Helicobacter pylori quantity and HP-NAP gene expression were evaluated in the faeces of healthy and asthmatic children.
MethodsH. pylori DNAs and RNAs were isolated from the stool samples of 92 asthmatic children (AC; 3–8 years) and 88 healthy controls (HC). Quantitative PCR was used to determine the quantity of H. pylori and HP-NAP expression relative to the 16S rRNA (reference gene). Gene expression was analysed using the delta delta-Ct method.
ResultsH. pylori DNA was detected in the stool samples of 18 (20.4%) of the 88 HC (p<0.0001, OR=0.79) and none of AC. No meaningful statistical differences were found between individuals with positive and negative family histories for asthma in AC and HC (p>0.05). H. pylori quantity was higher in seven of 18 H. pylori-positive samples, but HP-NAP expression levels were low in four of these seven samples. Based on a multivariate logistic regression analysis of these three variables together, only males displayed a significant difference based on gender differences (p<0.02) and it was determined that, based on the OR value of 0.46 and the 95% CI range of 0.241–0.888, male gender was an independent protective factor in asthma.
ConclusionsHP-NAP levels vary to the relative concentrations of bacteria in the stationary or late logarithmic phases. Different napA expression levels may be caused by different endogenous napA gene expression or different environmental conditions.
Asthma is a common chronic disease in childhood that is defined by chronic inflammation of the lower airways and the development of wheezing, cough, shortness of breath, and chest tightness.1 Asthma increased from 235 to 334 million cases between 2011 and 2014, while 14% of the world's children experience asthma symptoms and the burden of asthma is greatest for children aged 10–14 and the elderly aged 75–79.2 The hygiene hypothesis, an inverse association between infection and atopy, was first proposed by David Strachan in 1989. In this hypothesis, reduced childhood exposure to microorganisms shifts Th1/Th2 immune responses to Th1, reducing the number of allergic disorders. Strachan suggested that fewer family members would result in unhygienic contact, which would be protective and avoid cross-infection from siblings.3
Several studies support an inverse association between Helicobacter pylori infection and the frequency of allergic asthma.4,5H. pylori is a Gram-negative, microaerophilic gastric bacterium that colonises much of the world's population and is commonly acquired in childhood. In recent years, H. pylori prevalence has decreased substantially in developing countries due to socioeconomic development, effective therapy, and hygiene improvements.6,7 An inverse correlation was observed between prior acquisition of H. pylori and the possibility of being diagnosed with asthma or allergy, and the decreasing incidence of H. pylori in the developed world paralleled an increase in the incidence of childhood allergies and autoimmune diseases. This observation came from the third National Health and Nutrition Examination Survey (NHANES).8 Another suggestion was the relation of reduced risk of childhood-onset (≤15 years) asthma with cagA+ strains of H. pylori.9,10 In a study from Finland, a three-fold increase in the incidence of allergy was reported, with a 30% decrease in H. pylori prevalence, between 1973 and 1994.11H. pylori infections are usually acquired in early childhood, when the immune system is not mature. H. pylori infections stimulate a Th1 response in the gastric mucosa and also in peripheral blood. Cytokines of Th1 cells can suppress a Th2 response, which is related to the H. pylori neutrophil-activating protein (HP-NAP), an important Th1-promoting virulence factor that inhibits Th2 cytokine release in humans and mice.5
We suggested that the dominant factor of the hygiene hypothesis may possibly be the immunomodulation function of H. pylori by HP-NAP. In our study, we aimed to determine the relationship between allergic asthma and H. pylori's protective effect by quantifying the number of H. pylori bacteria in the stool samples of diagnosed asthmatic children (allergic asthma) and matched healthy control children, and also by quantifying the expression of the HP-NAP gene implicated in the disease.
Materials and methodsPatient and control groupsOur study was conducted as a cross-sectional, case control study conducted from March 2014-January 2015. Ninety-two children diagnosed with clinical asthma, as defined by the international diagnostic criteria,12 between the ages of three and eight who applied to the Allergy Polyclinic of the Pediatrics Department of Istanbul University Cerrahpasa Medical Faculty were included in the study. Fifty-four (58.6%) of the cases were boys and 38 (41.3%) were girls. The mean age was 5.67±1.24 years. All clinical symptoms and laboratory data (immunological, biochemical, etc.) belonging to the patients that were consistent with allergic asthma were taken from the patients’ files. The control group was composed of 88 healthy children with a similar mean age, gender distribution, and standard of living who applied to the healthy children polyclinic of the same medical centre in the same timeframe. Of the control group, 52 (59%) were boys and 36 (40.9%) were girls. The mean age was 5.43±1.61 years (p>0.05). While the control and study groups were being assembled, those who had used antibiotics at any time during the two weeks prior to the beginning of the study, as well as those who had a history of an infectious disease in the past month, were excluded from the study.
Collection of the stool sampleIn order to investigate H. pylori quantity and gene expression, 20g stool samples were collected from cases consistent with the study criteria who applied to the Allergy and Healthy Children Polyclinics of Cerrahpasa Medical Faculty. Stool samples were divided into two parts: one part stored at −20°C for H. pylori DNA isolation and the other stored at −80°C for RNA isolation until molecular investigations began. The location of storage was inside the MagNA PureLC DNA isolation kit in the Medical Microbiology department.
Molecular testsBacterial DNA isolation from stool samples was performed using a Stool DNA isolation kit (cat: 27600, Norgen Biotek Corp, Canada); bacterial RNA isolation was performed using a Stool Total RNA isolation kit (cat: 49500, Norgen Biotek Corp, Canada) in accordance with the manufacturer's instructions. The DNA and RNA concentrations in the specimens following all isolations were quantified spectrophotometrically using a Nanodrop instrument (Thermo Scientific, USA). The obtained DNA samples were stored at −20°C until the H. pylori quantitative PCR (qPCR) study began. The RNA samples obtained were reverse-transcribed into cDNA using the Transcriptor First Strand cDNA synthesis kit (cat: 04896866001, Roche Diagnostics GmBH, Germany) in accordance with manufacturer instructions. The obtained cDNAs were stored at −20°C until the HP-NAP A qPCR study began.
In order for the obtained DNA to be detected by H. pylori qPCR, the Genesig H. pylori kit (cat: Path-Hpylori, Genesig, PrimerDesign Ltd, UK) and the oasig lyophilised qPCR MasterMix kit (cat: Precision-oasig150, Genesig, PrimerDesign Ltd, UK) were used and the studies were performed on a LightCycler 96 (Roche Diagnostics GmBH, Germany) qPCR instrument in accordance with manufacturer instructions. The PCR protocol of the study was as follows: denaturation for 2min at 95°C, followed by 50 cycles of 10s at 95°C and 60s of 60°C. The primer and probe series were created for the HP-NAP (GenBank Accession No.: U16121.1) and 16S rRNA genes using the NCBI primer design and primer blast tool websites. The 16S rRNA gene was used in our study as a reference gene. The primers and the probes were ordered from IDT (Integrated DNA Technology, USA) and used for gene expression studies. As primer-probe pairs, cDNAs and the Lightcycler 480 Probe master kit (cat: 04707494001, Roche Diagnostics GmBH, Germany) were used in accordance with manufacturer instructions. The gene expression studies were performed using a LightCycler 96 (Roche Diagnostics GmBH, Germany) system. For each run, to normalise the amount of sample cDNA added to each reaction, the Cq value of the HP-NAP gene was subtracted by the Cq value of the housekeeping (reference) 16SrRNA gene (delta Ct (ΔCT)=Cq HP-NAP−Cq 16SrRNA), and then for a comparison between the expression of HP-NAP in vitro and in vivo, the delta Cq values of the in vivo were subtracted by the delta Cq value of the in vitro [delta−delta Ct (ΔΔCT)]=delta Cq in vivo−delta Cq in vitro). The relative fold changes were calculated by the formula of 2−ΔΔCt method published by Livak and Schmittgen.13
Statistical analysisBiostatistics statementThe statistical methods of this study were reviewed by Suphi Vehid from the Department of Biostatistics, Istanbul University Cerrahpasa Faculty of Medicine, Istanbul University, Istanbul, Turkey. For the evaluation of the data obtained from the study, IBM SPSS Statistics version 20 package software was used for statistical analysis and calculations. As complementary statistical methods, the mean average and the standard deviation values were calculated. The Mann–Whitney U test was used for the comparison of the averages between the two groups. For the p values that were obtained from these tests, p<0.05 was meaningful, p<0.01 was highly meaningful, and p<0.001 was very highly meaningful. The chi-squared test was used for the association of categorical variables. The effects of independent variables (the existence of H. pylori, a positive family history of asthma, being a male, etc.) on the existence of asthma were evaluated using logical regression analysis, which is a multivariate analysis.
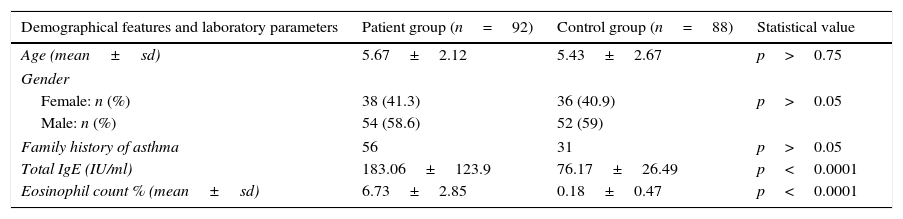
ResultsNo meaningful statistical differences were found between individuals with positive and negative family histories for asthma in the patient and control groups (p>0.05). The differences in both the mean IgE levels and the mean eosinophil percentage rates between the patient and control groups were very highly meaningful (p<0.0001) (Table 1).
Demographical features and laboratory parameters of patient and control groups.
| Demographical features and laboratory parameters | Patient group (n=92) | Control group (n=88) | Statistical value |
|---|---|---|---|
| Age (mean±sd) | 5.67±2.12 | 5.43±2.67 | p>0.75 |
| Gender | |||
| Female: n (%) | 38 (41.3) | 36 (40.9) | p>0.05 |
| Male: n (%) | 54 (58.6) | 52 (59) | |
| Family history of asthma | 56 | 31 | p>0.05 |
| Total IgE (IU/ml) | 183.06±123.9 | 76.17±26.49 | p<0.0001 |
| Eosinophil count % (mean±sd) | 6.73±2.85 | 0.18±0.47 | p<0.0001 |
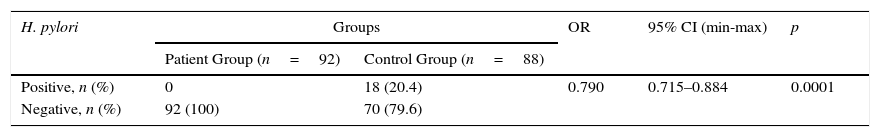
H. pylori DNA was detected in the stool samples of 18 (20.4%) of the 88 healthy controls (p<0.0001, OR=0.79) and none of the asthmatic samples (Table 2).
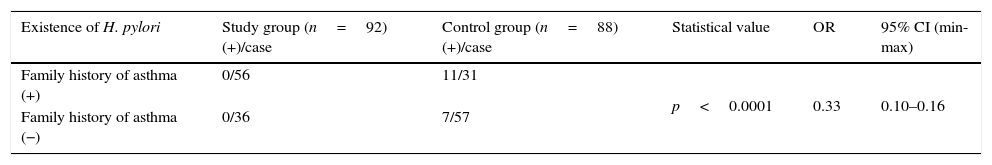
When the distribution of H. pylori was analysed based on family history for asthma in the patient and control groups, the distribution in the control group was statistically significantly higher than in the patient group (p<0.0001) and the OR value was 0.33 (Table 3).
The distribution of H. pylori was analysed based on family history for asthma in the patient and control groups.
| Existence of H. pylori | Study group (n=92) (+)/case | Control group (n=88) (+)/case | Statistical value | OR | 95% CI (min-max) |
|---|---|---|---|---|---|
| Family history of asthma (+) | 0/56 | 11/31 | p<0.0001 | 0.33 | 0.10–0.16 |
| Family history of asthma (−) | 0/36 | 7/57 |
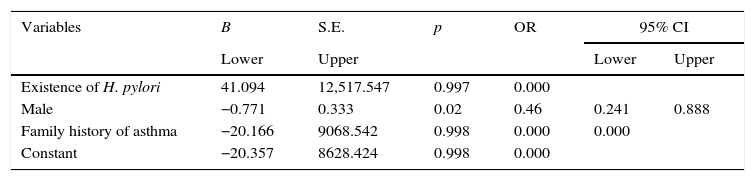
In our research, based on the univariate analysis of variables, such as the differences in H. pylori distribution according to the presence of H. pylori, gender and family history for asthma in the patient and control groups, we found that differences in H. pylori distribution between the two groups and the presence of H. pylori according to family history for asthma were significantly higher in control group (p<0.05); male control group had a statistically significantly higher presence of H. pylori than the male patient group. On the other hand, based on a multivariate logistic regression analysis of these three variables together, only males displayed a significant difference based on gender differences (p<0.02) and it was determined that, based on the OR value of 0.46 and the 95% CI range of 0.241–0.888, male gender was an independent protective factor in asthma (Table 4).
Multivariate logistic regression analysis of variables that could be risk factors for asthma.
| Variables | B | S.E. | p | OR | 95% CI | |
|---|---|---|---|---|---|---|
| Lower | Upper | Lower | Upper | |||
| Existence of H. pylori | 41.094 | 12,517.547 | 0.997 | 0.000 | ||
| Male | −0.771 | 0.333 | 0.02 | 0.46 | 0.241 | 0.888 |
| Family history of asthma | −20.166 | 9068.542 | 0.998 | 0.000 | 0.000 | |
| Constant | −20.357 | 8628.424 | 0.998 | 0.000 | ||
B; beta regression coefficient, S.E.; standard error, OR; odds ratio, CI; confidence interval.
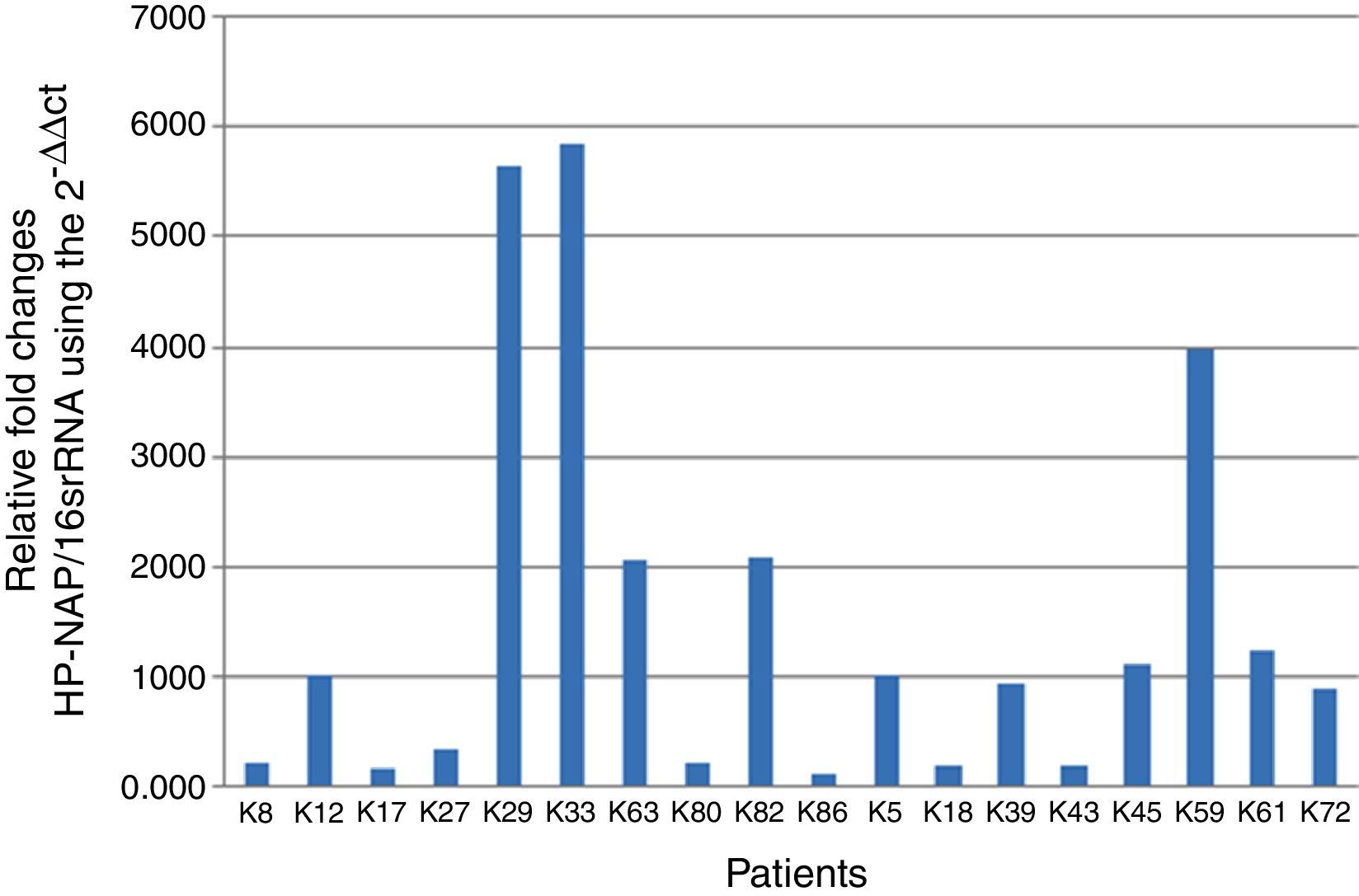
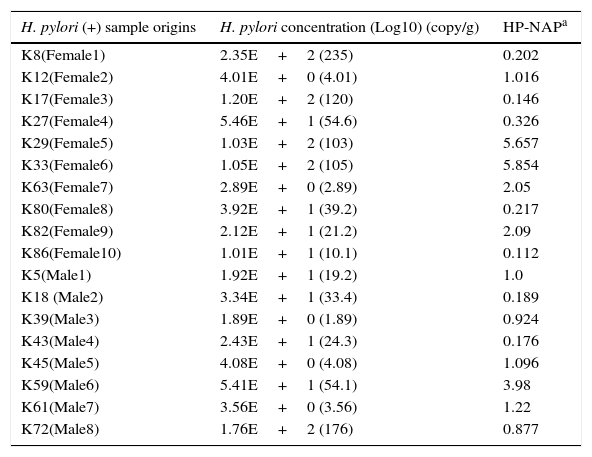
H. pylori was detected in 18 stool samples taken from healthy children. The log10 quantities of H. pylori in these 10-g samples and the relative gene expression levels of HP-NAP compared to 16S rRNA in the same samples are expressed in Table 5. H. pylori quantities in seven of the H. pylori-positive stool samples were higher than those in eleven of the samples (Table 5).
H. pylori and HP-NAP quantities in H. pylori positive cases.
| H. pylori (+) sample origins | H. pylori concentration (Log10) (copy/g) | HP-NAPa |
|---|---|---|
| K8(Female1) | 2.35E+2 (235) | 0.202 |
| K12(Female2) | 4.01E+0 (4.01) | 1.016 |
| K17(Female3) | 1.20E+2 (120) | 0.146 |
| K27(Female4) | 5.46E+1 (54.6) | 0.326 |
| K29(Female5) | 1.03E+2 (103) | 5.657 |
| K33(Female6) | 1.05E+2 (105) | 5.854 |
| K63(Female7) | 2.89E+0 (2.89) | 2.05 |
| K80(Female8) | 3.92E+1 (39.2) | 0.217 |
| K82(Female9) | 2.12E+1 (21.2) | 2.09 |
| K86(Female10) | 1.01E+1 (10.1) | 0.112 |
| K5(Male1) | 1.92E+1 (19.2) | 1.0 |
| K18 (Male2) | 3.34E+1 (33.4) | 0.189 |
| K39(Male3) | 1.89E+0 (1.89) | 0.924 |
| K43(Male4) | 2.43E+1 (24.3) | 0.176 |
| K45(Male5) | 4.08E+0 (4.08) | 1.096 |
| K59(Male6) | 5.41E+1 (54.1) | 3.98 |
| K61(Male7) | 3.56E+0 (3.56) | 1.22 |
| K72(Male8) | 1.76E+2 (176) | 0.877 |
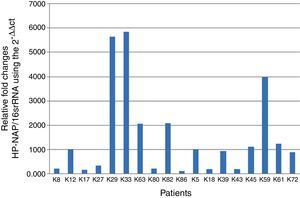
The bar graph shows the ratios of HP-NAP to the reference 16S rRNA gene using the 2−ΔΔCt method, where the vertical bars represent H. pylori-positive patients (18 selected samples of H. pylori positive control group). The standard deviation is visible due to repeating the experiment three times (Fig. 1).
DiscussionIn 1998, Chow et al.14 reported an inverse relation between cagA+ strains of H. pylori infection and the risk of oesophageal and gastric cardia adenocarcinoma. They suggested that the protection from oesophageal and gastric cardia adenocarcinomas was related to a reduced gastric acidity due to cagA+ H. pylori lowering the gastric acidity. Even though a certain relationship did not yet exist, cumulative reports related to the positive effects of H. pylori drew attention to the positive effects of this bacterium. In an extensive literature survey, we found many studies that suggested an inverse correlation between the risk of developing extra-gastric diseases and the presence of H. pylori infection. These diseases include gastro-oesophageal reflux disease and esophagitis, asthma and allergy, several autoimmune disorders (including coeliac disease, systemic lupus erythematosus, rheumatoid arthritis, and multiple sclerosis), inflammatory bowel disease, and irritable bowel syndrome.4,15–19
In 2000, Matricardi et al.20 reported that, compared to controls, there was a lower prevalence of T. gondii, hepatitis A virus, and H. pylori (p=0.325) in atopic participants. Allergic asthma and allergic rhinitis were infrequent among the participants exposed to H. pylori, T. gondii, and hepatitis A virus in their study. In another study, Chen and Blaser,10 evaluated the associations of H. pylori status with a history of asthma and allergy and with skin sensitisation using data from 7663 adults; they concluded that the presence of cagA+ H. pylori strains was inversely related to having asthma (OR: 0.79), and the inverse association of cagA positivity with childhood-onset (age ≤15 years) asthma was stronger (OR: 0.63) than that with adult-onset asthma (OR: 0.97). In our study, similar to the above study, a statistically significant difference was found between these groups (p<0.0001, OR=0.79). On the other hand, all the stool samples of our study group were negative for H. pylori DNA, while 18 stool samples of the 88 (20.4%) control group samples were positive for H. pylori DNA, and the difference between the two groups was very highly significant (p<0.0001, OR=0.79). In a study including 300 children (5–18 years old), Khamerchian et al.21 demonstrated that among 138 H. pylori-positive patients, eight cases (5.8%) were asthmatic while 28 (17.3%) of the 162 H. pylori-negative patients were asthmatic. This difference was statistically significant (p=0.002). They suggested that there was an inverse correlation between H. pylori infection and asthma in childhood.
The most important immunological mechanism discovered in the last years was the stimulation of Treg cells by H. pylori. Arnold et al.22 demonstrated that H. pylori infection protected mice against the clinical and histopathological symptoms of asthma in an experimental model of allergic airway disease induced by ovalbumin-specific sensitisation.
Oertli et al.23 also indicated that dendritic cells (DCs) exposed to H. pylori are programmed to become tolerogenic, driving Treg differentiation and protection from asthma by the production of IL-18. They suggested that direct contact between H. pylori and DCs was needed to induce tolerogenic DCs. In their experiments, they also observed that IL-18 derived from DCs has an important role in the conversion of naive T cells to Tregs, which will inhibit Th2 responses. Oertli et al.24 reported recently two H. pylori virulence determinants, VacA and γ-glutamyl transpeptidase (GGT), that promote the efficient induction of Tregs, and VacA is required to prevent allergen-induced asthma. H. pylori infection was needed to reprogram dendritic cells (DCs) into a tolerogenic phenotype and H. pylori infection induces regulatory T cells (Tregs) with highly suppressive activity in models of allergen-induced asthma.
It was proposed that the IL-10 released from Treg cells stimulated by H. pylori has an important role in suppressing Th2 activity. IL-10 suppresses mast cell activation and cytokine and eosinophil production in these cells.5,25,26 There are research attempts to find a plausible mechanism for the inverse relationship between H. pylori infection and asthma. In the last few years, emphasis was placed on HP NAP. HP-NAP application increased IFN-γ producing T-cells and decreased IL-4 secreting cells when HP-NAP was added to allergen-induced T-cell lines.27 Both systemic and mucosal administration of HP-NAP effectively prevented allergic asthma. It was reported that H. pylori infections acquired in childhood, which is usually considered the time of onset for asthma, may cause a Th1-mediated immune response in the gut mucosa and in peripheral circulation, and consequently cause the release of cytokines from Th1 cells that inhibit the Th2 response, which is the most important T-cell response related to allergy.5 In order to explain why this protection by H. pylori was only seen in children, Matsushima and Nagai28 also showed the importance of neonatal infection and explained why adults infected with H. pylori do not respond similarly to neonatally infected mice when exposed to allergens. This may be due to differences in IL-18 production in neonates and adults or may be due to barrier differences between the neonatal gut and adult gut.
It was shown that HP-NAP, a 150-kDa oligomeric protein, increases the synthesis of tissue factors in monocytes and the release of oxygen radicals from neutrophils. It also increases the expression of TNF-α, IL-23, IL-18, IFN-γ, and IL-12; IL-12 is the most important cytokine in the differentiation of Th cells into the Th1 phenotype and triggers neutrophils and monocytes via an agonistic interaction with TLR2. Thereafter, Th2 response, which is dominant in allergies, shifts towards the Th1 response. In another in vitro study, HP-NAP inhibited bronchial inflammation related to allergic asthma.29–32 It was also reported that H. pylori infections can alter the Th1/Th2 balance by affecting gastric hormones and that a reduction in somatostatin levels and an increase in gastrin production inhibit the release of Th2 cytokines and modulate a Th1 immune response.5,27,33
HP-NAP, by acting on both neutrophils and monocytes following the engagement of TLR2, significantly contributes to creating an IL-12- and IL-23-enriched milieu, and as such it represents a key bacterial factor that can drive the differentiation of antigen-stimulated T cells towards a polarised Th-1 phenotype.27 Furthermore, HP-NAP triggers the production of reactive oxygen species (ROS) inside neutrophils, thus affecting the adhesion of neutrophils to endothelial cells. HP-NAP activates neutrophils by stimulating ROS production and myeloperoxidase release, and HP-NAP regulates β2-integrin expression by stimulating chemotaxis.34 Amedei et al.35 showed that the addition of HP-NAP to allergen-induced T-cell lines derived from allergic asthmatic patients led to a drastic increase in IFN-γ-producing T cells and to a decrease in IL-4-secreting cells, thus resulting in a redirection of the immune response from a Th-2 to a Th-1 phenotype. Dundon et al.36 later demonstrated, in a growth medium that provides the normal reproduction conditions of H. pylori, that HP-NAP accumulates in the stationary phase of its reproduction. Thompson et al.37 showed that the levels of expression for many of the virulence genes of H. pylori, including HP-NAP, cagA, flaA and pfr, peak especially in the stationary or late logarithmic phase of bacterial reproduction. Researchers have named this critical level where gene expression attains a certain dynamic the “Log-Stat switch.” HP-NAP on iron starvation might serve as a means of releasing iron from host cells.38 Based on our findings, which indicate that in H. pylori-positive cases, HP-NAP levels are never constant, but rather are related to the relative concentrations of bacteria in the stationary or late logarithmic phases. Therefore, we suggest that different napA expression levels in our H. pylori-positive cases may be caused by different endogenous napA gene expression levels or different environmental conditions that affect the expression of the napA gene. In order to confirm this hypothesis, while the total DNA quantity of the K27 sample was 5.46E, the expression level of napA was 0.326. On the other hand, the total DNA quantity of K29 sample was 1.03E and the expression level of napA was 5.657. As seen from the above results, the expression level of napA is independent of the H. pylori quantity. In the present study, the inverse association between H. pylori and current asthma status in children was statistically evaluated and the presence of H. pylori was inversely related to ever having had asthma (OR, 0.79; 95% CI, 0.715–0.884). At the same time, the inverse association with being male was stronger (OR, 0.46; 95% CI, 0.24–0.88). These results suggest that there is an inverse relation between H. pylori infection and the asthma status of the children in our study groups.
As a limitation of the study, we could not detect cagA+ strains and H. pylori factors, such as gamma-glutamyl transpeptidase (GGT) and H. pylori vacuolating cytotoxin A (VacA), which also produce a potent Treg cell response. Measuring the expression of the H. pylori napA gene may be important for the development of new strategies (like vaccines) against H. pylori diseases. HP-NAP may play a role in mechanisms that underlie such a negative association. HP-NAP can be considered an important candidate for novel strategies to prevent and treat asthma and allergic diseases by inhibiting Th2 responses.31 As a conclusion, HP-NAP was suggested as an important protein to protect children from allergic diseases (asthma) by shifting Th1/Th2 immune responses to Th1, which shifts away from allergic disorder formation.
We believe that that more serial and comprehensive studies that include cagA+ and VacA+ H. pylori strains and also HP-NAP vaccine studies to protect from childhood asthma are needed and could bring new perspectives on this matter.
Ethical disclosuresConfidentiality of dataThe authors declare that they have followed the protocols of their work centre on the publication of patient data and that all the patients included in the study have received sufficient information and have given their informed consent in writing to participate in that study.
Right to privacy and informed consentThe authors have obtained the informed consent of the patients and/or subjects mentioned in the article. The author for correspondence is in possession of this document.
The study was approved by the Clinical Research Ethics Board of Istanbul University, Cerrahpasa Faculty of Medicine, and all patients gave their informed consent before participating in the study. Ethical approval: 83045809/6030 06 March 2014.
Protection of human and animal subjectsThe authors declare that the procedures followed were in accordance with the regulations of the responsible Clinical Research Ethics Committee and in accordance with those of the World Medical Association and the Helsinki Declaration.
Conflict of interestThe authors declare that there are no conflicts of interest.
Supported by Research Fund of the Istanbul University, project number: 1404.
This research was presented as a poster in the 17th International Congress On Infectious Diseases (ICID) Hayderabad, 2016, Abstract Number: 41.138.
We also thank Harika Oykü Dinç (Cerrahpasa Medical Faculty, Turkey), Hayriye Kırkoyun (Istanbul Medical Faculty, Turkey), Esad Bonabi (Aydin University, Turkey) for their technical assistance related to the poster presentation in the 17th ICID.