Background. The 20S proteasome is the proteolytic core of the major intracellular protein degradative system, the ubiquitin-proteasome system. Since little is known about proteasomes of human liver, we have investigated the proteasome spectrum in adult human liver.
Material and methods. 20S proteasomes were chromatographically purified from adult human liver and from HuH7 cells. They were divided into subpopulations and subtypes and characterized with regard to their proteolytic activities using short fluorogenic oligo- and long poly-peptide substrates. Their subunit composition was studied by immunoblotting.
Results. Proteasomes from adult human liver tissue can be separated into three subpopulations (I, II, III), each of which is composed of several subtypes, which total to a spectrum of 14 different subtypes. Two minor subtypes contain only the immuno-subunits β1i and β5i but not their standard counterparts; all others are intermediate subtypes containing β1 and β5 standard- and β1i and β5i immuno-subunits in various compositions. With regard to the proteolytic activities we observed that a decreasing content of subunit β1i in the subtypes goes along with a decreasing ratio of chymotrypsin-like/caspase-like activity, whereas the degradation rate of a 30 mer polypeptide substrate increased with decreasing β1i content. By comparison, 20S proteasomes from HuH7 cells do not contain immuno-subunits but are pure standard proteasomes, which can be separated into three subtypes.
Conclusion. These findings suggest that adult human liver contains a spectrum of 14 different 20S proteasome subtypes with different enzymatic properties reflecting most probably an adaptive response of liver cell functions to challenging factors during lifetime.
The liver is a central organ for whole body protein metabolism since it is the main tissue for nitrogen-fixation from nutrient derived compounds and for nitrogen-elimination by means of urea biosynthesis as well as the site of synthesis and breakdown of the majority of blood plasma proteins. Therefore, it is not surprising that this tissue has been used for many basic studies of protein turnover1 and especially intracellular protein degradation within the endosomal-lysosomal compartment (for a review see2). Moreover, the phenomena that all proteins have distinct and different half-lifes3 and that the process of protein breakdown is energy-dependent have also been detected in liver tissue.4 These findings induced several investigators to search for an ATP-dependent proteolytic system in this tissue. Actually, a neutral, ATP-dependent protease was identified, characterized and later on also purified from rat liver extracts.5–8 It was designated proteasome and found to be the core protease of the ubiquitin-proteasome system that is responsible for the highly regulated breakdown of the majority of intracellular proteins.9 The 20S proteasome is a multicatalytic proteinase with three different active site-containing β-subunits, β1, β2, and β5, which are integrated in a ring of seven β-subunits that is attached to a ring formed by seven α-subunits. The whole enzyme is a hetero-dimer with a subunit configuration of a7β7 β7α7. The active sites are burrowed in the interior of the barrel-shaped complex and approachable for substrate proteins only through a small diameter central pore in the outer α-ring.10 For the three β-subunits that catalyze peptide-bond hydrolysis alternative subunits exist and are designated immuno-subunits β1i, β2i and β5i, because their genes are located in the major histocompatibility complex. These subunits are constitutively expressed in cells of the immune system but are also expressed in most other cells, when exposed to cytokines like interferons. Proteasomes containing these immuno-subunits were designated immuno-proteasomes and were originally thought to be mainly responsible for protein processing to generate major histocompatibility complex class I epitopes but are now known to play a more general role in protein homoeostasis.11,12 Many cells contain proteasomes comprising both standard- and immuno-subunits, which are designated intermediate-type proteasomes.13 Control of 20S proteasome activity is ensured by association of various regulatory complexes like the 19S regulator (19S Reg), PA28, PA200 and PI31.14 Association of some of these regulators to the 20S proteasome leads to the formation for instance of the 26S and hybrid proteasomes, respectively, which are able to degrade substrate proteins marked for degradation by polyubiquitination.15 Although many investigations on the function of the ubiquitin-proteasome system have been performed with liver tissue and the proteasomes of liver have been characterized from species like rat,6,7,16–19 mice,20,21 calf,22,23 chicken,24 and ostrich,25 the knowledge about the 20S proteasome from human liver is very poor. 20S proteasomes purified from human normal and neoplastic liver tissue were tested on their susceptibility to various protease inhibitors and monovalent cations at a time, when neither the proteolytic mechanism nor the subunit complexity of proteasomes were known.26,27 Therefore, we have purified 20S proteasomes from human liver and have characterized its subunit composition and protease activity. Since hepatocytes amount to 80% of liver volume, we have also purified 20S proteasomes from the human hepatocarcinoma cell line HuH7 and compared their properties with those of adult liver tissue.
Material and MethodsMaterialHuman liver and spleen tissue was obtained from three medico-legal autopsies with postmortem intervals ranging between 6 to 12 h postmortem including at least a cooling period of 1 to 5 h prior to autopsy. While proteasome-regulator complexes (e.g. 26S proteasomes) may partially dissociate under these conditons, 20S proteasomes are known to be stable. The cause of death varied from different natural to unnatural causes. The liver was neither injured nor affected by any pathology prior to death in any of the cases. Consent to use the human postmortem samples was given by the prosecutor in charge.
Human erythrocyte concentrates were bought from the German Red Cross (Berlin, Germany). Antibodies towards subunits β5, β1i and β5i were from Abcam (UK) and against α1 from Cappel (Belgium). Rabbit antiserum K43 against subunit β1 was laboratory stock. HuH7 cells were grown in DMEM medium supplemented with 4.5% glucose, 10% FCS, 1% 200 mM glutamine.
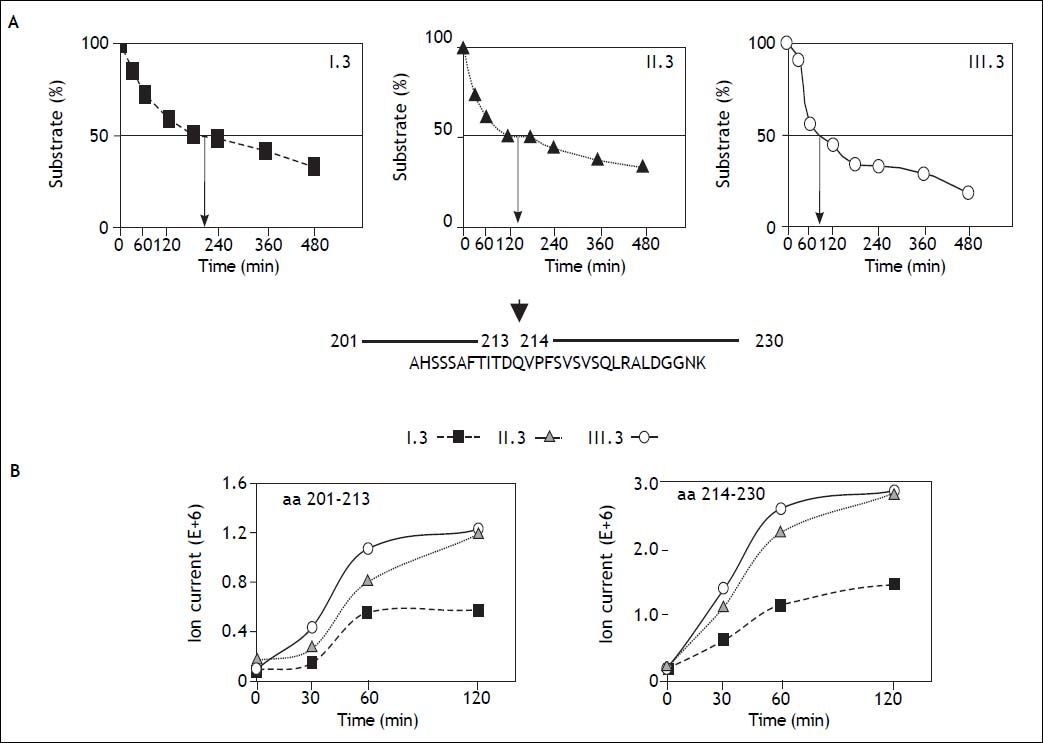
Measurement of proteolytic activityFluorogenic peptide substrates were purchased from Bachem AG (Switzerland). Substrate for the chymotrypsin-like activity was Suc-LLVY-MCA (succinyl-LLVY-methylcoumarylamide), for trypsinlike activity Bz-VGR-MCA (benzoyl-VGR-methylcoumarylamide) and for caspase-like activity was Z-LLE-MCA (carbobenzoxy-LLE-methylcoumarylamide). All substrates were dissolved in TEAD buffer (20 mM Tris/HCl, 1 mM EDTA, 1mM NaN3, 1 mM DTT, pH 7) at concentrations of 200 (Suc-LLVYMCA) and 400 (Bz-VGR- and Z-LLE-MCA), respectively. For measurement of proteasome activity 20 enzyme solution was incubated with 20µL of substrate solution at 37 °C and enzymatically released MCA was fluorometrically measured after 30 min at 355 nm excitation and 460 nm emission. The polypeptide substrate gp100201–230 (sequence: AHSSSAFTITDQVPFSVSVSQLRALDGGNK) referred to the human gp100PMEL17 sequence) was synthesized by using Fmoc solid phase chemistry.28 For digestion 20S proteasome (0.5 µg) was incubated with 40 of the polypeptide dissolved in TEAD buffer at 37 °C over a period of 8 h. Digestion was stopped at time points indicated in figure 1 by adding trifluoroacetic acid at a final concentration of 0.3% and frozen at −20 °C. The digestion mixture (substrate and its degradation fragments) was then separated by liquid chromatography and analyzed by electrospray ionization mass spectrometry (ESI-MS).29 The quantity of detected ions is proportional to the produced signal for each fragment. Peak areas defined by mass to charge and retention time illustrate the relative rate of product formation as described in detail elsewhere.30
Degradation of a 30mer polypeptide by three different 20S human liver proteasome subtypes. A. The 30mer polypeptide (AHSSSAFTITDQVPFSVSVSQLRALDGGNK) (m/z = 1040.6; charge +3) derived from the human melanocyte protein GP100 was incubated with equal amounts of the three major 20S proteasome subtypes I.3, II.3, and III.3. At the time points indicated the amount of non-degraded substrate was measured by mass spectrometry. Arrows indicate the incubation time needed to obtain 50% substrate degradation. B. The amount (ion intensity) of the two degradation fragments aa201-213 (m/z = 682.3; charge +2) and aa214-230 (m/z = 887.4; charge +2) generated by the 20S proteasome subtypes I.3, II.3, and III.3 are plotted versus incubation time. The result corroborate the data in panel A showing that subtype I.3 works most slowly.
Liver as well as spleen tissue was homogenized in a 4-fold (w/v) volume of TEAD-buffer by means of 10 strokes in a Potter-Elvejhem homogenizer. HuH7 cells were disrupted by repeated freeze-thawing and additional sonifying. The suspensions obtained were centrifuged for 60 min at 20,000g and the resulting supernatants were used to isolate and purify 20S proteasomes according the methodology described by.31 Proteasome from erythrocytes were purified in a similar way. Separation of purified proteasomes by hydrophobic interaction chromatography and high resolution anion-exchange chromatography into subpopulations and subtypes, respectively, was performed as detailed in.32
Electrophoretic techniquesFor non-denaturing polyacrylamide gel electrophoresis (native PAGE) 5 of purified proteasome was loaded to NativePage 3-12% Bis-Tris Gels (Invitrogen, Germany) and run at 4 °C for 14 hours with 40 V. To detect proteasome activity by substrate overlay technique the gels were incubated in 100 μΜ Suc-LLVY-MCA dissolved in TEAD buffer at 37 °C for 15 min. Fluorescence was detected under UV illumination. The gel was then stained with Coomassie brilliant blue. SDS polyacrylamide gel electrophoresis (SDS-PAGE) was performed on 12.5% gels and the first dimension of the two-dimensional gel-electrophoresis (2D-PAGE) was performed under non-equilibrium pH gradient conditions with pH-gradient from 3 to 10.33 After electrophoresis proteasomes were blotted onto polyvinylidene difluoride-membranes, which were blocked with 5% milk powder dissolved in 20 mM Tris, 0.5 M NaCl, 1 mM NaN3, pH 7.5, incubated with subunit-specific antibodies, the binding of which were detected by enhanced chemiluminescence technique.
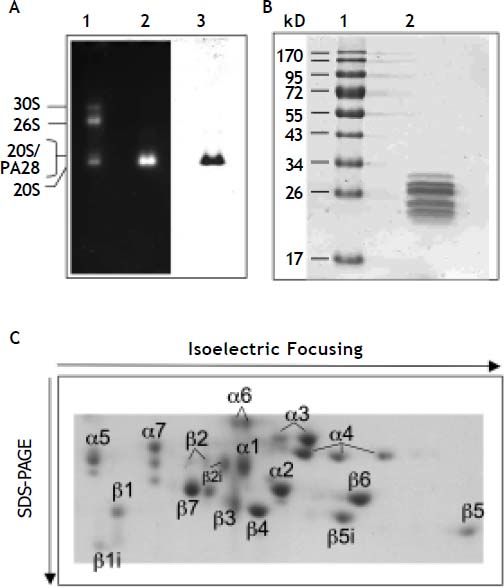
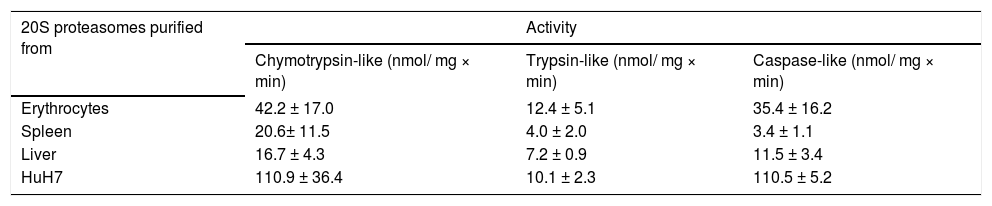
ResultsPurity and specific activity of 20S proteasomesAccording to analyses by SDS- and native-PAGE the 20S proteasomes were purified to apparent homogeneity (Figure 2A and B). Their specific activity was measured with fluorogenic peptide substrates and compared to 20S proteasomes purified from human spleen and erythrocytes. Erythrocytes contain standard proteasomes with high caspase-like activity, whereas spleen proteasomes contain predominantly immuno-subunits and exhibit low caspase-like activity. The caspase-like activity of liver proteasomes is higher than in spleen proteasomes but lower than in erythrocyte proteasomes (Table 1). This data suggest that human liver proteasomes are not pure standard proteasomes but may also contain immuno-subunits. Therefore, we analyzed the subunit composition of 20S proteasomes by 2DPAGE (Figure 2C). In fact both standard-subunits β1, β2 and β5 as well as the immuno-subunits β1i, β2i and β5i are present in human liver proteasomes and this subunit pattern is different from that found in proteasomes from erythrocytes and spleen.34 Therefore, we have employed hydrophobic interaction chromatography that is known to separate pro-teasomes according to their content of standard and immuno-subunits.29,35
Native-, SDS- and 2D-PAGE of 20S proteasome purified from human liver. A. A mixture of 30S, 26S, 20S and 20S/PA28 proteasomes from human erythrocytes (lane 1) and 5 μg 20S proteasome purified from human liver (lane 2) was subjected to non-denaturing PAGE. After electrophoresis proteasome activity was visualized by substrate overlay technique with Suc-LLVY-MCA (lane 1 and 2). Afterwards the part of the gel containing lane 2 was stained with Coomassie (lane 3). B. 0.5 μg of purified human liver 20S proteasome was subjected to SDS-PAGE and the gel stained with Coomassie. Molecular mass standards (lane 1); human liver 20S proteasome (lane 2). C. 30 μg of human liver 20S proteasome was subjected to 2D-PAGE and the proteasome subunits then stained with Commassie. Designation of proteasome subunits corresponds to.13
Proteolytic activities of 20S proteasomes purified from different human tissues and cells.
| 20S proteasomes purified from | Activity | ||
|---|---|---|---|
| Chymotrypsin-like (nmol/ mg × min) | Trypsin-like (nmol/ mg × min) | Caspase-like (nmol/ mg × min) | |
| Erythrocytes | 42.2 ± 17.0 | 12.4 ± 5.1 | 35.4 ± 16.2 |
| Spleen | 20.6± 11.5 | 4.0 ± 2.0 | 3.4 ± 1.1 |
| Liver | 16.7 ± 4.3 | 7.2 ± 0.9 | 11.5 ± 3.4 |
| HuH7 | 110.9 ± 36.4 | 10.1 ± 2.3 | 110.5 ± 5.2 |
Chymotrypsin-, trypsin- and caspase-like activities were measured with fluorogenic peptide substrates. Data are means ± SD of three (two, HuH7) separate enzyme preparations.
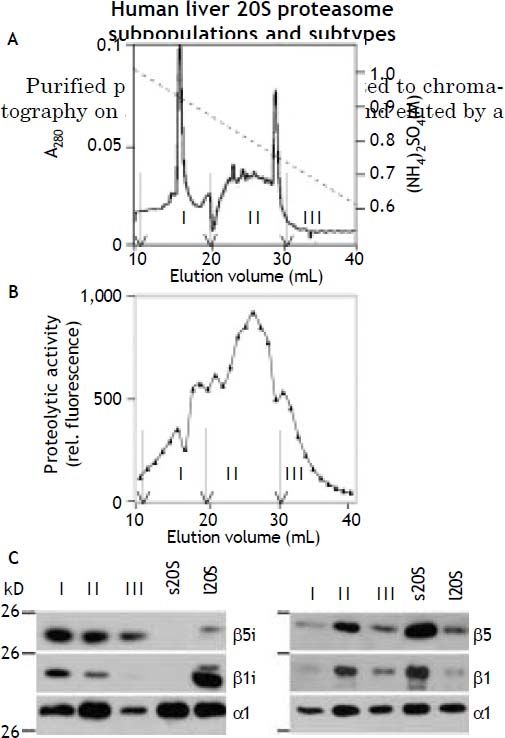
linear decreasing ammonium sulfate gradient. Thus they were separated into three protein peaks containing proteolytic activity, which were designated proteasome subpopulation I, II, and III (Figure 3A and 3B). Fractions comprising each of the three peaks were pooled and analyzed for their content of standard- and immuno-subunits. As shown by immunoblotting all three subpopulations contained standard-subunits β1, β5 as well as the immunosubunit β5i; the subunit β1i was clearly present only in subpopulation I and II and only a very faint band to be seen in subpopulation III (Figure 3C). As outlined above the presence of subunit β1i entails a low caspase-like activity a feature mirrored here in the fact that the ratio of the chymotrypsin- to caspase-like activity was 3.9, 1.9, and 1.4 in subpopulation I, II and III, respectively (data not shown).
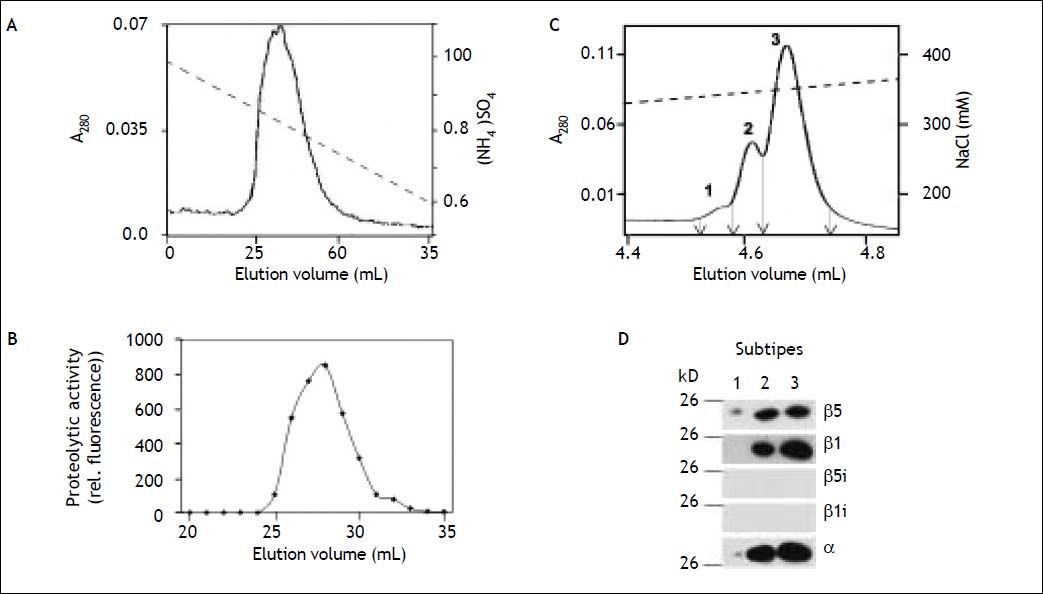
Separation of human liver 20S proteasomes into subpopulations. A. Purified 20S proteasome was subjected to hydrophobic interaction chromatography on Phenyl-Superose and eluted from the column by a decreasing concentration-gradient of (NH4)2SO4 (dashed line). B. The eluant was collected in fractions of 1 mL and their proteasome activity measured with Suc-LLVY-MCA as substrate. C. As indicated by arrows fractions were pooled to form three subpopulations: subpopulation I (11-19 mL), subpopulation II (20-30 mL), and subpopulation III (31-40 mL). C. Aliquots of the three subpopulations were subjected to SDS-PAGE and then tested by immuno-blotting using antibodies to proteasome subunits α1 (loading control), β1, β1i, β5, and β5i, respectively. The specificity of the antibodies was checked by use of 20S proteasomes from human erythrocytes (standard proteasome, s20S) and from human spleen (immunoproteasome, i20S). Note the β1i antibody shows slight crossreactivity with β1, which is present in small amounts also in 20S proteasome from spleen.34
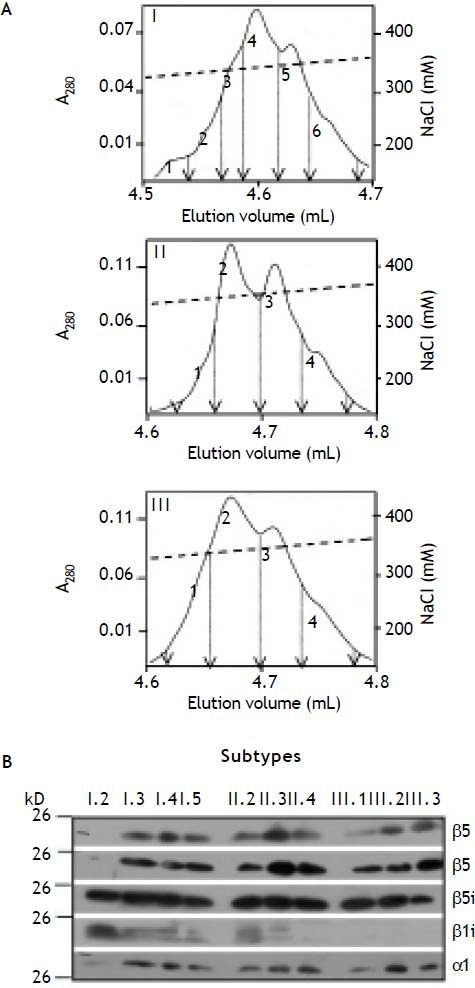
Proteasomes have been found to show great diversity depending on cell- and tissue-type of their origin. This is not only due to their different content in standard- and immuno-subunits, but also due to the fact that they are subjects of several post-translational modifications (for review see36). Hence, 20S proteasome subpopulations can be further subdivided into proteasome subtypes by highresolution anion exchange chromatography. As shown in Figure 4A each proteasome subpopulation was subdivided into 4-6 proteasome subtypes. All subtypes were analysed for their content of standard- and immuno-subunits except of subtypes I.1, I.6, II.1, and III.4, the amounts of which were too small for further analysis. The minor subtype I.2 contains β1i and β5i and no standard subunits. The presence of β2 and β2i was not investigated here, because antibodies showed mutual crossreac- tivity to these subunits. All other subtypes contain standard- as well as immuno-subunits and thereby are intermediate-type proteasomes. However, subtypes II.4, III.1, III.2 and III.3 differ from the other intermediate-subtypes since subunit β1i was not detectable in these proteasomes by immunoblotting (Figure 4B).
Separation of 20S proteasome subpopulations from human liver into subtypes. A. The subpopulations of human 20S proteasomes were subjected to high resolution anion exchange chromatography on Mini Q. Subpopulation III was subjected onto the column as a whole, while material of I and II were chromatographed in several runs, each. Proteasomes bound were eluted by a linear increasing gradient of NaCl (dashed line) resolving each of the three subpopulations (I, II, III) into subtypes (designated 1-4 or 6). Only, the section containing the proteasome subtypes is shown. Fractions separating the subtypes and their pools are indicated by arrows. B. Peak fractions of each proteasome subtype (except of I.1, and III.4) were subjected to SDS-PAGE and then tested by immunoblotting for their content of proteasome subunit α1 (loading control), β1, β5, β1i, and β5i, respectively.
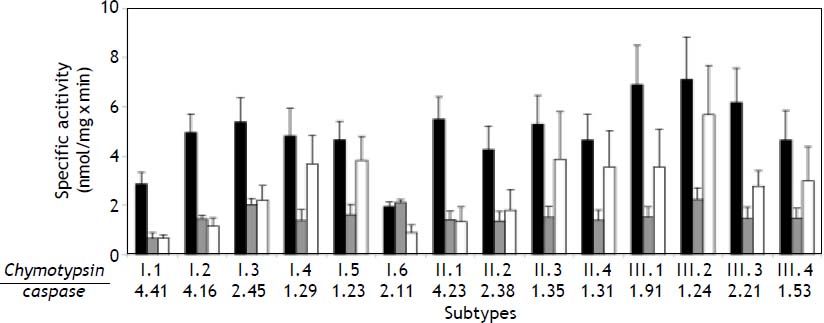
When measuring the proteolytic activities of the 14 proteasome subtypes with fluorogenic peptide substrates, we observed that the caspase-like activity increased when comparing the four to six subtypes in both subpopulations I and II, respectively. With exception of I.6 this resulted in a decreased ratio of chymotrypsin- to caspase-like activity in the order of their elution from the column. This feature was not found in the subtypes of subpopulation II, indicating that these activities are not exclusively determined by the ratio of subunit β1/ β1i but posttranslational modifications like subunit phosphorylation and acetylation have shown to affect the specific activities, too.37,38 There was no simple pattern of differences in the trypsin-like activity (Figure 5).
Proteolytic activities of human liver 20S proteasome subtypes. 20S proteasome subtypes were tested with the fluorogenic peptide substrates Suc-LLVY-MCA, Bz-VGR-MCA, and Z-LLE-MCA for their chymotrypsin- (black column), trypsin- (grey column) and caspase-like (white column) activity, respectively. Values are means ± SD of three separate preparations. The series of figures below the diagram represent the ratio of chymotrypsin-/caspase-like activity measured in each proteasome subtype.
A lthough short fluorogenic peptide substrates are generally used to measure proteasomal activity, they have only limited similarity to the physiological polypeptide and protein substrates. Therefore, we also measured the degradative activity of liver proteasome subtypes towards a polypeptide derived from the melanocyte protein gp100 that was found to be a useful model substrate for 20S proteasomes.39 The 30mer polypeptide gp100201–230 was incubated with the same amounts of microgram of one of the major subtypes of the three subpopulations and the amount of non-degraded substrate was measured at the times indicated in Figure 1A. The time required for degradation of 50% of the substrate was about 200 min, 140 min, and 90 min for subtype I.3, II.3, and III.3, respectively. This result suggests that a high content of immuno-subunit β1i in a proteasome subtype slows down the degradation rate for this polypeptide substrate. Similar differences are also reflected by the generation kinetics of two major degradation fragments, gp100201–213 and gp100214–230, which are clearly faster for subtypes II.3 and Ш.3 as compared to I.3 (Figure 1B). However, this phenomenon may be determined by the ratio of β5/ β5i in the three subtypes, since this cut is probably catalyzed by β5, which preferentially binds valine or alanine, whereas β5i prefers cleavage after large nonpolar residues like Tyr.40 On the other hand, the various proteasome subtypes degraded another model polypeptide substrate, a 25mer polypeptide derived from the murine cytomegalovirus immediate early protein pp8916–40,28 at about the same rate (data not shown).
20S proteasomes from the hepatocyte carcinoma cell line HuH7Hepatocytes constitute about 60% of the total hepatic cell population. To study the involvement of the ubiquitin/proteasome-system in hepatocytes during various patho-physiological processes HuH7 hepatocarcinoma cells were used as a model cell line.41–45 However, nothing is known about the feature of proteasomes in this cell line. Therefore, we have purified 20S proteasomes from HuH7 cells to apparent homogeneity and subjected them to hydrophobic interaction chromatography. In contrast to 20S proteasomes from human liver and as shown in Figure 6A/B proteasomes from these cells were eluted from the column as a single subpopulation. Measurement of their hydrolytic activity to- wards fluorogenic peptide substrates revealed high chymotrypsin- and caspase-like activity of similar size and a low trypsin-like activity (Table 1). Subsequent high-resolution anion exchange chromatography of purified HuH7 proteasomes resolved them into three subtypes (Figure 6C), all of which lack immuno-subunits and contain just standard proteasome subunits β1 and β5 (Figure 6D). The pattern of activity and of subtypes is very similar to that of proteasomes from erythrocytes and differs essentially from proteasomes of adult human liver tissue (Table 1).46
20S proteasomes from HuH7 cells. A and B. 20S proteasomes were purified from HuH7 cells and subjected to hydrophobic interaction chromatography. Proteasomes bound were eluted by a linear decreasing gradient of (NH4)2SO4 (dashed line) as a single protein peak and activity peak. C. After hydrophobic interaction chromatography 20S proteasomes were subjected to anion exchange chromatography and eluted from the column by an increasing gradient of NaCl (dashed line). Proteasomes were separated into three different subtypes, indicated 1-3. Fractions separating the subtypes and their pools are indicated by arrows. D. The three 20S proteasome subtypes from HuH7 cells were tested by SDS-PAGE and immunoblotting for their content of standard and immuno-subunits. The three subtypes contained the standard subunits β1 and β5, but no immuno-subunits.
Our observation that HuH7 hepatocarcinoma cells contain solely standard-proteasomes corroborates the results of histochemical studies with human fetal liver, where no histochemical reaction of antibodies specific for the immuno-subunits β1i and β5i was observed. Similarly, when mice are kept in a specific pathogen-free environment, their proteasomes in liver tissue contain exclusively standardproteasomes. Only after the animals were infected by viral and bacterial pathogens, standard-proteasomes are replaced by immuno-proteasomes.47 These data indicate that standard-proteasomes are sufficient for the basic functions in hepatocytes.48 Accordingly, liver tissue of normal adult human donors has experienced many immunological challenges effecting the expression and incorporation of proteasome immuno-subunits and thus could histochemically be localized in addition to standard subunits in hepatocytes as well as lymphocytes, monocytes and Kupffer cells.48
In our present investigation we show that both immuno-subunits, β1i and β5i, are found almost exclusively in intermediate-type proteasomes, i.e. proteasomes containing both standard- and immuno-subunits. They exist in three major subpopulations separable due to their different surface hydrophobicity: subpopulations I and II containing subunits β1, β1i, β5 and β5i and subpopulation III containing subunits β1, β5 and β5i (Figure 3). Moreover, each of these three subpopulations is composed of several proteasome subtypes, which can be separated by high-resolution anion exchange chromatography. Only after this separation step and subsequent immunoblot analysis proteasomes containing no standard subunits but β1i and β5i could be identified in the minor subtype I.2; subtype I.1 probably has a similar subunit composition (Figure 4B). By means of a fractionated im- munoprecipitation procedure with subunit-specific antibodies Guillaume and coworkers classified 15 % of human liver proteasomes as immunoproteasomes and 31% as standard proteasomes, the majority were also intermediate-type proteasomes.49 No standard proteasomes were detectable in liver tissues of the present investigation, which is consistent with our findings on proteasomes from rat liver tissue.29
Proteasomes from HuH7 cells do not contain any immuno-subunit but are pure standard proteasomes (Figure 6). The fact that these proteasomes could yet be separated into several subtypes by anion exchange chromatography indicates that the charge differences of proteasome subtypes may be due to differences in posttranslational modifications like phosphorylation, which are known to be present in proteasomes also from liver50 and were shown to have an impact on the separation of proteasome subtypes.51 Posttranslational modifications might also determine or result from the intracellular localization of proteasomes, which have been detected in the cytoplasm, nucleoplasm and attached to the endoplasmic reticulum of hepatocytes.48,52,53
When comparing the chymotrypsin- and caspaselike activity of proteasome subtypes within subpopulation I and subpopulation II, a clear decrease in their ratio is observed due to an increase in caspaselike activity (Figure 5). This might well be due to an apparently declining content of subunit β1i in the sequence of subtypes from 1 to 4 (or 6) (Figure 4), since the presence of β1i attenuates the caspase-like activity.10 Activity alterations due to posttranslational modifications have also been described and should be taken into consideration to explain this observation.37,38,54 In any case the different proteas-ome subtypes seem to have different specific activities towards fluorogenic peptide substrates, which most likely result in different degradative rates of cellular protein substrates. Therefore, we have investigated the degradation of the 30mer polypeptide gp100201–230 derived from the melanoma-specific protein gp100 by one of the major subtypes of each of the three subpopulations. The degradation rates increased as follows: I.3 < II.3 < III.3. Considering the decreasing presence of subunit β1i in I.3 > II.3 > III.3 it seems that β1i slows down the degradation of this substrate, indicating that the multiple proteasome subtypes may have different affinities to protein substrates and may thus generate degradation fragments in different amounts. Binding and unfolding of substrate proteins as well as their translocation into the 20S core enzyme are control- led by proteasome regulators like the 19S Reg, PA28, etc., but the molecular differences of 20S proteasome subtypes with respect to their substrate binding channel inside the proteolytic chamber determine their substrate affinity and degradation efficiency as well.40 Similarly, whether a substrate protein enters the proteasome with its N- or C-terminus has shown to determine the spectrum of degradation fragments.55 Supportive of this is the finding that some immuno-epitopes are generated predominantly by intermediate-type proteasomes.49 Likewise, we have shown previously that different intermediate-type proteasomes from rat liver degrade various polypeptide substrates at different rates.29 Thus, the adaptation of the proteasome system by diverging into a large spectrum of different subtypes is comprehensible when considering the many harmful and toxic challenges the liver is subjected to during its lifetime and has to defend the organism effectively.56,57 Consequently, strong expression of immuno-subunits and changes in proteasome regulators have been found in liver of patients suffering from hepatitis, cirrhosis, steatohepatitis and hepatocarcinoma.48,58
Abbreviations- •
2D: two-dimensional.
- •
Bz: benzoyl.
- •
MCA: methylcoumarylamide.
- •
PAGE: polyacrylamidegel electrophoresis.
- •
Sue: succinyl. •Z:carbobenzoxy.
S.G. was supported by a PhD studentship from the Sonnenfeld-Stiftung, Berlin. B.D. was supported by the Berliner Krebsgesellschaft (SEFF200907).