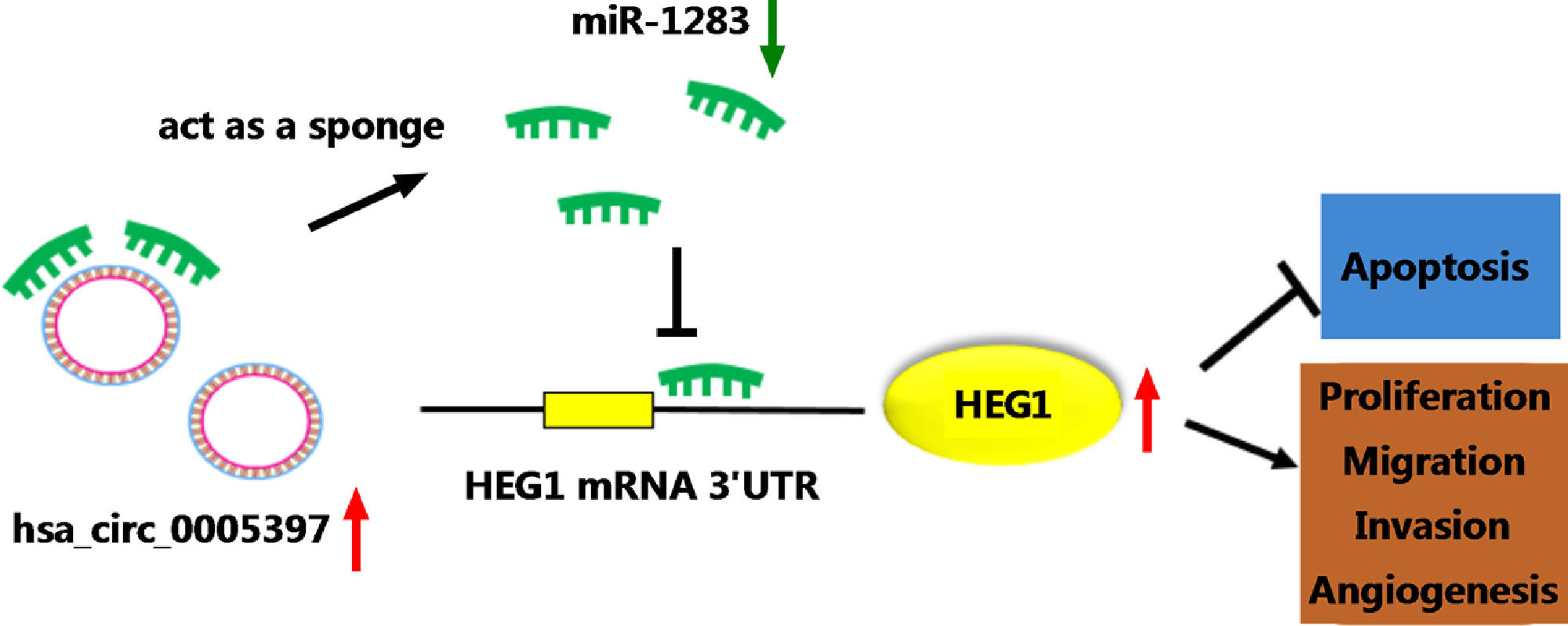
Circular RNA (circRNA) has been confirmed to be an important regulator for the progression of hepatocellular carcinoma (HCC). However, the role and regulatory mechanism of circ_0005397 in HCC are not completely clear.
Patients and methodsFifty HCC patients were included in this study. Reverse transcription-qPCR analysis was used to appraise circ_0005397, microRNA (miR)-1283, HEG homolog 1 (HEG1) mRNA expression levels, while western blot was used to identify HEG1, PCNA, Bax and Bcl-2 protein expression levels. Furthermore, cell proliferation, apoptosis, migration, invasion and angiogenesis were judged through cell counting kit-8 assay, EdU assay, Caspase3 activity test, flow cytometry, transwell assay and tube formation experiment. Dual-luciferase reporter assay and RIP assay were used to verify the targeting relationship between miR-1283 and circ_0005397 or HEG1. Finally, the effect of circ_0005397 on HCC tumor development was detected by mice experiments in vivo.
ResultsCirc_0005397 was highly expressed in HCC tissues and cells, in HCC cell lines. Circ_0005397 silencing inhibited proliferation, migration, invasion and angiogenesis, while induced apoptosis in HCC cells. Circ_0005397 could sponge miR-1283, and miR-1283 could target HEG1. MiR-1283 inhibitor incompletely counteracted the effect of si-circ_0005397 on HCC cell progression, while HEG1 overexpression partially overturned the effect of miR-1283 on HCC cell progression. Circ_0005397 regulated the expression of HEG1 through targeting miR-1283. Moreover, circ_0005397 silencing blocked the growth of HCC tumors in vivo.
ConclusionsCirc_0005397 regulated HEG1 by targeting miR-1283, thereby promoting HCC development.
Hepatocellular carcinoma (HCC) is one of the most common and dangerous malignant tumors in the world [1]. At present, many novel therapies for HCC have been identified, including metronomic capecitabine [2], lenvatinib plus pembrolizumab [3] and immune checkpoint inhibitors [4]. Unfortunately, the recurrence rate and mortality rate of HCC still show an upward trend as a result of its difficulty in eradication [5]. Therefore, it is urgent to identify the molecular mechanisms that influence the progression of HCC to further explore potential therapeutic targets for HCC.
Circular RNAs (circRNAs) are a special class of noncoding RNA molecules which widely exist in human eukaryotic transcriptome [6]. The circular structure makes its expression more stable than linear RNA [7–9]. There is increasing evidence that circRNAs are inseparable from the occurrence of HCC. For example, circ_0003288 could promote the EMT process and invasion in HCC [10], and circ_0006916 had an enhancing effect on HCC growth and invasion [11]. In addition, there is evidence that circ_0005397 is a novel and effective biomarker for treating HCC, which can relieve the diagnostic stress of HCC [12].
In most cases, circRNAs serve as microRNA (miRNA) sponge, thereby regulating the occurrence of malignant tumors [13,14]. MiR-1283 is a recently discovered miRNA that can directly target ATF4 to block glioma cell proliferation and invasion [15]. The study also has shown that miR-1283/ATF4 axis may participate in hypertension progression [16]. However, the function of miR-1283 in HCC progression has not been demonstrated. HEG homolog 1 (HEG1) is a cardiac developmental protein with EGF-like domain 1, which is closely associated with tumor progression [17,18]. Clinical evidence suggests that the expression level of HEG1 can be regarded as an evaluation index of HCC [19,20].
Here, our objective is to elucidate the function and potential molecular mechanism of circ_0005397 in HCC. We discovered that there were specific binding sites between miR-1283 and circ_0005397 or HEG1. Therefore, we proposed and verified the hypothesis of circ_0005397/miR-1283/HEG1 axis in regulating HCC progression.
2Materials and methods2.1SamplesHCC tumor tissues and adjacent normal tissues were collected from 50 HCC patients (stage I+II=22 and stage III=28) at Yantai Qishan Hospital and stored at -80°C until use.
2.2Cell culture and transfectionThe HCC cells (Huh-7 and SNU-387) and liver epithelial cells (THLE-2) were acquired from Biovector NTCC (Beijing, China). HCC cell lines were placed in RPMI-1640 medium (Gibco, Waltham, MA, USA), and THLE-2 cells were placed in BEGM Bullet Kit (Lonza, Walkersville, MD, USA) at 37°C with 5% CO2. All medium containing 10% FBS (Gibco) and 1% streptomycin (Invitrogen, Carlsbad, CA, USA).
Circ_0005397 small interfering RNA (si-circ_0005397) or overexpression vector, miR-1283 mimic or inhibitor (anti-miR-1283), pcDNA HEG1 overexpression vector, and their corresponding negative controls were gained by RiboBio (Guangzhou, China). Cell transfection was performed with Lipofectamine 2000 (Invitrogen).
2.3Reverse transcription-qPCR (RT-qPCR)RNA extraction was performed using TRIzol reagent (Invitrogen). Next, cDNA reverse transcription was carried out using cDNA Synthesis Kit (Vazyme, Nanjing, China). Finally, the expression of circRNA, miRNA and mRNA was detected by SYBR Green (Ambion, Carlsbad, CA, USA) on PCR System. Data were calculated by 2−ΔΔCt method with GAPDH or U6 as internal control. Primer sequences are shown in Table 1.
The primer sequences for RT-qPCR
| Name | Primers (5’-3’) | |
|---|---|---|
| circ_0005397 | Forward | TGCCGTTAACAACAAGCATTC |
| Reverse | TGGAACTCTCTCTGGGGTGA | |
| HEG1 | Forward | TGGAGAACGTTCGACCTTGG |
| Reverse | TTCCCCGTTTCCAGTGACAG | |
| miR-1283 | Forward | GCCGAGTCTACAAAGGAAAGCG |
| Reverse | CTCAACTGGTGTCGTGGAG | |
| GAPDH | Forward | GACAGTCAGCCGCATCTTCT |
| Reverse | GCGCCCAATACGACCAAATC | |
| U6 | Forward | CTCGCTTCGGCAGCACA |
| Reverse | AACGCTTCACGAATTTGCGT | |
| RHOT1 | Forward | CTCCTGGTGAGAGGAGTCCA |
| Reverse | TCCCAACTCTAGGTTCTCCCA |
HCC cells were inoculated into the 96-well plates. At indicated time point, cells were treated with CCK-8 solution (Beyotime, Shanghai, China). After 4 hr, the absorbance at 450 nm was measured by microplate reader.
2.5EdU assayCell proliferation ability was measured by EdU Cell proliferation Kit (RiboBio). In brief, HCC cells were inoculated into 96-well plates. Then, cell was treated with EdU solution and DAPI solution. EdU positive cells were analyzed by fluorescence microscope.
2.6Caspase 3 activity detectionCaspase-3 activity was obtained by a Caspase 3 Activity Assay Kit (Beyotime). HCC cells were collected, fixed, and added fluorescein to label anti-active Caspase-3 antibody. After cells were stained for 30 min, the absorbance was finally measured at 405 nm.
2.7Flow cytometryAnnexin V/FITC Apoptosis Detection Kit (Beyotime) was used. HCC cells were stained with Annexin V/FITC and propidium iodide in binding buffer. After 20 min, the cells were analyzed by flow cytometer.
2.8Western blotA protein extraction kit (Beyotime) was used to extract the total protein. The protein was subjected to SDS-PAGE gel, and then the protein band was transferred to the PVDF membrane (Millipore, Billerica, MA, USA). After adding 5% non-fat milk to the membrane, it was sealed for 2 hrs. It was incubated with the primary antibodies (Abcam, Cambridge, MA, USA) at 4°C overnight and the secondary antibody (Abcam) for 1 hr. Finally, imaging analysis was displayed with Clarity™ Western ECL Substrate Kit (Bio-Rad, Shanghai, China). Antibodies used were shown as below: anti-PCNA (1:1,000, ab18197), anti-Bax (1:1,000, ab32503), anti-Bcl-2 (1:1,000, ab32124), anti-HEG1 (1:1,000, ab121343), anti-GAPDH (1:2,500, ab9485), and Goat-anti-rabbit igG (1:50,000, ab205718).
2.9Transwell assayTranswell assay was used to observe the migration and invasion of tumor cells. Cells were seeded in the upper chamber (Corning, Tewksbury, MA, USA) pre-coated with or without Matrigel (for detecting cell invasion and migration), and 600 μL of medium containing 10% FBS were added to the lower chamber. Next, 24 hrs later, cells on the lower surface were fixed with methanol and stained by crystal violet. The numbers of migrated and invaded cells were counted with Image J software.
2.10Tube formation experimentThe medium of HCC cells was collected and mixed with fresh medium to prepare condition medium. HUVECs were suspended with condition medium. The suspension of HUVECs was added to a 96-well plate coated with Matrigel (Corning). After 12 h of cultivation, photographs were carried out to count the number of branches.
2.11Bioinformatics analysisCircinteractome and starbase software were respectively used to predict the miRNAs, which could combine with HEG1 or circ_0005397.
2.12Dual-luciferase reporter assayWild-type or mutant-type sequences of circ_0005397 or HEG1 3’UTR were inserted into pmirGLO vectors (Promega, Madison, WI, USA) to generate the circ_0005397WT/MUT vectors or HEG1 3’UTRWT/MUT vectors. The vectors were transfected into HCC cells with miR-1283 mimic or miR-NC. Luciferase activity was detected using Dual-luciferase Reporter Assay System (Promega).
2.13RIP assayRIP assay was determined by RIP Kit (Millipore). The cell lysates were cultivated with magnetic beads conjugated with anti-Ago2 or anti-IgG. Finally, the immunoprecipitated RNA was extracted and RNA enrichments were detected by RT-qPCR.
2.14Transplantation model in vivoThe nude mice (male BALB/c, 5 weeks old) were obtained from Vital River (Beijing, China). Huh-7 cells (4 × 106) stably transfected with sh-circ_0005397 or sh-NC were injected into the mice. The tumor volume was recorded every 7 days. The mice were euthanized on 28 days, and the tumor weight was weighed.
2.15Immunohistochemistry assay (IHC)Paraffin sections of mice tumor tissues were prepared. After dewaxing and hydration, the sections were incubated with 3% hydrogen peroxide and dropped with 5% goat serum blocking solution. The sections were hatched with anti-Ki67 (1:200, ab16667, Abcam) and HRP-polymer. Finally, DAB was used to immune for observation.
2.16Statistical analysisThe statistical data was presented by GraphPad Prism 7 in the form of mean ± standard deviation. Student's t-test or ANOVA was used to analyze the differences between groups. P < 0.05 was considered significant difference.
2.17Ethical statementWritten informed consent was obtained from each patient included in the study and the study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the Ethics Committee of Yantai Qishan Hospital.
The animal experiments were approved by the Animal Ethics Committee of Yantai Qishan Hospital.
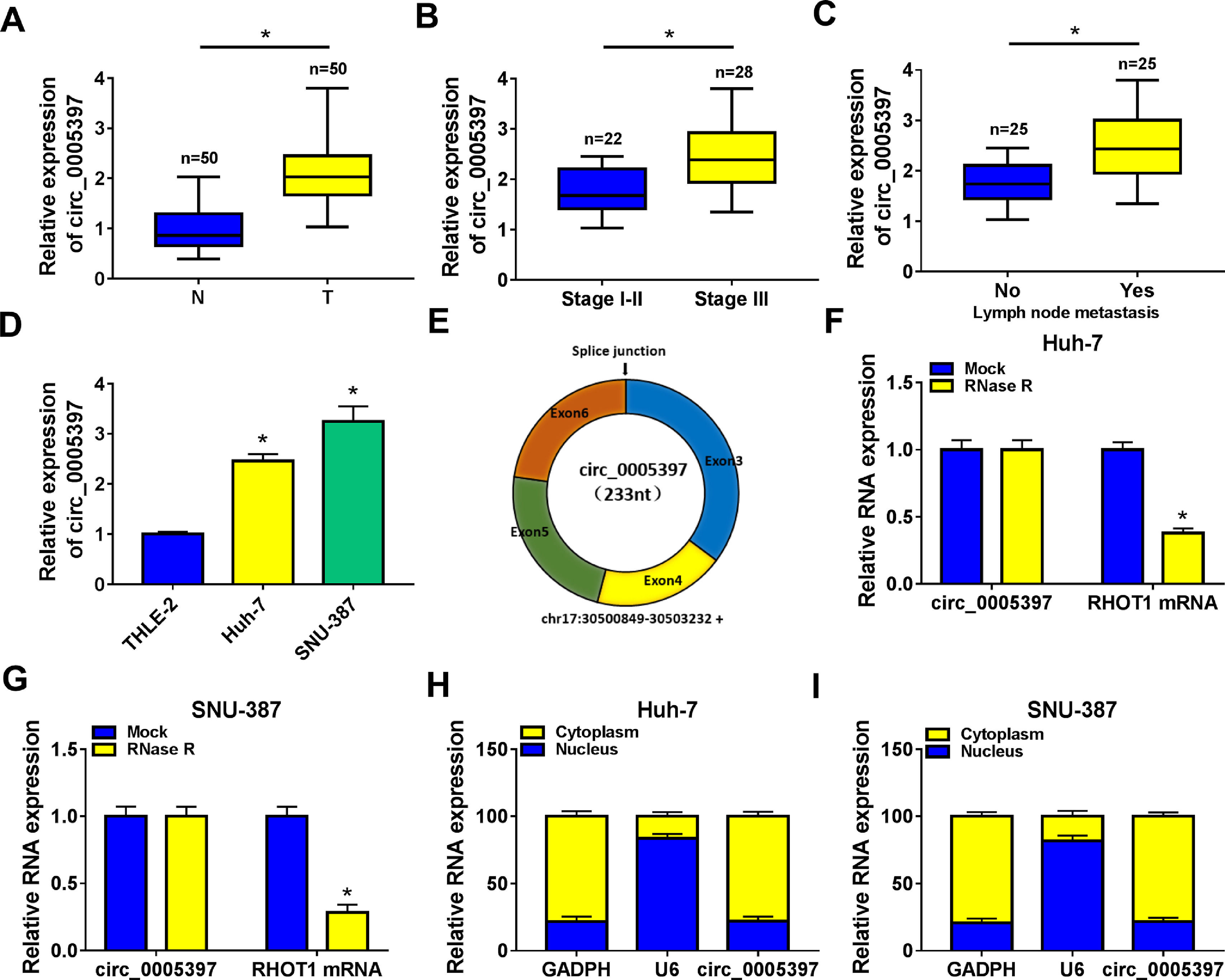
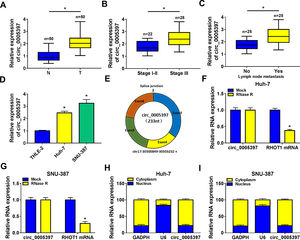
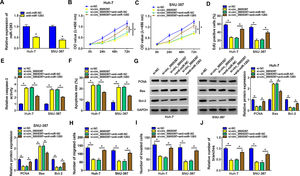
3Results3.1Circ_0005397 was upregulated in HCC tissues and cellsFirst, RT-qPCR showed that the expression level of circ_0005397 in HCC tissues (n=50) was significantly increased compared with normal tissues (n=50) (Fig. 1A). Circ_0005397 expression in HCC patients with stage III was greatly higher than that in stage I-II (Fig. 1B), and hsa_circ_0005397 expression level with lymph node metastasis in HCC patients was enhanced compared to those patients without lymph node metastasis (Fig. 1C). Moreover, our studies indicated that circ_0005397 was upregulated in two HCC cell lines (Huh-7 and SNU-387) with respect to the liver epithelial cell line (THLE-2) (Fig. 1D). The structure of circ_0005397 was shown in Fig. 1E, circRHOT1 (circ_0005397) with a sequence length of 233 nt was generated from chr17:30500849-30503232. To observe the stability of circ_0005397 in Huh-7 and SNU-387 cells, we conducted RNase R degradation assay, and the results showed that circ_0005397 could resist RNase R digestion while RHOT1 could be digested by RNase R (Fig. 1F and G). We identified the location of circ_0005397 in two HCC cells, cytoplasmic isolation and RT-qPCR showed that circ_0005397 mainly existed in the cytoplasm of HCC cells (Fig. 1H and I).
Upregulation of circ_0005397 in HCC tissues and cells. A. Relative expression of circ_0005397 in HCC tissues (n=50) compared with normal tissues (n=50) by RT-qPCR. B. Relative expression of circ_0005397 in various periods of the clinic by RT-qPCR. C. Relative expression of circ_0005397 with lymph node metastasis in HCC tissues compared to tissues without lymph node metastasis by RT-qPCR. D. Relative expression of circ_0005397 in HCC cells (Huh-7 and SNU-387) compared with normal cells (THLE-2) by RT-qPCR. E. Biological characteristics of circRHOT1 (circ_0005397). F, G. The degradation ability of circ_0005397 and RHOT1 in HCC cells on RNase R. H, I. The localization of circ_0005397 in HCC cells by cytoplasmic separation. *P < 0.05.
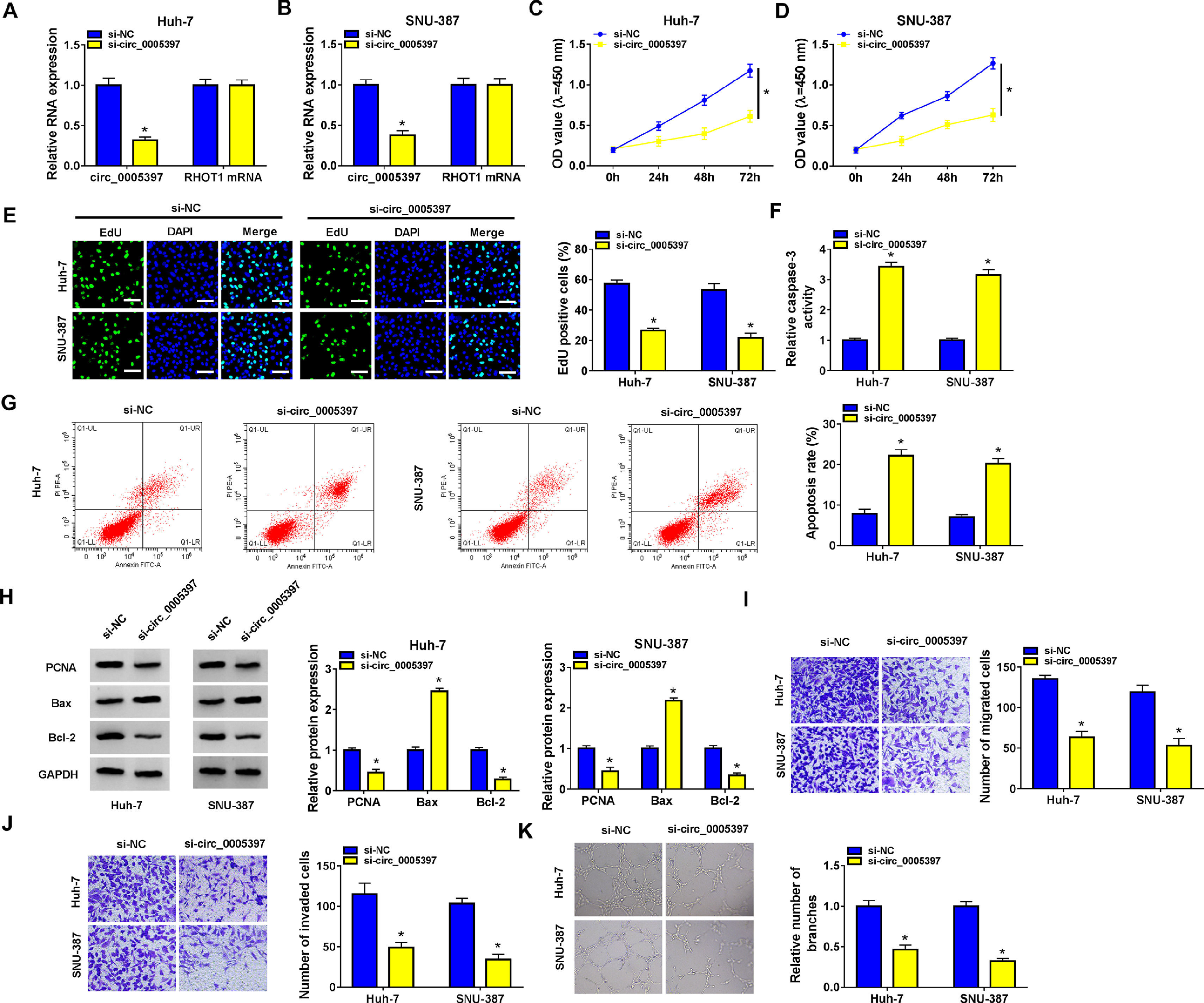
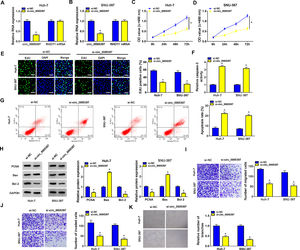
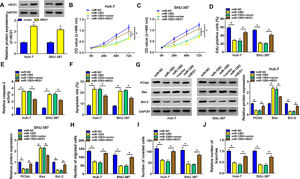
We identified the expression of circ_0005397 in Huh-7 and SNU-387 cells transfected with si-circ_0005397 via RT-qPCR, and the results showed that the expression of circ_0005397 was sharply decreased by si-circ_0005397, while the expression of RHOT1 was not changed (Fig. 2A and B). The results of CCK-8 and EdU assays showed that the activity and proliferation of HCC cells were blocked by circ_0005397 silencing (Fig. 2C-E). Compared with the control group, the Caspase 3 activities of Huh-7 and SNU-387 cells were increased and the apoptosis rates were conspicuously enhanced after transfection with si-circ_0005397 (Fig. 2F and G). Then, the protein levels of proliferative protein (PCNA), pro-apoptotic protein (Bax) and anti-apoptotic protein (Bcl-2) were examined by western blot; the data revealed that si-circ_0005397 could significantly hinder the expression of PCNA and Bcl-2, and impel the expression of Bax (Fig. 2H). Finally, si-circ_0005397 obviously impeded cell migration, invasion, and angiogenesis in vitro (Fig. 2I-K). In short, silencing circ_0005397 could confine cell viability, proliferation, migration, invasion and angiogenesis, and elevate cell apoptosis rate in Huh-7 and SNU-387 cells.
The effect of circ_0005397 silencing on cell viability, migration, invasion and angiogenesis in HCC cells. A, B. The RNA expression levels of circ_0005397 or RHTO1 mRNA after si-NC and si-circ_0005397 were transfected in Huh-7 and SNU-387 cells. C, D. The detection of cell viability of Huh-7 and SNU-387 cells was transfected with si-NC and si-circ_0005397 by CCK-8 assay. E. The cell proliferation of Huh-7 and SNU-387 cells transfected with si-NC and si-circ_0005397 by EdU assay. F, G. The determination of Caspase 3 activity and cell apoptosis rate was determined when Huh-7 and SNU-387 cells were transfected with si-NC and si-circ_0005397. H. PCNA, Bax and Bcl-2 protein levels were texted by western blot when Huh-7 and SNU-387 cells were transfected with si-NC and si-circ_0005397. I-K. The migration, invasion and angiogenesis of cells were observed after transfection of si-NC or si-circ_0005397 in Huh-7 and SNU-387 cells. *P < 0.05.
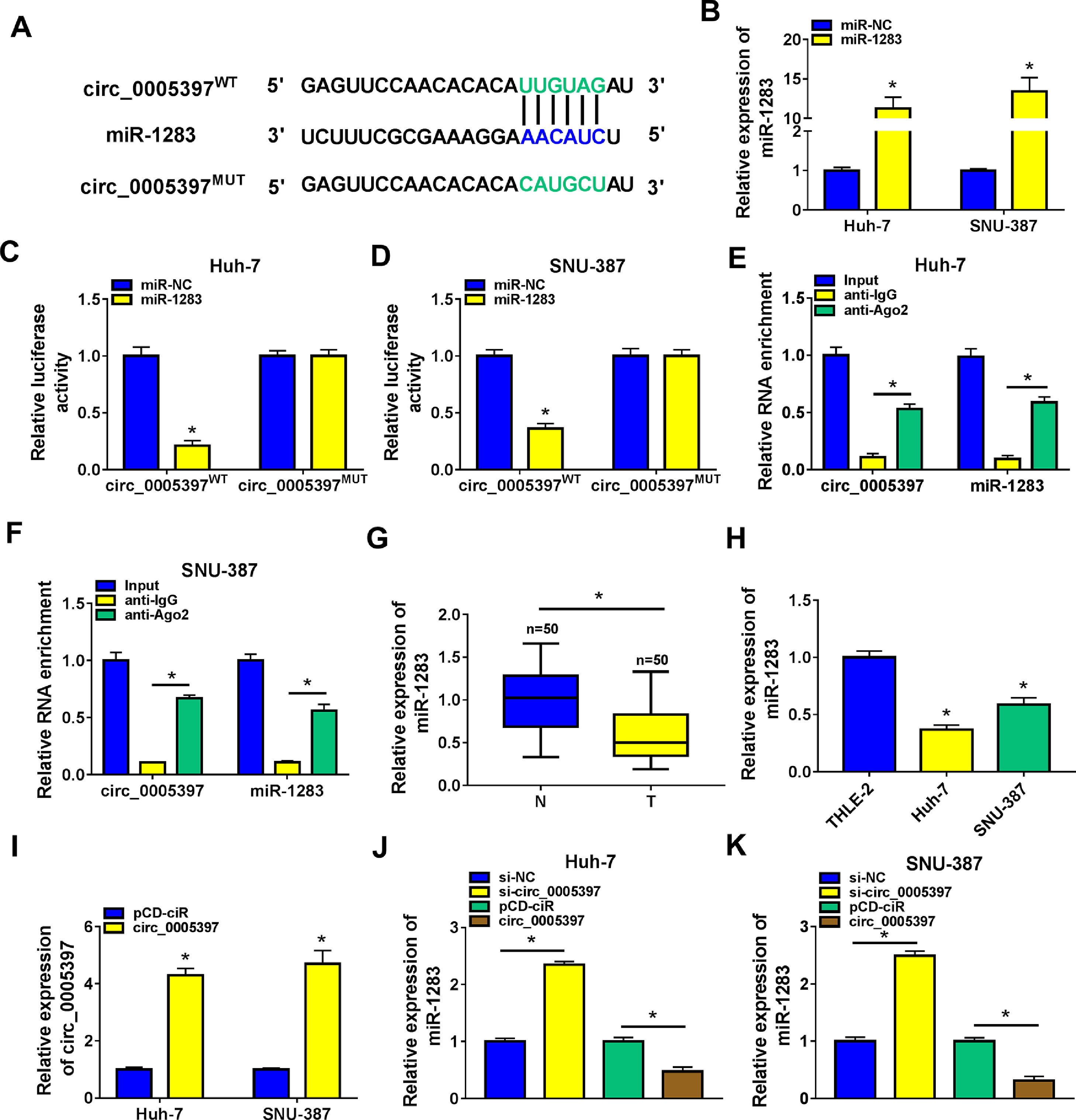
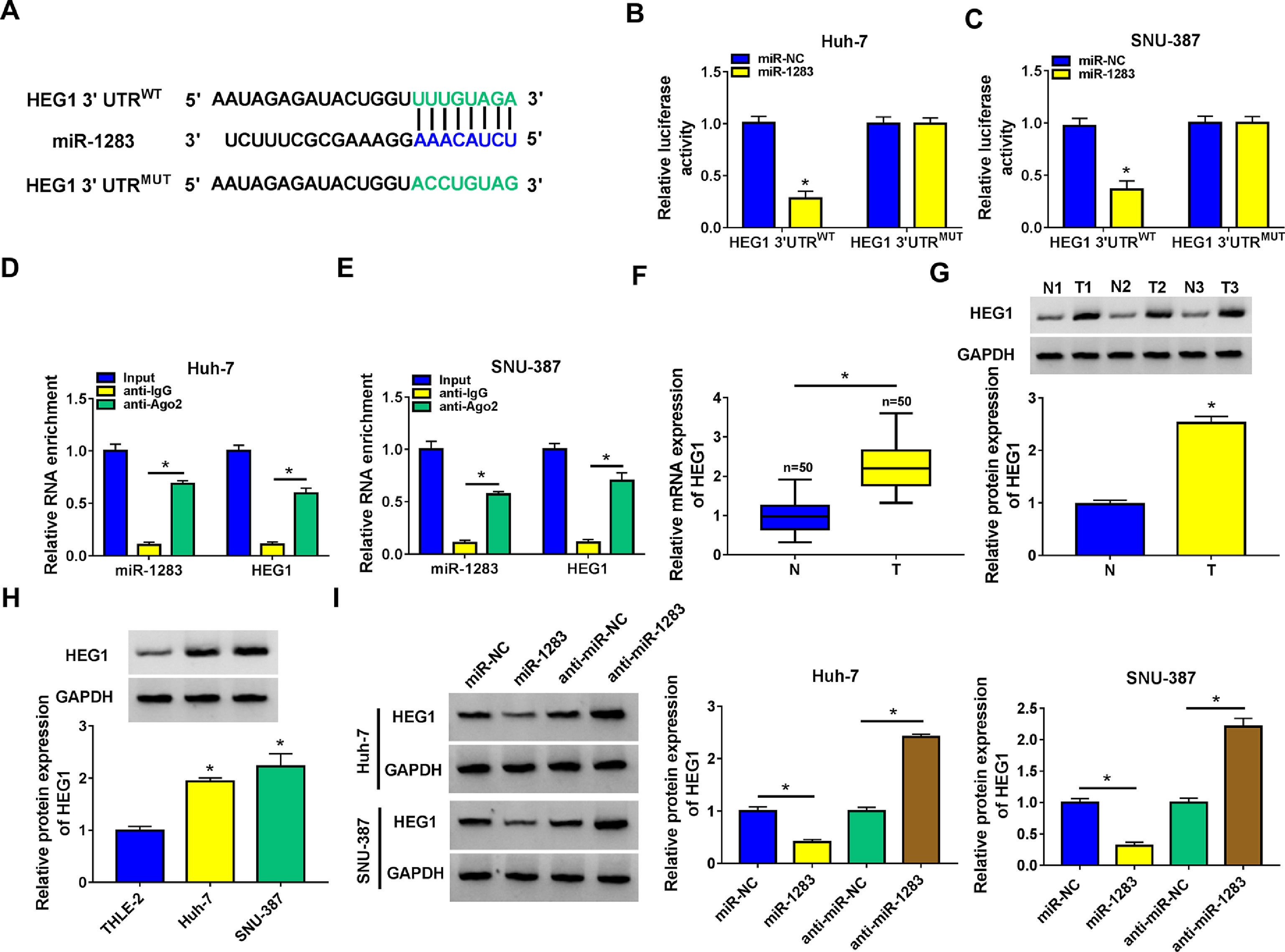
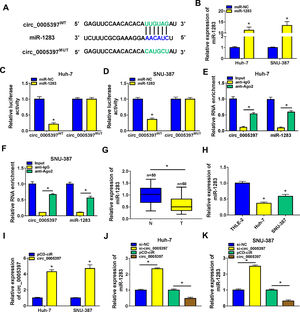
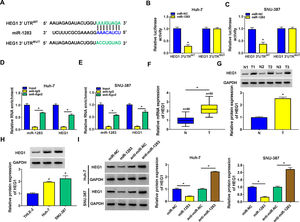
In order to identify the molecular mechanism of circ_0005397 in HCC, we investigated its interaction with miRNA. CircInteractome predicted that circ_0005397 had some binding sites with miR-1283 (Fig. 3A). Overexpression of miR-1283 was achieved in Huh-7 cells and SNU-387 cells transfected with miR-1283 (Fig. 3B). Next, we used dual-luciferase reporter gene assay to confirm the relationship between circ_0005397 and miR-1283. The results showed that after co-transfecting HCC cells with circ_0005397WT and miR-1283, the luciferase activities were significantly lower than the circ_0005397WT and miR-NC group, while circ_0005397MUT and miR-1283 group found no differences (Fig. 3C and D). And RIP assay further verified the direct interaction between circ_0005397 and miR-1283 (FIG. 3E and F). RT-qPCR dates showed low expression of miR-1283 in HCC tumor tissues and cells (Fig. 3G and H). Circ_0005397 realized high expression in HCC cells transfected with circ_0005397 overexpression vector (Fig. 3I). Moreover, in Huh-7 cells and SNU-387 cells, the expression of miR-1283 in si-circ_0005397 group was effectively rose compared with the si-NC group, and the expression of miR-1283 in circ_0005397 group was effectively debased compared with the control group (Fig. 3J and K). It was confirmed that circ_0005397 negatively regulated miR-1283. Overall, it meant that miR-1283 was the target of circ_0005397 in HCC cells.
Circ_0005397 interacted with miRNA in HCC cells. A. The binding sites between circ_0005397 and miR-1283 were estimated by circInteractome. B. The expression level of miR-1283 after transfection with miR-1283 in Huh-7 cells and SNU-387 cells. C, D. The interaction between circ_0005397 and miR-1283 was confirmed by dual-luciferase reporter assay. E, F. The RNA enrichment situation of circ_0005397 and miR-1283 was confirmed by RIP assay. G, H. The expression level of miR-1283 in HCC tumor tissues and cells by RT-qPCR. I. The expression level of circ_0005397 after transfection with circ_0005397 in Huh-7 cells and SNU-387 cells. J, K. The expression level of miR-1283 was assessed by RT-qPCR assay in Huh-7 cells and SNU-387 cells transfected with si-NC, si-circ_0005397, vector and circ_0005397. Each experiment was independently repeated three times at least. *P < 0.05.
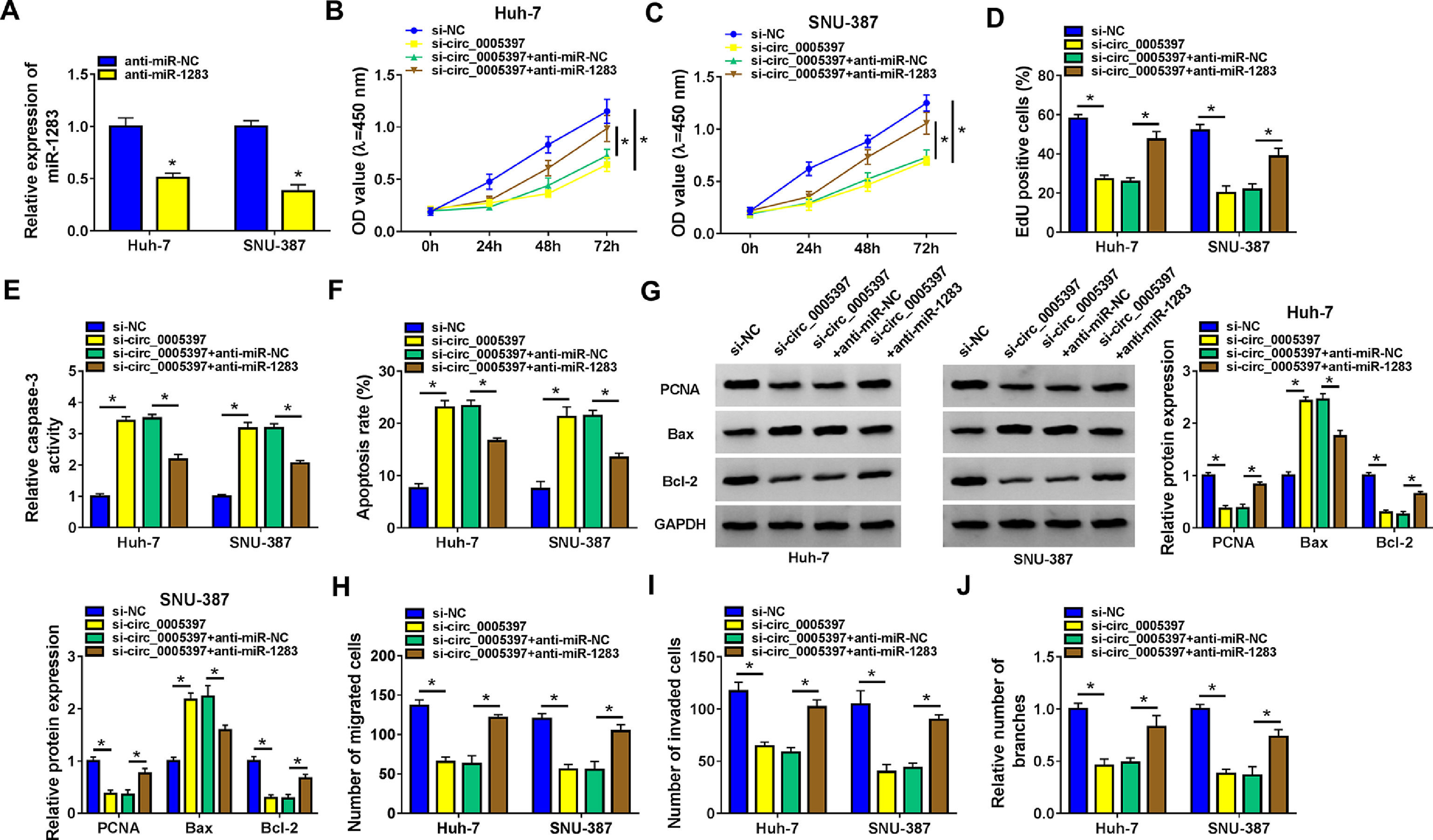
To determine the inhibitory effect, we measured the expression of miR-1283 by RT-qPCR. Low expression of miR-1283 was achieved in Huh-7 cells and SNU-387 cells transfected with anti-miR-1283 (Fig. 4A). Next, we revealed the effect of miR-1283 and circ_0005397 in the progression of HCC through several experiments. In each experiment, si-circ_0005397 and anti-miR-1283 were transfected into Huh-7 and SNU-387 cells, respectively. CCK-8 and EdU assays showed that si-circ_0005397 inhibited the viability and proliferation of Huh-7 and SNU-387 cells, while the introduction of anti-miR-1283 alleviated the effect of si-circ_0005397 on Huh-7 and SNU-387 cells (Fig. 4B-D). Then, we found that si-circ_0005397 accelerated the Caspase 3 activity and cell apoptosis in Huh-7 and SNU-387 cells by the Caspase 3 activity assays and apoptosis assays; however, the outcome could be reverted by si-circ_0005397 and anti-miR-1283 co-treatment in both Huh-7 and SNU-387 cells (Fig. 4E and F). Subsequently, we comprehensively detected the expression of cell proliferation and apoptosis related proteins in multiple groups in order to study the mechanism of circ_0005397 regulating HCC cell apoptosis. Western blot data demonstrated that si-circ_0005397 remarkably suppressed the expression of PCNA and Bcl-2, and impelled the expression of Bax. However, anti-miR-1283 co-transfection completely reversed changes in protein expression induced by si-circ_0005397 (Fig. 4G). At the same time, transwell analysis showed that the inhibitory effects of migration, invasion and angiogenesis caused by circ_0005397 silencing were alleviated by down-regulation of miR-1283 in HCC cells (Fig. 4H-J). These experiments implied circ_0005397 controlled the development of HCC cells by regulating miR-1283. Co-transfection of anti-miR-1283 could reverse the negative effects of single defect of circ_0005397 on the functions of HCC cells.
The recovery function of miR-1283 for circ_0005397 in Huh-7 cells and SNU-387 cells. A. The expressions of miR-1283 transfected with anti-miR-1283 in HCC cells were detected by RT-qPCR. B, C. The detections of cell viability of HCC cells were transfected with si-NC, si-circ_0005397, si-circ_0005397+anti-miR-NC, and si-circ_0005397+anti-miR-1283 by CCK-8 assay. D. The cell proliferation of HCC cells was texted by EdU assay. E, F. The determination of Caspase 3 activity and cell apoptosis rate was to be seen in HCC cells. G. PCNA, Bax and Bcl-2 protein levels in HCC cells were measured by western blot. H-J. The migration, invasion and angiogenesis of transfected Huh-7 and SNU-387 cells. *P < 0.05.
Then, starbase software predicted that HEG1 3’UTR was found to possess some binding sites with miR-1283 (Fig. 5A). Dual-luciferase reporter assay was performed in Huh-7 and SNU-387 cells to check the predicted interactions. The data supported that the upregulation of miR-1283 blocked the luciferase activity of HEG1 3’UTRWT reporter vector in HCC cells, while the luciferase activity of HEG1 3’UTRMUT reporter vector did not change (Fig. 5B and C). And RIP assays further verified the direct interaction between HEG1 and miR-1283 in Huh-7 cells (Fig. 5D) and SNU-387 cells (Fig. 5E). Furthermore, RT-qPCR and western blot suggested that HEG1 expression levels were greatly heightened in HCC tissues (Fig. 5F and G) and cells (Fig. 5H) compared with their own control. Whereafter, the protein level of HEG1 was measured in Huh-7 and SNU-387 transfected cells. As shown in Fig. 5I, the protein expression of HEG1 was clearly decreased when miR-1283 was overexpressed, but its expression level was significantly promoted when miR-1283 was silenced (Fig. 5I). Collectively, these discoveries explained that miR-1283 could suppress HEG1 expression by interacting with HEG1 in HCC cells.
MiR-1283 targeted regulation HEG1 in HCC cells. A. The binding sites between HEG1 3’UTR and miR-1283 were found by starbase. B, C. The luciferase activities of HEG1 3’UTRWT and HEG1 3’UTRMUT were detected by dual-luciferase reporter assay. D, E. The direct interaction between HEG1 and miR-1283 in HCC cells through RIP assay. F. The mRNA expression levels of HEG1 in HCC tumor tissues. G, H. The protein expression levels of HEG1 in HCC tumor tissues and cells. I. The HEG1 protein level was detected while HCC cells were transfected with miR-NC, miR-1283, anti-miR-NC and anti-miR-1283. Each experiment was independently repeated three times at least. *P < 0.05.
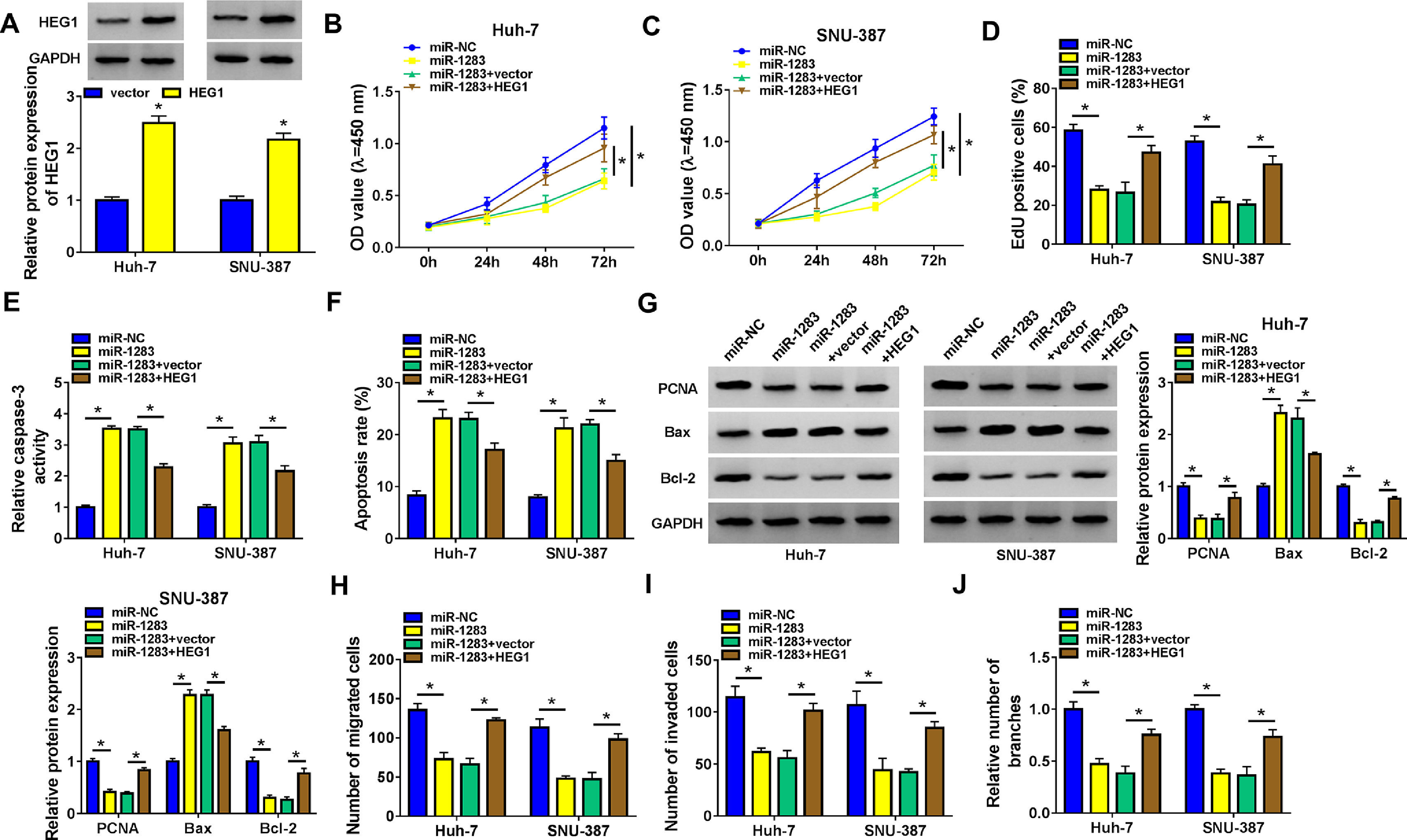
To further investigate whether miR-1283 induced HCC growth is regulated by HEG1, we conducted recovery experiments in Huh-7 and SNU-387 cells. As illustrated in Fig. 6A, the protein expression of HEG1 was elevated in Huh-7 cells and SNU-387 cells transfected with overexpression vector of HEG1. CCK-8 and EdU assays showed that miR-1283 mimics confined cell viability (Fig. 6B and C), and proliferation (Fig. 6D) of HCC cells, which were moderated by the overexpression of HEG1. And co-transfection of HEG1 overturned the effects of miR-1283 overexpression on Caspase 3 activity (Fig. 6E), cell apoptosis (Fig. 6F) and some signature proteins of cell proliferation and apoptosis (Fig. 6G) in HCC cells. MiR-1283 confined cell migration (Fig. 6H), invasion (Fig. 6I) and angiogenesis (Fig. 6J) of HCC cells through adjusting HEG1 expression.
The recovery functions of HEG1 against miR-1283 in HCC cells. A. The protein expressions of HEG1 transfected with HEG1 in HCC cells were detected. B, C. The viability of HCC cells was detected after transfection with miR-NC, miR-1283, miR-1283+vector, and miR-1283+HEG1 by CCK-8 assay. D. The cell proliferation of HCC cells was measured by EdU assay. E, F. The determination of Caspase 3 activity and cell apoptosis rate was to be seen in HCC cells. G. PCNA, Bax and Bcl-2 protein levels in HCC cells were texted by western blot. H-J. The migration, invasion and angiogenesis of transfected HCC cells. *P < 0.05.
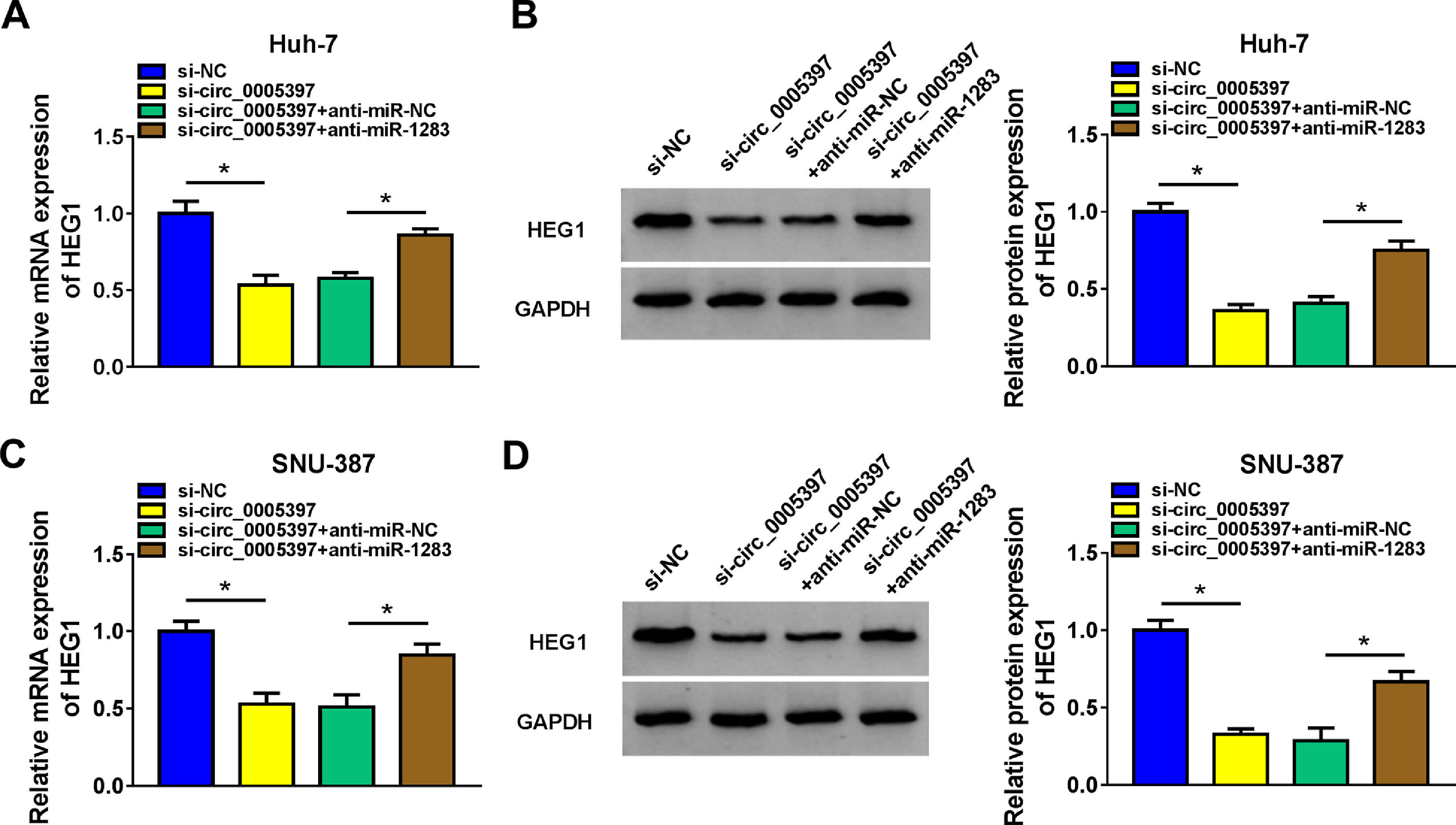
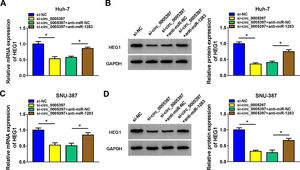
To validate whether circ_0005397 could affect the expression of HEG1 in HCC cells through miR-1283, we determined the expression levels of HEG1 in Huh-7 and SNU-387 cells. Recovery experiments showed that circ_0005397 silencing reduced both the mRNA and protein levels of HEG1, whereas anti-miR-1283 co-transfection in Huh-7 cells alleviated the negative effects of si-circ_0005397 on HEG1 levels (Fig. 7A and B). As can be seen in Fig. 7C and D, there were similar results in SNU-387 cells to Huh-7 cells. In a nutshell, circ_0005397 could play a role through the miR-1283/HEG1 axis in HCC cells.
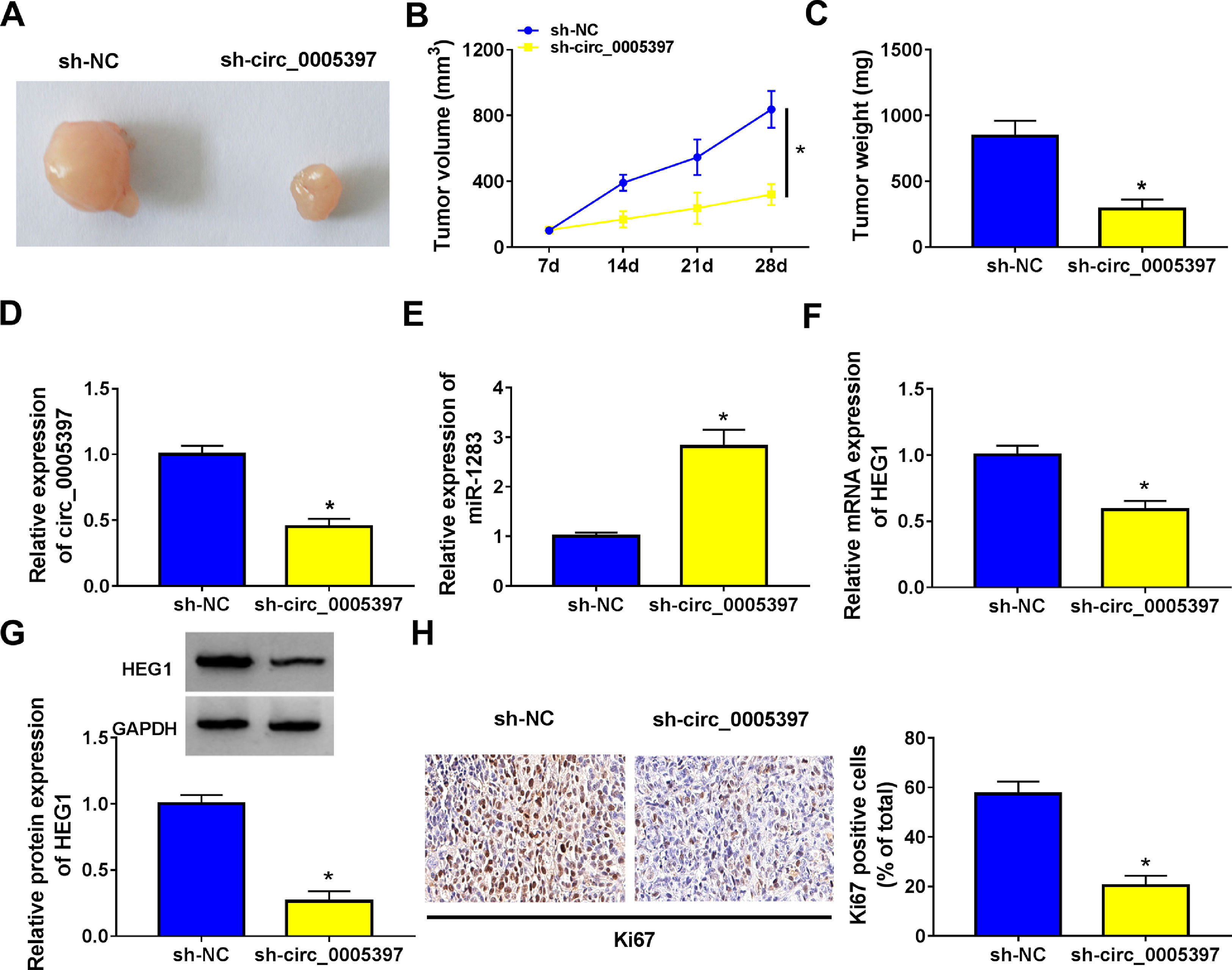
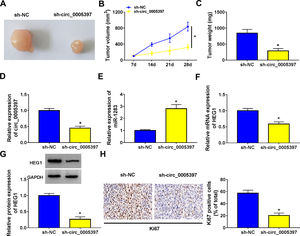
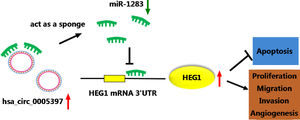
3.8Silencing circ_0005397 in vivo suppressed tumor growthTo validate whether circ_0005397 had an effect in HCC cells in vivo, Huh-7 cells with sh-circ_0005397 or negative control (sh-NC) were subcutaneously injected into immunodeficient mice, and the tumor growth was observed. Tumors from injected sh-circ_0005397 grew slowly, as indicated by reduced tumor volume (Fig. 8A and 8B) and reduced tumor weight (Fig. 8C). In addition, the tumor tissues obtained from nude mice in sh-circ_0005397 group had low expression of circ_0005397 compared to the sh-NC group (Fig. 8D), accompanied with high expression of miR-1283 (Fig. 8E). But both the mRNA and protein levels of HEG1 were decreased in sh-circ_0005397 group (Fig. 8F and G). Immunohistochemistry assay further confirmed sh-circ_0005397 silencing hampered HEG1 positive cells in mice tumor tissues (Fig. 8H). The above tests illustrated that circ_0005397/miR-1283/HEG1 axis controlled the growth of xenograft tumors. In general, circ_0005397 could accelerate HCC growth by promoting the expression of HEG1 through targeting interaction with miR-1283 (Fig. 9).
The effect of circ_0005397 silencing on tumor growth in vivo. A, B. The changes of tumor volume after subcutaneous injection of Huh-7 cells containing sh-circ_0005397 or sh-NC in nude mice. C. The changes of tumor weight after subcutaneous injection of Huh-7 cells containing sh-circ_0005397 or sh-NC in nude mice. D. The expression level of circ_0005397 in tumor tissue of nude mice in sh-circ_0005397 and sh-NC group. E. The expression level of miR-1283 in tumor tissue of nude mice in sh-circ_0005397 and sh-NC group. F, G. The expression of HEG1 mRNA and protein in tumor tissue of nude mice in sh-circ_0005397 and sh-NC group. H. The determination of Ki67 in sh-circ_0005397 group was carried out by IHC assay. *P < 0.05.
With the rapid development of biotechnology, the research on the molecular mechanism of circRNA in malignant tumors had attracted great attention [21]. In terms of mechanism, circRNA has been recognized as a kind of tumor suppressor, which regulated the expression of a variety of oncogenes [22]. Here, we explored an upregulated circRNA, circ_0005397, in HCC malignant behaviors. A recent study showed that circ_0005397 had elevated expression in HCC and it played an oncogene role in HCC [23]. Consistent with this research, we confirmed the highly expressed circ_0005397 in HCC tissues and cells. In terms of function, circ_0005397 silencing prevented the proliferation, metastasis and angiogenesis of HCC, and induced cell apoptosis. In addition, circ_0005397 knockdown also had an inhibitory effect on HCC tumor growth. These evidences provide a theoretical basis for circ_0005397 to become a potential therapeutic target for HCC.
We first identified that circ_0005397 could sponge miR-1283 in HCC. Many evidences suggest that miR-1283 may be a tumor suppressor to participate in regulating tumor development [15]. In glioma, circ-TTBK2 had been found to sponge miR-1283 to reduce miR-1283 expression, thereby enhancing glioma cell proliferation and metastasis [24]. A previous study indicated that upregulation of miR-1283 suppressed HCC proliferation and promoted apoptosis [25]. Consistent with those results, our data showed that miR-1283 was underexpressed in HCC, and its overexpression contributed to the inhibition of HCC cell proliferation, metastasis and angiogenesis. Furthermore, miR-1283 inhibitor overturned the inhibitory effect of si-circ_0005397 on malignant behavior of HCC cells, which confirmed that circ_0005397 indeed sponged miR-1283 to accelerate HCC progression.
Additionally, we identified that HEG1 was a downstream target gene of miR-1283. Zhao et al.[20] found that HEG1 and Wnt/β-Catenin signaling pathway had a deep connection to promote the development of HCC. We observed the overexpression of HEG1 in HCC cells, which was consistent with the previous results [19,20,26]. Importantly, HEG1 overexpression weakened the negative impact of miR-1283 up-regulation on cell proliferation, metastasis and angiogenesis. Besides, our data indicated that circ_0005397, as a miR-1283 sponge, played an active role in regulating HEG1 expression. This completes our hypothesis about the circ_0005397/miR-1283/HEG1 axis in HCC.
Our study may provide a potential theoretical target for HCC therapy. Of course, our current study has some limitations, and we have not yet carried out rescue trials in vivo to further confirm the conclusion that the circ_0005397/miR-1283/HEG1 axis regulates HCC tumor growth. This is a further problem we need to solve. In future studies, we will carry out functional tests to further confirm the influence of circ_0005397/miR-1283/HEG1 axis on HCC malignant progression to enrich our research results.
5ConclusionsIn conclusion, our results showed that circ_0005397 sponged miR-1283 to regulate HEG1, thus promoting HCC proliferation, metastasis and angiogenesis. These findings further shed light on the molecular mechanisms of HCC and had drawn an important stroke for the capture of HCC.
FundingThis research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
AuthorshipLijuan Liu, Yanna Wang and Yanfang Li: the conception and design of the study, or acquisition of data, or analysis and interpretation of data. Haifeng Yu, Youde Liu and Jing Sun: drafting the article or revising it critically for important intellectual content. The manuscript has been read and approved by all named authors.
None