | 0.69 | 0.38 |
| CD4+ T cell (cell/μL) | 952 (842–1075) | 694 (443–1022) | 0.1 | 585 (375–770) | <0.01* | 0.77 |
| CD4+ T cell (% of lymphocyte) | 46.7 (42.9–50.6) | 24.4 (19.5–45.7) | <0.01* | 35.3 (16.1–39.3) | <0.01* | 0.81 |
| Severe infectious manifestation; N (%)a | 0 (0.0) | 12 (100) | – | 12 (100) | – | 1.0 |
| Autoimmunity; N (%) | 0 (0.0) | 8 (66.7) | – | 0 (0.0) | – | <0.01* |
| Enteropathy; N (%) | 0 (0.0) | 9 (75.0) | – | 1 (8.3) | – | <0.01* |
| Splenomegaly; N (%) | 0 (0.0) | 9 (75.0) | – | 4 (33.3) | – | 0.04* |
| Allergy; N (%) | 0 (0.0) | 2 (16.7) | – | 0 (0.0) | – | 0.4 |
HC: healthy control; LRBA: LPS-responsive-beige-like anchor; CVID: common variable immunodeficiency; M: male; F: female.
IQR: range with 75th and 25th percentiles. N: count. Mann–Whitney U test was used.
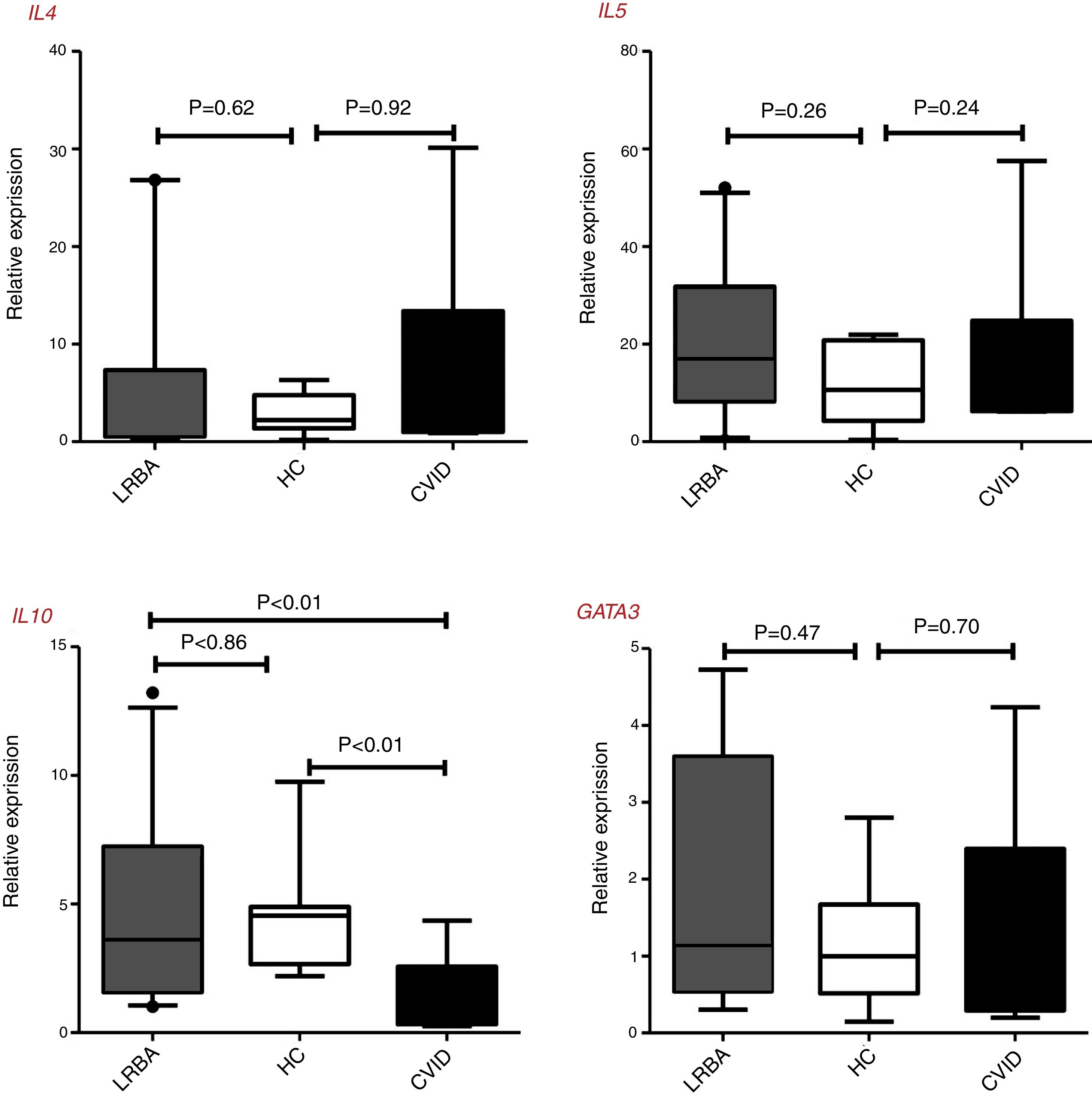
There were no statistically significant differences of GATA3, IL4, and IL5 gene expressions by CD4+ T cells of patients and controls, however, the IL10 expressions in CVID patients [0.72 (0.32–2.57) was significantly lower than in LRBA patients [3.62 (1.57–7.23), p<0.01 and HCs [4.54 (2.67–4.88), p<0.01] (Fig. 1). There was a significant positive correlation between GATA3 and IL4 gene expression (p<0.01, r=0.893) and between IL5 and IL10 (p=0.03, r=0.736) in the stimulated CD4+ T cells of CVID patients. Two of the LRBA patients have a history of asthma and/or allergy (one with asthma and the other asthma and allergic dermatitis), the mean (SD) of GATA3 gene expression was higher in these two patients than other patients with LRBA deficiency [4.35 (0.52) vs. 1.17 (1.01) (p=0.04)].
Th2 cells genes expression in patients with CVID, and LRBA deficiency and healthy controls (HC). Comparison of IL4, IL5, IL10, and GATA3 genes expression in the stimulated CD4+ T cells of the CVID (N=12) and LRBA (N=12) patients and HC (N=12) by the quantitative RT-PCR. The median is represented by a horizontal line, the interquartile range (IQR) by box, and the 10th and 90th percentiles by whiskers. Outlier symbol (●) showed data beyond the end of the whiskers. N: number; LRBA: LPS responsive beige-like anchor protein; CVID: common variable immunodeficiency; HC: healthy controls.
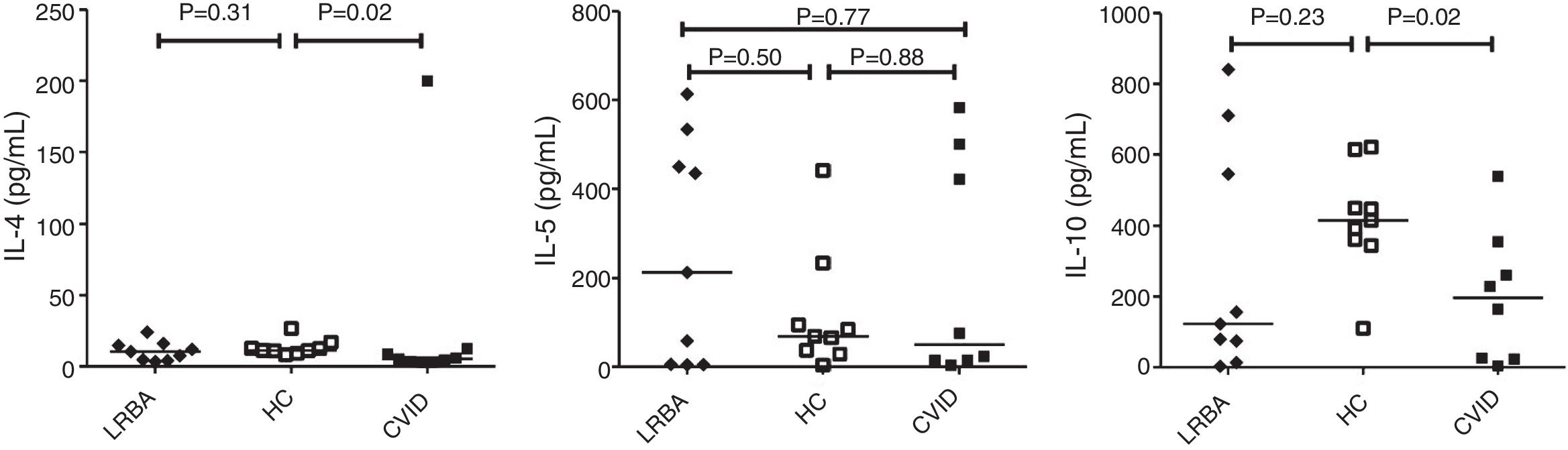
By comparison with HCs, CVID patients showed a prominent decrease in IL-4 [median (IQR): 11.4 (10.1–14.8) vs. 5.3 (3.3–10.6)pg/mL; p=0.02] and IL-10 [median (IQR): 415 (353–532) vs. 196 (24–331)pg/mL; p=0.02] production by CD4+ T cells (Fig. 2). There was a significant positive correlation between IL-5 with IL-4 (p=0.03, r=0.700) and IL-10 (p=0.02, r=0.750) concentration in the stimulated CD4+ T cells of LRBA patients and between IL-4 and IL-10 (p=0.02, r=0.786) in CVID patients. There was no significant association between cytokine mRNA and their relevant protein level, except for IL-5 in patients with LRBA deficiency (p=0.01, r=0.767). There was no significant association among the concentration of IL4, IL-5 and IL-10 with GATA3 mRNA expression in CVID and LRBA patients. There was no association between cytokines production by CD4+ T cells of patients with CVID and LRBA deficiency with IgG, IgA, IgM and IgE serum levels at the time of diagnosis of immunodeficiency. A negative association was also found between IL-4 (p=0.45, r=−0.310) and IL-5 (p=0.3, r=−0.738) production by CD4+ T cells and IFN-γ production (unpublished data) in patients with LRBA deficiency.
Cytokine secretion by CD+ T cells in CVID and LRBA patients and HC. Comparison of IL-4, IL-5 and IL-10 production by CD4+ T cells of CVID (N=8) and LRBA (N=9) patients and HC (N=9), with stimulation by anti-CD3 and anti-CD28 mAbs. The median is represented by a horizontal line; each dot represents an individual patient or HC. N: number; LRBA: LPS responsive beige-like anchor protein; CVID: common variable immunodeficiency; HC: healthy controls.
Immunological abnormalities in CVID and LRBA deficiency include late B cell defects, hypogammaglobulinemia, and T-cell deficiency.6 Several T-cell abnormalities including decreased CD4+ T-cell count (especially reduction on Treg frequency and function), increased T-cell activation and apoptosis, and abnormalities in cytokine production have been reported in both CVID and LRBA deficiency.6,16 In this study, we found that the percentages of CD4+ T cells in the CVID and LRBA group were significantly lower than that in the HC. Our results were similar to previous reports on CD4+ T cell deficiency on CVID14 and LRBA deficiency.6 Several studies reported that patients with CVID and LRBA deficiency have lower Treg cell counts and higher level of effector CD4+ T cell.2,6,17 In CVID and LRBA patients, Treg cell defects are associated with dysregulated CD+ T cell activation and skewing toward effector and memory phenotype.18 This is synchronous with the high frequency of inflammatory and autoimmune diseases, and further with Treg deficiency and low secretion of IL-10 by CD4+ T cells in patients with CVID and LRBA deficiency. This may be one of the probable causes of the high frequency of inflammatory and autoimmunity in these patients. Although in the current study there was a high frequency of inflammatory and autoimmune diseases in LRBA patients, this was not shown in CVID patients. The fewer clinical symptoms (other than infections) in CVID patients in our study are evident in comparison with the previous study. We suggest that the lower frequency of clinical symptoms in our study in comparison with the previous study is due to the inclusion of CVID patients with no gene defects in our study. Obviously, during mutation analysis, the monogenic disorders like LRBA, CTLA-4, CD27 and CD70 deficiencies and IPEX syndrome that have more severe clinical presentations including autoimmunity, enteropathy and lymphoproliferative diseases are detected,2,19,20 therefore, the rest of the CVID patients in whom mutations are not found, are those patients with milder clinical symptoms. Therefore, the pathogenesis of autoimmunity and inflammation in immunodeficiency is multifactorial and not only dependent to Treg defects.
Apart from inflammatory and autoimmune disorder, asthma and allergic diseases were also reported in patients with CVID and LRBA deficiency albeit with a lower frequency.6,21 In our previous study of 187 CVID patients, 22 (11.7%) had asthma and allergic diseases, and seven (3.7%) presented more than one allergic manifestation.21 In another study asthma had been diagnosed in 14.5% and atopy had been identified in 9.7% of CVID patients.22 In the current study similar to autoimmunity and inflammatory complications, atopic diseases were not observed in the CVID patients but only reported in two of the LRBA patients. The low frequency of asthma and allergic diseases in our study may be due to the exclusion of patients with monogenic defects from the CVID population. Transcription factor GATA3 and IL-4 was reported to be associated with Th2 response and atopic diseases.23 In the current study there was no statistical difference in the GATA3 and IL-4 level between HC with CVID and LRBA patients (the higher GATA3 expression was only shown in two LRBA patients with atopic diseases). This was in agreement with a lower frequency of atopic diseases in these patients. We proposed that the unchanged Th2 response in our LRBA patients may be due to the dysregulation of immune homeostasis. Consistent with immune dysregulation and Treg cell disturbance, LRBA patients encounter higher episodes of infectious complications and overload of microbial antigens due to hypogammaglubolinemia.7 The antigen derived from pathogens can activate naive CD4+ T cells, which can differentiate into effector CD4+ T cells particularly Th1, Th17 and Th22 cells. The results of our recent study showed that the frequencies of Th1, Th1-like Th17 and Th22 cells along with the expression of their transcription factors were significantly increased in patients with LRBA deficiency. Moreover, IFN-γ and IL-22 production in LRBA-deficient CD4+ T cells were elevated after lymphocyte stimulation.15 This Th subset and their pro-inflammatory cytokines have been shown to participate in the antibody production fate and pathogenesis of autoimmune diseases.7,24,25 Our result showed that the CD4+ T cell of CVID patients produce lower levels of IL-10 and IL-4, but review of the literature showed a heterogeneous and contradictory Th2 cytokine signature in CVID patients.10,26 We proposed that the altered Th2 cytokine profile in various studies could be attributed to the assessment of cytokine production or gene expression in PBMC, or purified CD4+ T cells, use of different stimulators and different inclusion criteria for CVID patients; additionally differences in patients’ clinical status and associated complications which may lead to activation of myeloid and lymphoid lineages that especially could be driven by the high prevalence of inflammatory and/or bacterial infections in the gastrointestinal or respiratory tract in the majority of CVID patients. There is no relevance between our obtained results in CVID with other studies since the inclusion criteria for CVID patients (those patients who had no known monogenic disease included in the study) in our study were different from previous studies. Notably, there is no similar study in patients with LRBA deficiency, to compare obtained results with those from other studies.
ConclusionAlthough it was proposed that the imbalance of CD4+ T cell subsets (particularly Th1, Th17, and Treg cells) and their corresponding cytokines might be a milestone in the immunopathogenesis of autoimmunity and inflammatory futures in patients with CVID and LRBA deficiency, it does not however seem that the Th2 subset and its relevant cytokines play an important role in the pathogenesis of these two diseases.
Conflicts of interestThe authors declare that they have no conflicts of interest.
This work was supported by the vice chancellor for research, Alborz University of Medical Sciences, under Grant No. 1396-02-00-1521. The authors would like to thank the Clinical Research Development Center of Imam Ali-Karaj Hospital.