Circular RNAs (circRNAs) are identified to show important regulatory functions in cancer biology. We attempted to analyze the role of circ_0000291 in hepatocellular carcinoma (HCC) progression and its related mechanism.
MethodsThe circular characteristic of circ_0000291 was tested using exonuclease RNase R. Cell proliferation was analyzed by 5-Ethynyl-2’-deoxyuridine (EdU) incorporation and colony formation assays. Cell apoptosis was measured by flow cytometry and a caspase 3 activity assay kit. Transwell assays were performed to analyze cell migration and invasion abilities. Sphere formation assay was conducted to analyze cell stemness. Dual-luciferase reporter and RNA-pull down assays were conducted to verify the interaction between microRNA-1322 (miR-1322) and circ_0000291 or ubiquitin conjugating enzyme E2 T (UBE2T).
ResultsCirc_0000291 was markedly up-regulated in HCC tissues and cell lines. HCC patients with high expression of circ_0000291 displayed a low survival rate. Circ_0000291 knockdown restrained the proliferation, migration, invasion, and stemness and induced the apoptosis of HCC cells. Circ_0000291 directly interacted with miR-1322 and negatively regulated miR-1322 expression. Circ_0000291 knockdown-mediated anti-tumor impacts in HCC cells were largely overturned by the interference of miR-1322. miR-1322 directly paired with the 3’ untranslated region (3’UTR) of UBE2T, and UBE2T was negatively regulated by miR-1322. UBE2T overexpression largely reversed circ_0000291 silencing-induced effects in HCC cells. Circ_0000291 positively regulated UBE2T expression by absorbing miR-1322 in HCC cells. Circ_0000291 silencing notably reduced the tumorigenic potential in vivo.
ConclusionCirc_0000291 facilitated HCC progression by targeting miR-1322/UBE2T axis, which provided novel potential biomarkers and targets for HCC patients.
Hepatocellular carcinoma (HCC) is a common malignancy globally [1]. The most common risk factors for HCC are chronic infection with hepatitis B virus (HBV) or hepatitis C virus (HCV), excess body weight, heavy alcohol intake, aflatoxin-contaminated foods, and type 2 diabetes [2]. Although the wide use of advanced therapeutic strategies, including hepatectomy, liver transplantation, and radiofrequency ablation (RFA), the outcome of HCC is still dismal with high recurrence and metastasis [3,4]. Recent studies have shown that many novel therapeutic strategies, such as metronomic capecitabine, lenvatinib plus pembrolizumab, and biochemical predictors in response to immune checkpoint inhibitors, have potential anti-HCC activity [5–7]. More intriguingly, in terms of molecular basis, the tumorigenesis of HCC is a multi-step process that involves the dysregulation of many oncogenes and tumor suppressor genes and epigenetic changes [8–10]. Therefore, identifying novel biomarkers and targets based on the epigenetic regulators is urgently needed for HCC.
Circular RNAs (circRNAs) are natural RNA circles that are generated by back-splicing of pre-mRNAs [11]. Recently, some studies have confirmed that circRNAs can undergo a translational process [12,13]. Accumulating evidence has demonstrated that circRNAs are mainly localized in the cytoplasmic fraction, where they can directly bind to microRNAs (miRNAs) to adjust miRNA downstream gene expression, thereby involving in the modulation of multiple biological processes such as human tumorigenesis [14,15]. In HCC, Ding et al. found that circ_0001955 facilitates cell growth and motility by binding to miR-145-5p to up-regulate NRAS proto-oncogene, GTPase (NRAS) expression [16]. Gao et al. found that circ_0001178 contributes to HCC progression by sponging miR-382 to elevate vascular endothelial growth factor A (VEGFA) level [17]. However, the modulatory networks of circRNAs in HCC development remain largely unknown.
We analyzed the circRNA expression profiles in three GEO datasets (GSE97332, GSE94508, and GSE164803) and selected circ_0000291 for further study because previous articles reported that circ_0000291 plays an oncogenic role in gastric cancer [18] and breast cancer [19]. However, its role in HCC is still undetermined. We intended to assess circ_0000291 function in HCC tumorigenesis and attempted to investigate its working mechanism in HCC.
2Materials and methods2.1Patients and tissue samplesFifty pairs of HCC tissue specimens and adjacent normal tissue specimens were harvested from 50 HCC patients who underwent surgical resection at the Beilun Branch of the First Affiliated Hospital of Medical College of Zhejiang University. The key risk factors of the 50 patients were chronic HBV infection (31 cases), aflatoxin exposure (9 cases), or both (10 cases). All tissue specimens were preserved in liquid nitrogen.
2.2Cell cultureFour HCC cell lines (Huh-7, LM3, LM6, and MHCC97H) and human normal THLE-2 cell line acquired from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) were cultured in DMEM (Invitrogen, Carlsbad, CA, USA) enriched with 10% FBS (Invitrogen) and 1% antibiotic mixture (Sangon Biotech, Shanghai, China) at 37°C in a humidified incubator containing 5% CO2.
2.3RNase R digestion assayTotal RNA samples were treated with RNase R (100 μg/mL; Applied Biological Materials, Vancouver, Canada) at room temperature for 30 min and subsequently subjected to RT-qPCR assay.
2.4RT-qPCRThe Trizol reagent (Invitrogen) was used for RNA isolation. Reverse transcription was implemented via M-MLV reverse transcriptase kit (for circRNAs and messenger RNAs (mRNAs); Invitrogen) and miRNA cDNA Synthesis Kit (for miRNAs; GeneCopoeia, Rockville, MD, USA). qPCR was conducted using the specific primers (Table 1) and the Power SYBR-Green PCR master mix (Applied Biosystems, Foster, CA, USA). The relative abundance was examined by the 2−ΔΔCt formula [20], and β-actin or U6 served as the housekeeping gene.
Primer sequences used for qPCR
The short hairpin (sh)RNAs, containing sh-circ_0000291 and sh-NC, were purchased from Genechem (Shanghai, China). miR-1322 mimics and inhibitors (miR-1322 and anti-miR-1322) and negative controls (miR-NC and anti-NC) were acquired from Ribobio (Guangzhou, China). For the ectopic expression of ubiquitin conjugating enzyme E2 T (UBE2T), the coding sequence of UBE2T was inserted into the pcDNA 3.1 vector (Genechem). Transient transfection was conducted using Lipofectamine 3000 (Invitrogen).
2.65-Ethynyl-2’-deoxyuridine (EdU) incorporation assayHCC cells were laid on sterile coverslips, and DNA synthesis was marked using an EdU kit (Ribobio) according to the user's instructions. Cell nuclei were marked with DAPI (Beyotime, Shanghai, China). Cell images were taken under laser confocal microscopy (Olympus, Tokyo, Japan).
2.7Colony formation assayHCC cells were laid on 12-well plates. Cells were cultivated for 14 d, and colonies were immobilized and then dyed with 0.1% crystal violet (Sangon Biotech). Colonies with at least 50 cells were counted.
2.8Flow cytometryTransfected HCC cells were collected and double-stained with Annexin V-FITC (Invitrogen) and propidium iodide (PI) (Invitrogen) for 15 min in the dark. Cell samples were loaded on a flow cytometer (Beckman Coulter, Brea, CA, USA) and the cell apoptosis rate was then analyzed.
2.9Caspase 3 activityCaspase 3 activity was examined using the caspase 3 activity assay kit (Beyotime). In brief, cell extracts were mixed with the assay buffer and Ac-DEVD-pNA at 37°C for 2h, and optical density was measured at 405 nm.
2.10Transwell assaysThe 24-well plate transwell chambers covered with or without Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) were adopted to analyze cell invasion and migration abilities, respectively. HCC cells were laid on the upper chambers in serum-free medium and were permitted to penetrate the membrane to the below chambers that were filled with complete medium. The migrated or invaded HCC cells were stained and then counted under an optical microscope (Olympus).
2.11Sphere formation assaySphere formation assay was implemented to evaluate cell stemness. Transfected HCC cells were laid on the 96-well Clear Round Bottom Ultra Low Attachment Microplate (Corning). After incubation for 7 d, tumor spheres were observed. The proportion of cells that could form tumor balls was considered sphere formation efficiency.
2.12Western blot assayTotal protein samples were isolated from tissues and cells using RIPA lysis buffer (Beyotime). Proteins (25 μg) were loaded onto 10% separating gel and shifted onto the PVDF membrane (Millipore, Billerica, MA, USA). After being blocked in 5% skimmed milk, the membrane was incubated with the primary antibodies (Abcam, Cambridge, MA, USA) specific for different proteins at 4°C overnight. After incubation with the secondary antibody (Abcam) for 2 h, protein bands were visualized using the ECL system (Pierce, Waltham, MA, USA). The intensities of protein bands were quantified using Image J software and β-actin served as the internal reference. The primary antibodies (Abcam) included anti-PCNA (catalog number: ab29), anti-Bax (catalog number: ab32503), anti-Slug (catalog number: ab27568), anti-SOX2 (catalog number: ab92494), anti-UBE2T (catalog number: ab179802), and anti-β-actin (catalog number: ab8226) antibodies.
2.13Bioinformatics analysisThe interacted miRNAs of circ_0000291 were foretold by circBank (http://www.circbank.cn) [21] and circInteractome (https://circinteractome.nia.nih.gov) [22] databases. TargetScan database (http://www.targetscan.org/vert_72) was adopted to foretell the interacted mRNAs of miR-1322.
Three Gene Expression Omnibus (GEO) datasets (GSE97332, GSE94508, and GSE164803) were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/). The top 40 up-regulated genes in HCC were obtained by the GEPIA database (http://gepia.cancer-pku.cn/detail.php).
2.14Dual-luciferase reporter assayThe wild-type reporter plasmids, including circ_0000291 WT and UBE2T 3’UTR WT, were constructed by inserting the partial fragment of circ_0000291 and UBE2T 3’UTR into the pmirGLO vector (Promega, Madison, WI, USA). Meanwhile, the mutant-type reporter plasmids, including circ_0000291 MUT and UBE2T 3’UTR MUT, were also constructed. HCC cells were laid on the 24-well plates and introduced with reporter vectors and small RNAs with Lipofectamine 3000 (Invitrogen). The luciferase intensities in four groups were evaluated via the commercial Dual-luciferase reporter assay system kit (Promega).
2.15RNA-pull down assayA total of 2 μg cell lysates were incubated with 100 pmol biotinylated RNA (Bio-NC, Bio-miR-1322, or miR-1322 MUT). Subsequently, 100 μL washed streptavidin agarose beads were added into the reaction system for 2 h. RT-qPCR was conducted to analyze RNA enrichment in the precipitated complex.
2.16Xenograft mouse modelBALB/c nude mice were acquired from Vital River Laboratory Animal Technology (Beijing, China) and were assigned to two groups (n=6/group): sh-NC and sh-circ_0000291. Huh-7 cell line with the stable infection of sh-NC or sh-circ_0000291 was used in animal experiments. A total of 100 μL lentivirus-infected Huh-7 cell suspension (1 × 106 cells) was subcutaneously inoculated into the back region of the nude mice. Tumor dimension was recorded weekly as length × width2 × 0.5. After injection for 4 weeks, all nude mice were sacrificed via CO2 asphyxia method, and xenograft tumors were excised. Immunohistochemistry (IHC) assays were implemented using standard methods [23] to measure PCNA abundance in tumor specimens using the antibody of PCNA (catalog number: ab29; Abcam).
2.17Statistical analysisAll in vitro assays were independently repeated at least three times. The data were represented as mean ± SD. The survival rate was analyzed via Kaplan-Meier methods and log-rank tests. Expression correlation was evaluated via Pearson correlation analysis. The differences were assessed by Student's t-test or one-way ANOVA. Differences were deemed as significant with P values of less than 0.05.
2.18Ethical statementThis clinical study was authorized by the ethics committee of the Beilun Branch of the First Affiliated Hospital of Medical College of Zhejiang University with written informed consent. The protocol was authorized by the Institutional Animal Care and Use Committee of the Beilun Branch of the First Affiliated Hospital of Medical College of Zhejiang University.
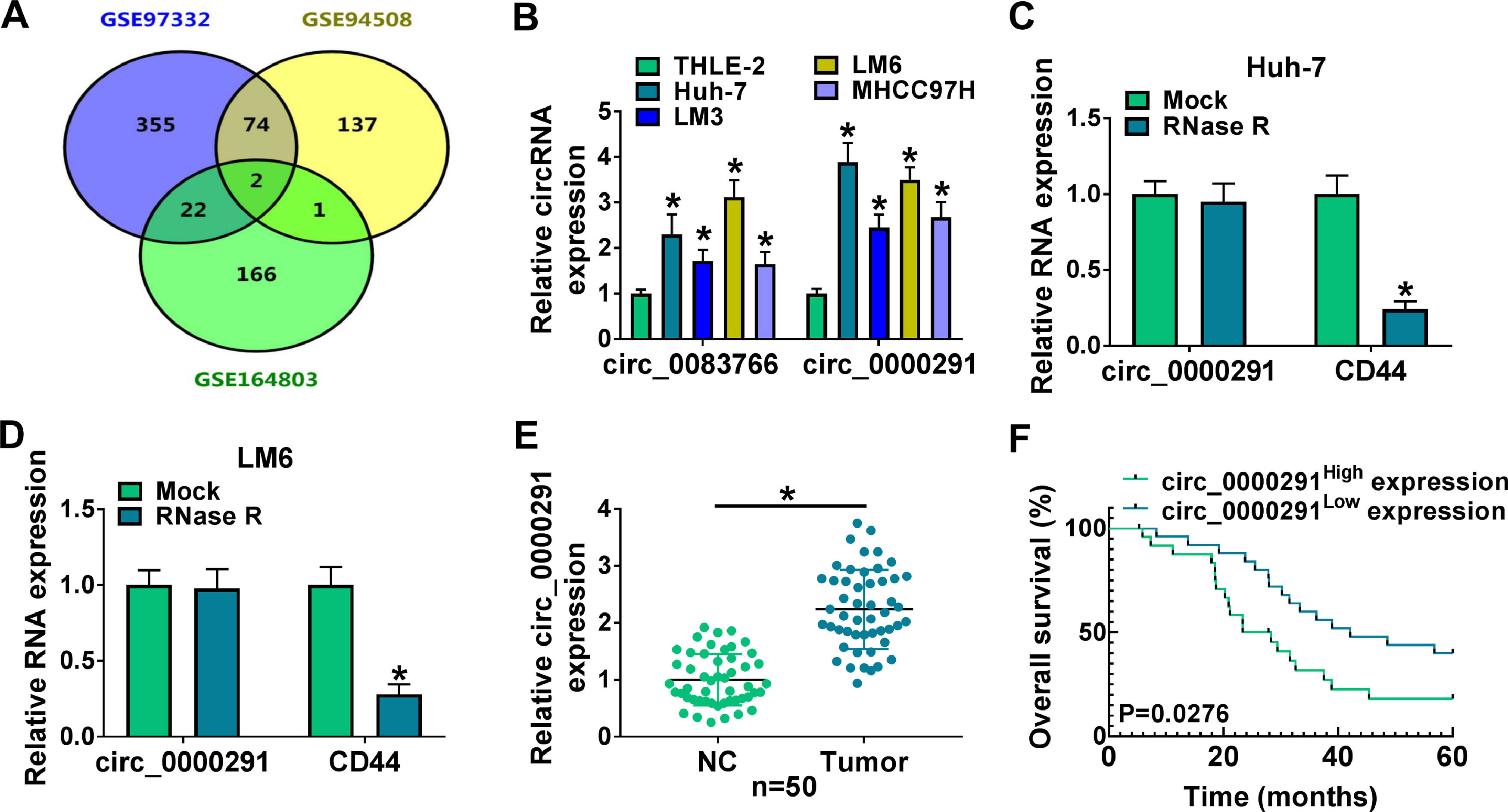
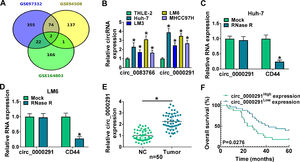
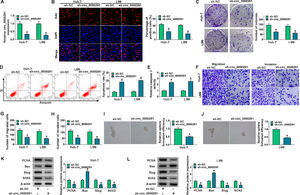
3Results3.1The expression of circ_0000291 in HCC tissues and cell lines and its clinical significanceWe analyzed the up-regulated circRNAs in HCC tissues compared with adjacent healthy tissues from the three GEO datasets (GSE97332, GSE94508, and GSE164803) that were downloaded from NCBI with log2(fold change)>1 and P-value<0.05 as the threshold values. Venn diagram showed two circRNAs (circ_0083766 and circ_0000291) that overlapped among these datasets (Figure 1A). We measured the expression of circ_0083766 and circ_0000291 in four HCC cell lines (Huh-7, LM3, LM6, and MHCC97H) and the normal liver cell line THLE-2. The data displayed that both circ_0083766 and circ_0000291 were up-regulated in HCC cell lines compared with the THLE-2 cell line, and circ_0000291 expression was more pronounced in HCC cell lines than circ_0083766 expression (Figure 1B). Therefore, circ_0000291 was selected as the follow-up research object. We found that circ_0000291, but not its linear CD44 mRNA, was resistant to the digestion of RNase R (Figure 1C and 1D), suggesting that circ_0000291 was an indeed a circular transcript. We analyzed 50 pairs of HCC tissues and adjacent normal tissues and found that circ_0000291 expression was elevated in HCC tissues (Figure 1E). To analyze the prognostic value of circ_0000291, we divided 50 HCC patients into two groups (n=25/group) with the median value of circ_0000291 expression as the cutoff. The results showed that the survival rate of HCC patients with high expression of circ_0000291 was lower than that with low expression of circ_0000291 (Figure 1F). Additionally, there was no a significant correlation between circ_0000291 expression and the etiology of the 55 HCC patients (Table 2). These data suggested the abnormal up-regulation of circ_0000291 in human HCC and its clinical prognostic value.
The expression of circ_0000291 in HCC tissues and cell lines and its clinical significance. (A) Three GEO datasets (GSE97332, GSE94508, and GSE164803) were downloaded from NCBI, and we screened the up-regulated circRNAs in HCC tissues compared to adjacent normal tissues with log2(fold change)>1 and P-value<0.05 as the threshold values. Circ_0083766 and circ_0000291 were located at the intersection of three databases. (B) RT-qPCR was conducted to determine the expression of circ_0083766 and circ_0000291 in four HCC cell lines (Huh-7, LM3, LM6, and MHCC97H) and normal liver cell line THLE-2. (C and D) Circ_0000291 was derived from the back-splicing of CD44 pre-mRNA. RNase R was used to test the circular characteristic of circ_0000291, and its linear form CD44 was regarded as the control. (E) RT-qPCR was conducted to measure the expression of circ_0000291 in 50 pairs of HCC tissues and adjacent normal tissues (normal control; NC). (F) Kaplan-Meier analysis was performed to analyze the prognostic role of circ_0000291 in HCC patients. HCC patients were divided into circ_0000291high expression group and circ_0000291low expression group with the median value of circ_0000291 expression as the cutoff. *P<0.05.
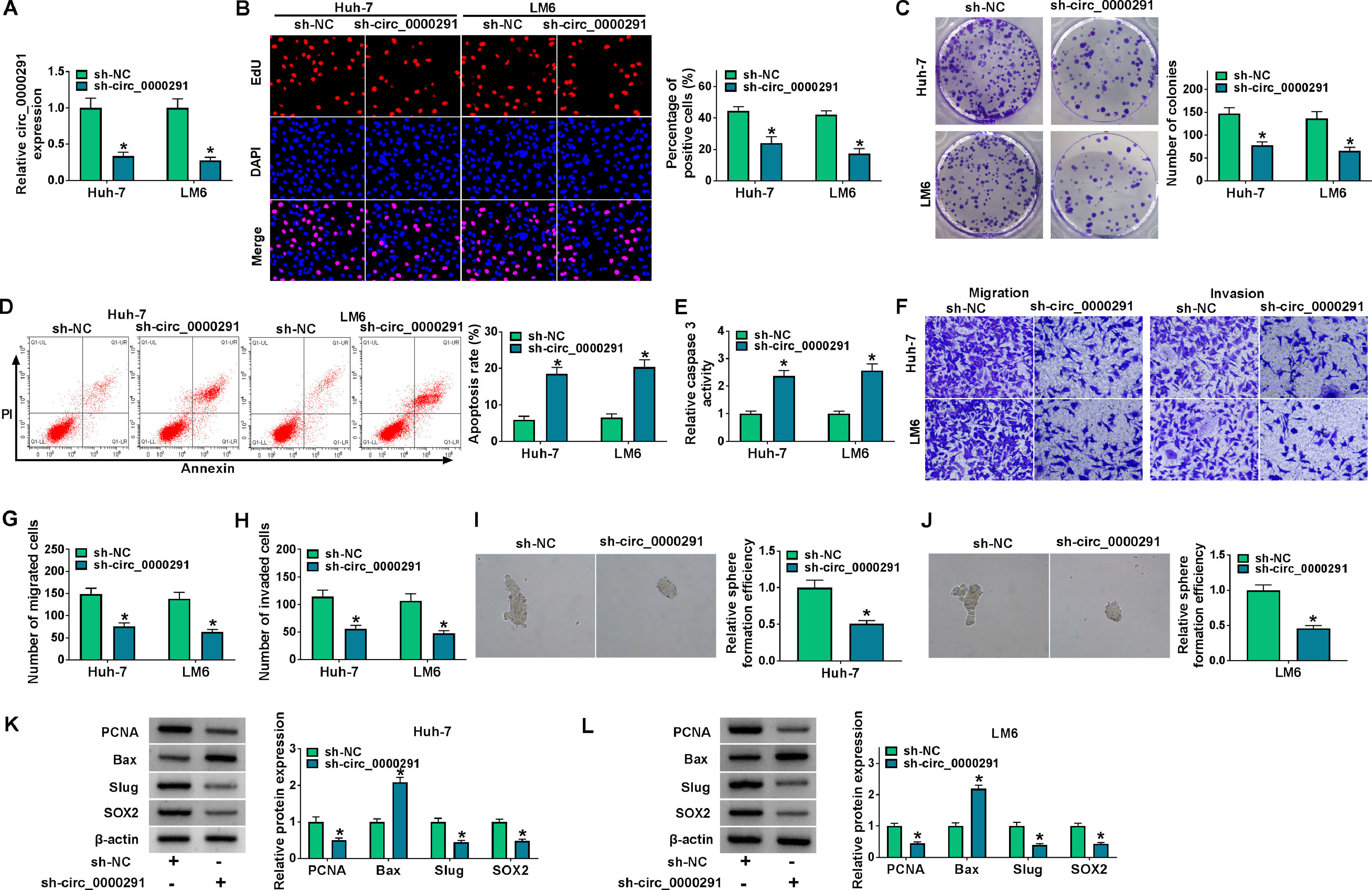
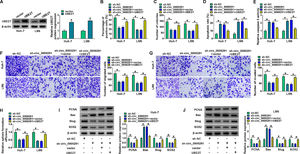
Considering that circ_0000291 was aberrantly up-regulated in HCC cells, we silenced circ_0000291 using sh-circ_0000291 to explore its biological function. RT-qPCR confirmed that the knockdown efficiency of sh-circ_0000291 was significant in HCC cells (Figure 2A). EdU assays revealed that circ_0000291 knockdown reduced the percentage of EdU+ cells (Figure 2B). Colony formation assays showed that circ_0000291 silencing decreased the number of colonies (Figure 2C). These data demonstrated that circ_0000291 knockdown suppressed the proliferation of HCC cells. Circ_0000291 interference elevated the apoptosis rate and the activity of caspase 3 (Figure 2D and 2E), indicating that circ_0000291 silencing induced the apoptosis of HCC cells. Transwell assays showed that the numbers of migrated and invaded cells were reduced by circ_0000291 knockdown (Figure 2F-2H), indicating that circ_0000291 silencing restrained the migration and invasion of HCC cells. Circ_0000291 knockdown also reduced the sphere formation efficiency of HCC cells (Figure 2I and 2J). Western blot assays were conducted to measure the protein levels of proliferation-, apoptosis-, motility-, and sphere formation-associated markers (PCNA, Bax, Slug, and SOX2) in HCC cells. The data showed that circ_0000291 knockdown reduced the expression of PCNA, Slug, and SOX2 and increased the level of Bax in Huh-7 and LM6 HCC cells (Figure 2K and 2L). These results showed that circ_0000291 knockdown suppressed the proliferation, migration, invasion, and stemness and induced the apoptosis of HCC cells.
Circ_0000291 plays an oncogenic role in HCC cells in vitro. (A-L) Huh-7 and LM6 cell lines stably transfected with sh-NC or sh-circ_0000291 were used to conduct a series of functional experiments. (A) The expression of circ_0000291 was examined by RT-qPCR. (B) EdU assay was conducted to analyze the proliferation ability of HCC cells. (C) Colony formation assay was performed to assess the proliferation ability of HCC cells. (D) Cell apoptosis was analyzed by flow cytometry. (E) The activity of caspase 3 was analyzed by a commercial caspase 3 activity assay kit. (F-H) Transwell assays were conducted to analyze the migration and invasion of HCC cells. (I and J) The sphere formation efficiency was assessed by sphere formation assay. (K and L) Western blot assay was conducted to measure the protein levels of proliferation-, apoptosis-, motility-, and sphere formation-associated markers (PCNA, Bax, Slug, and SOX2) in HCC cells. *P<0.05.
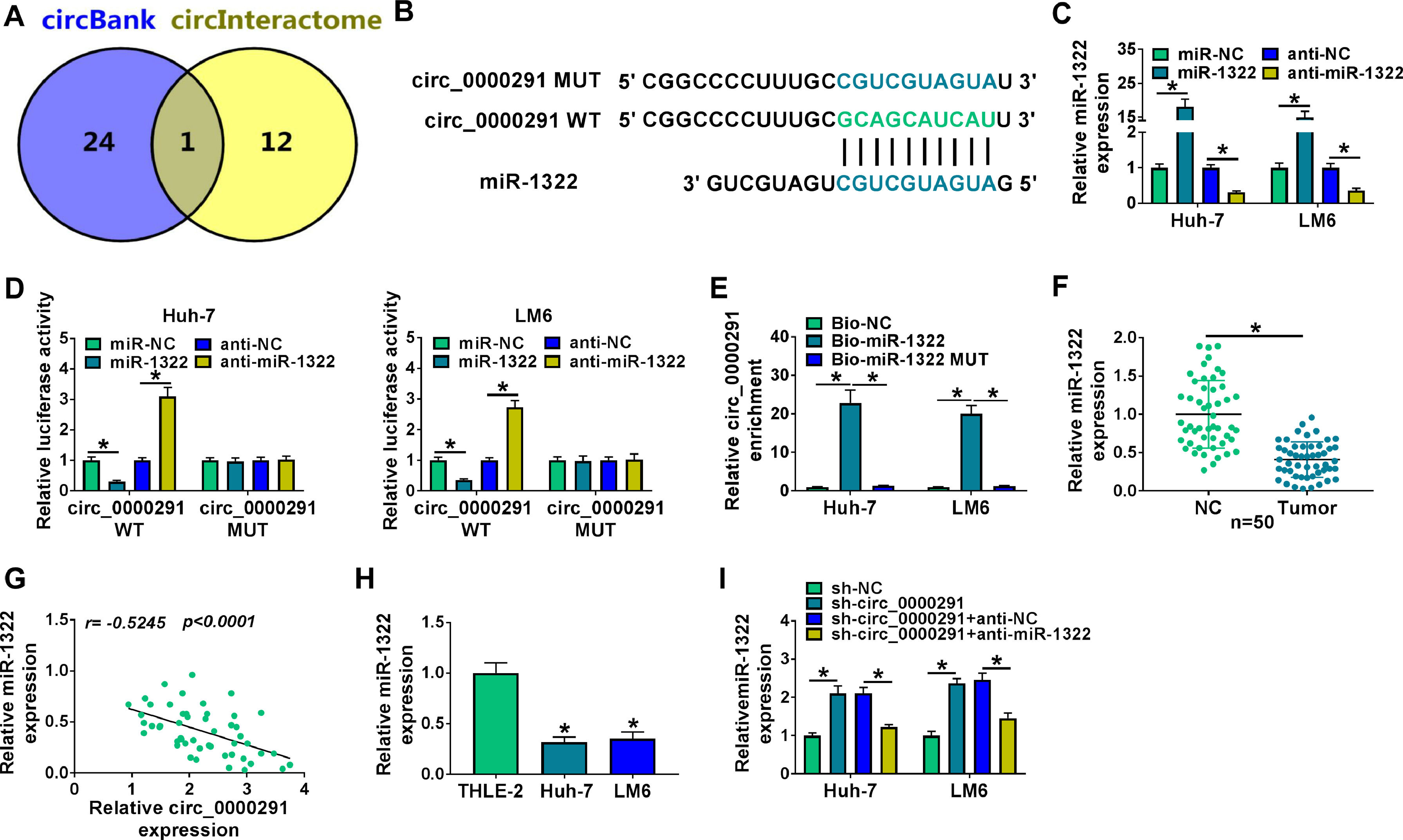
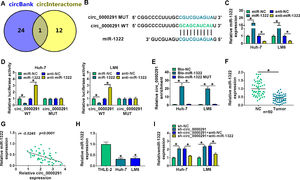
Through circBank [21] and circInteractome [22] databases, we found that miR-1322 was a potential target of circ_0000291 (Figure 3A). The putative binding sequence between circ_0000291 and miR-1322 was shown in Figure 3B. Subsequently, dual-luciferase reporter and RNA-pull down assays were carried out to verify the interaction between circ_0000291 and miR-1322 in HCC cells. RT-qPCR verified the significant transfection efficiencies of miR-1322 and anti-miR-1322 in altering miR-1322 expression in HCC cells (Figure 3C). Overexpression of miR-1322 significantly reduced the luciferase activity of the wild-type reporter vector (circ_0000291 WT), while this suppressive effect was lost when the miR-1322 binding sites were mutated (Figure 3D), suggesting that circ_0000291 directly interacted with miR-1322 via the putative sites. RNA-pull down assays revealed that circ_0000291 was pulled down by the biotinylated miR-1322 probe (Figure 3E), reinforcing the direct interaction between circ_0000291 and miR-1322. The expression of miR-1322 was down-regulated in HCC tissues compared with adjacent healthy tissues (Figure 3F). A significant negative correlation between the expression of circ_0000291 and miR-1322 was identified in HCC tissues (Figure 3G). Moreover, miR-1322 was down-regulated in HCC cell lines compared with the THLE-2 cell line (Figure 3H). Furthermore, circ_0000291 silencing resulted in a notable up-regulation in miR-1322 expression, and this effect was diminished by the addition of anti-miR-1322 in HCC cells (Figure 3I). These data demonstrated that miR-1322 was a direct target of circ_0000291 in HCC cells.
Circ_0000291 directly sponges miR-1322 in HCC cells. (A) The miRNA targets of circ_0000291 were predicted by circBank and circInteractome databases. Only miR-1322 was located at the intersection of two bioinformatics databases. (B) The putative or mutant binding sequence with miR-1322 in circ_0000291 was shown. (C) RT-qPCR was conducted to assess the overexpression efficiency of miR-1322 mimics (miR-1322) and the knockdown efficiency of miR-1322 inhibitor (anti-miR-1322) in HCC cells. (D and E) Dual-luciferase reporter assay and RNA-pull down assay were conducted to confirm the target relationship between circ_0000291 and miR-1322 in HCC cells. (F) RT-qPCR was implemented to examine the expression of miR-1322 in 50 pairs of HCC tissues and para-cancer healthy tissues. (G) Pearson's correlation analysis was performed to analyze the linear correlation between the expression of circ_0000291 and miR-1322 in HCC tissues. (H) The level of miR-1322 was determined in HCC cell lines (Huh-7 and LM6) and the THLE-2 cell line by RT-qPCR. (I) Huh-7 and LM6 cells were transfected with sh-circ_0000291 alone or together with anti-miR-1322. The expression of miR-1322 was examined by RT-qPCR. *P<0.05.
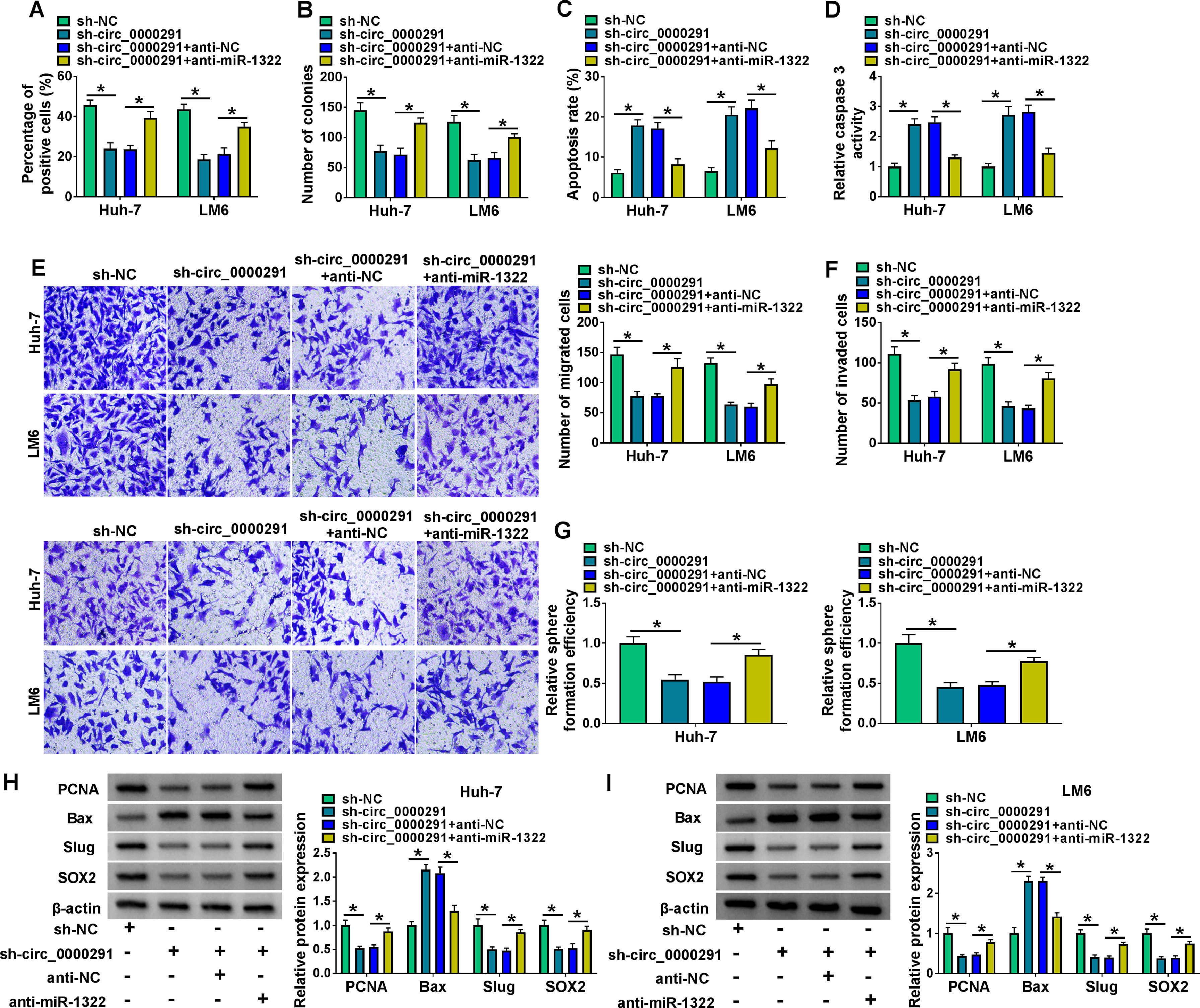
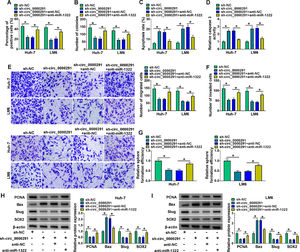
To verify whether circ_0000291 participated in regulating HCC progression by targeting miR-1322, rescue experiments were performed. Circ_0000291 knockdown-induced suppressive effect on the proliferation of HCC cells was largely rescued by the addition of anti-miR-1322 (Figure 4A and 4B). Circ_0000291 interference induced cell apoptosis and elevated the activity of caspase 3, which were largely diminished by miR-1322 knockdown (Figure 4C and 4D). Circ_0000291 absence also inhibited the migration and invasion of HCC cells, and these suppressive impacts were largely counteracted by the knockdown of miR-1322 (Figure 4E and 4F). Circ_0000291 silencing suppressed the stemness of HCC cells, which was largely rescued by the introduction of anti-miR-1322 (Figure 4G). Additionally, circ_0000291 knockdown reduced the protein levels of PCNA, Slug, and SOX2 and increased the protein expression of Bax, and these effects were largely reversed by miR-1322 knockdown (Figure 4H and 4I). These data revealed that circ_0000291 silencing suppressed HCC progression largely by up-regulating miR-1322 in vitro.
Circ_0000291 knockdown-induced anti-tumor effects in HCC cells are largely overturned by miR-1322 interference. (A-I) HCC cells were transfected with sh-circ_0000291 alone or together with anti-miR-1322. (A and B) Cell proliferation was analyzed by EdU assay and colony formation assay. (C and D) Cell apoptosis was assessed by flow cytometry and a commercial caspase 3 activity assay kit. (E and F) Cell migration and invasion abilities were examined by transwell assays. (G) The relative sphere formation efficiency was assessed by sphere formation assay. (H and I) The protein levels of PCNA, Bax, Slug, and SOX2 were determined by western blot assay. *P<0.05.
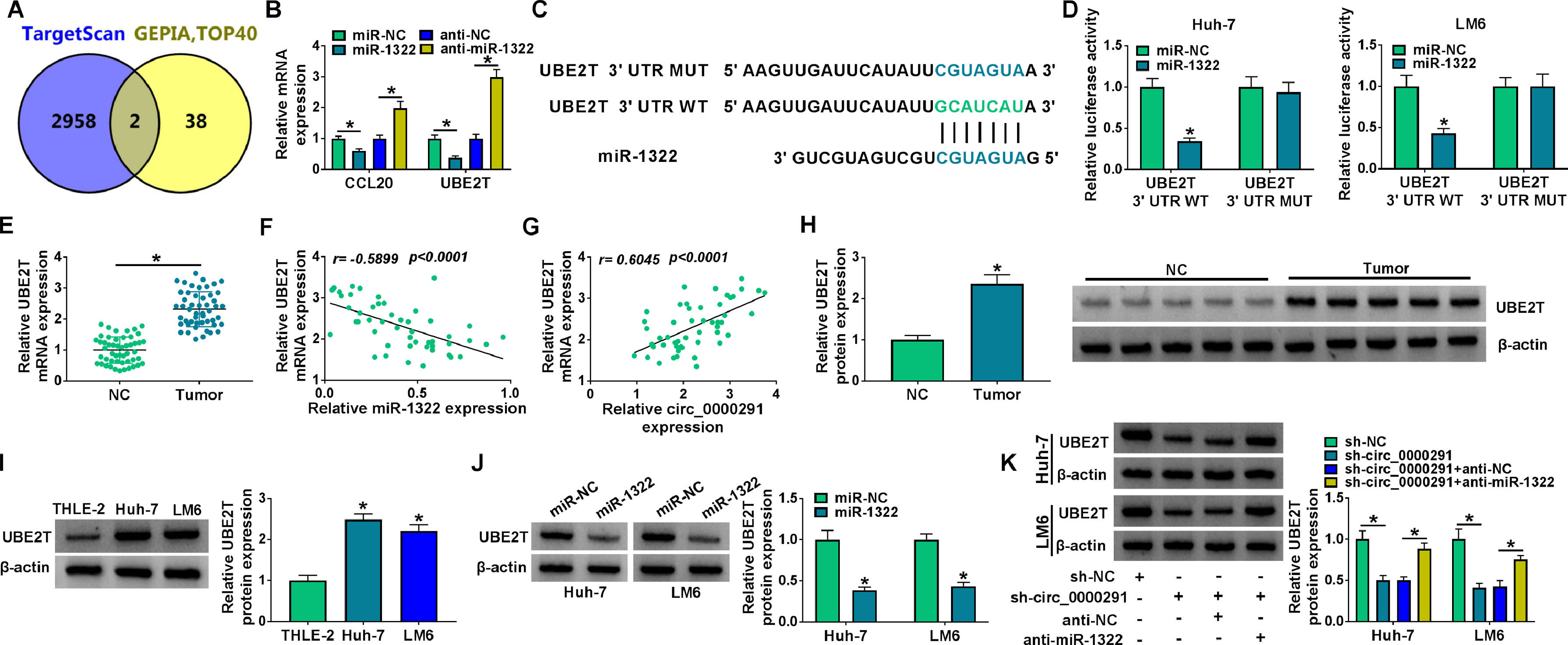
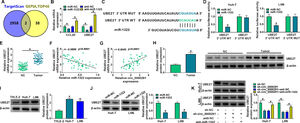
Increasing evidence has confirmed that miRNAs exert important functions in HCC development by inhibiting the expression of target mRNAs [24,25]. We predicted the possible mRNA targets of miR-1322 using the TargetScan database. Meanwhile, the top 40 up-regulated genes in HCC were obtained from the GEPIA database. Venn diagram showed two genes (CCL20 and UBE2T) that overlapped between the databases (Figure 5A). Overexpression of miR-1322 reduced the mRNA expression of CCL20 and UBE2T, while miR-1322 knockdown up-regulated the mRNA levels of CCL20 and UBE2T in HCC cells (Figure 5B). Since the negative regulatory relationship between miR-1322 and UBE2T was more obvious than that between miR-1322 and CCL20, we chose UBE2T for the following research. The predicted binding sites between miR-1322 and UBE2T 3’UTR were shown in Figure 5C. Overexpression of miR-1322 reduced the luciferase intensity of the UBE2T 3’UTR WT reporter vector, rather than the UBE2T 3’UTR MUT reporter vector (Figure 5D), suggesting the direct interaction between miR-1322 and UBE2T 3’UTR. UBE2T mRNA expression was up-regulated in HCC tissues compared with adjacent normal tissues (Figure 5E). miR-1322 expression was negatively correlated with UBE2T mRNA expression and was positively correlated with circ_0000291 expression in HCC tissues (Figure 5F and 5G). Additionally, the protein expression of UBE2T was higher in HCC tissues than that in adjacent normal tissues (Figure 5H). In line with tissue samples, UBE2T protein expression was up-regulated in HCC cell lines compared with the THLE-2 cell line (Figure 5I). Furthermore, miR-1322 overexpression led to a marked reduction in UBE2T protein expression in HCC cells (Figure 5J), supporting the targeting of UBE2T by miR-1322. Circ_0000291 absence reduced the protein level of UBE2T, and the addition of anti-miR-1322 largely rescued the expression of UBE2T in HCC cells (Figure 5K). These data showed that circ_0000291 can regulate UBE2T expression by acting as a miR-1322 sponge in HCC cells.
UBE2T is a direct target of miR-1322 in HCC cells. (A) We used the TargetScan database to predict the mRNA targets of miR-1322. Meanwhile, the top 40 up-regulated genes in HCC were obtained by GEPIA database. The Venn diagram showed that CCL20 and UBE2T were located at the intersection of the two databases. (B) The mRNA expression of CCL20 and UBE2T in HCC cells transfected with miR-NC, miR-1322, anti-NC, or anti-miR-1322 was examined by RT-qPCR. (C) The putative or mutant binding sequence with miR-1322 in the 3’UTR of UBE2T was shown. (D) The interaction between miR-1322 and UBE2T in HCC cells was verified by dual-luciferase reporter assay. (E) RT-qPCR was conducted to analyze the mRNA expression of UBE2T in 50 pairs of HCC tissues and adjacent normal tissues. (F and G) The linear correlation between the expression of miR-1322 and circ_0000291 or UBE2T in HCC tissues was analyzed by Pearson's correlation coefficient. (H) Western blot assay was performed to examine the protein expression of UBE2T in HCC tissues and adjacent normal tissues. (I) The protein level of UBE2T was determined in HCC cell lines and the THLE-2 cell line by western blot assay. (J) HCC cells were transfected with miR-NC or miR-1322, and western blot assay was performed to detect the protein level of UBE2T in transfected HCC cells. (K) HCC cells were transfected with sh-circ_0000291 alone or together with anti-miR-1322. Western blot assay was implemented to determine the protein expression of UBE2T in transfected HCC cells. *P<0.05.
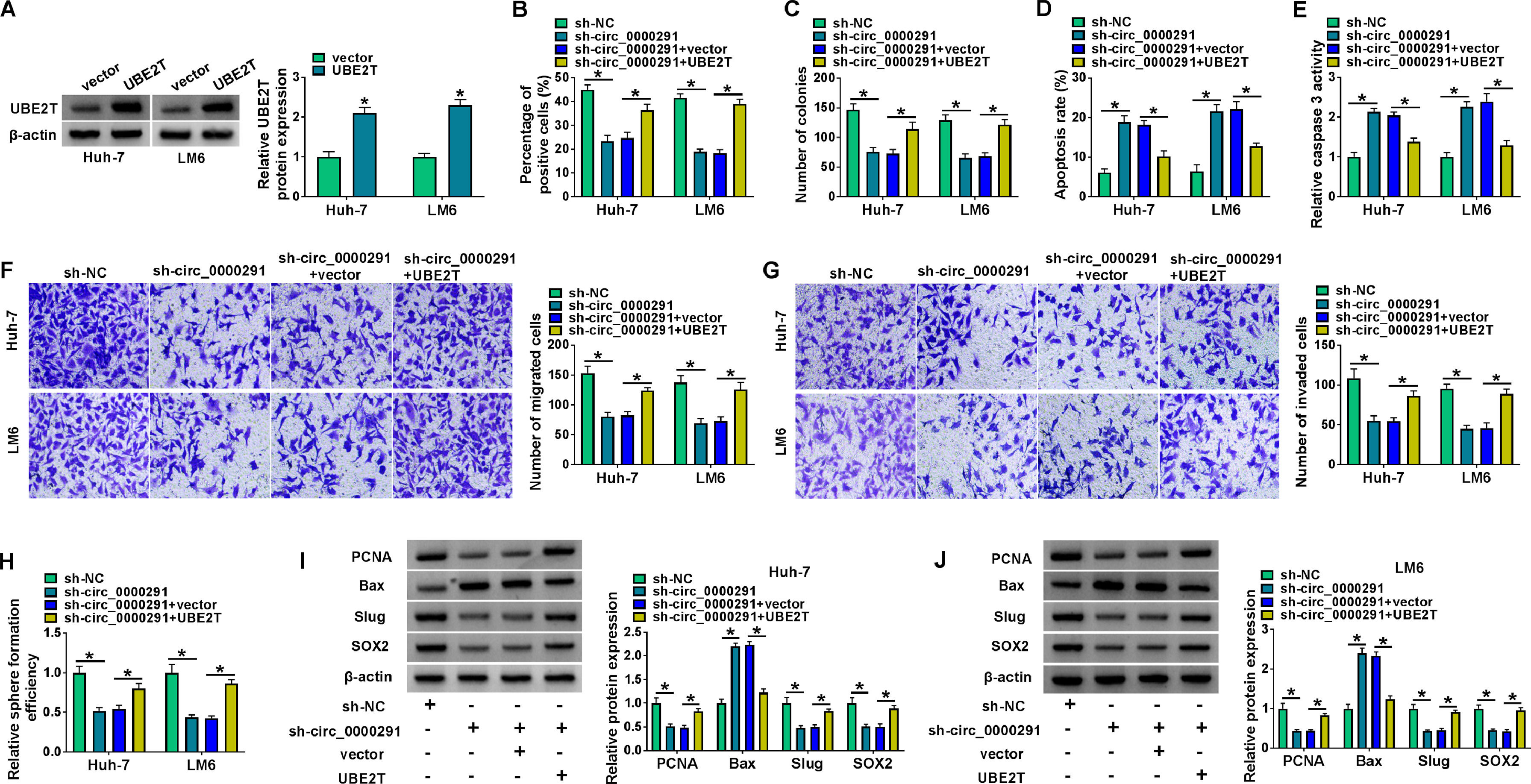
To analyze whether circ_0000291 silencing restrained HCC progression by regulating UBE2T in vitro, compensation experiments were performed. The overexpression efficiency of the UBE2T plasmid was confirmed by western blot in HCC cells (Figure 6A). We found that the addition of the UBE2T plasmid largely reversed circ_0000291 silencing-mediated alterations in cell proliferation, apoptosis, migration, invasion, and stemness (Figure 6B-6J). These data suggested that circ_0000291 silencing suppressed HCC progression largely by down-regulating UBE2T.
UBE2T overexpression largely offsets circ_0000291 knockdown-mediated anti-tumor effects in HCC cells. (A) Western blot assay was conducted to analyze the overexpression efficiency of UBE2T plasmid in HCC cells. (B-J) Huh-7 and LM6 cells were transfected with sh-circ_00002961 alone or together with UBE2T plasmid. (B and C) The proliferation capacity of transfected HCC cells was evaluated by EdU assay and colony formation assay. (D and E) The apoptosis of transfected HCC cells was assessed by flow cytometry and a commercial caspase 3 activity assay kit. (F and G) The migration and invasion abilities of transfected HCC cells were examined by transwell assays. (H) Sphere formation assay was performed to analyze the sphere formation efficiency of transfected HCC cells. (I and J) The protein expression of PCNA, Bax, Slug, and SOX2 in transfected HCC cells was examined by western blot assay. *P<0.05.
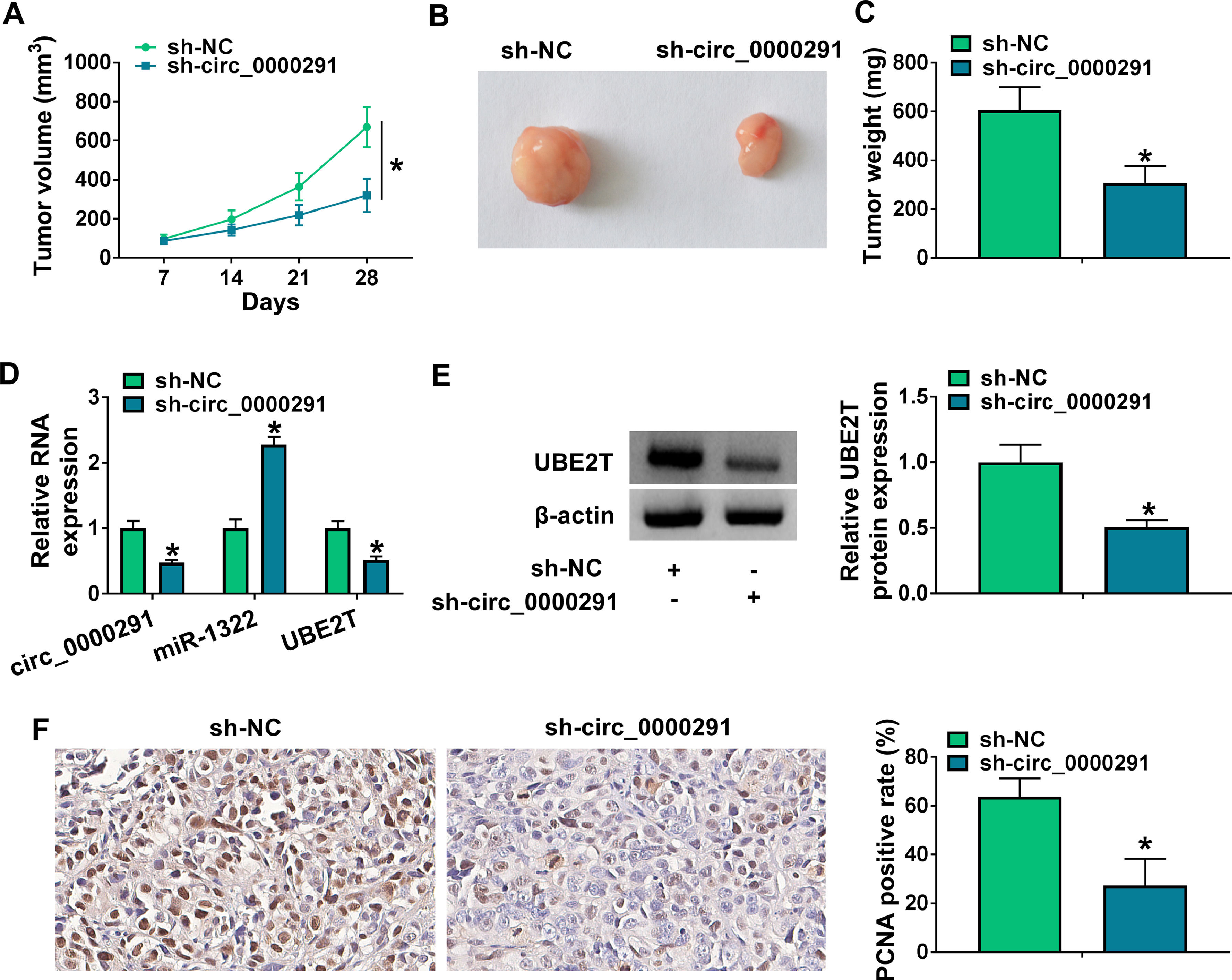
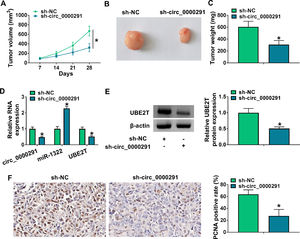
To investigate the role of circ_0000291 in tumor growth in vivo, a tumor-bearing mice model was established by subcutaneous injection of Huh-7 cells stably expressing sh-NC or sh-circ_0000291. Circ_0000291 knockdown significantly suppressed the growth of xenograft tumors compared with the sh-NC group (Figure 7A). Accordingly, the final tumor size and weight in the sh-circ_0000291 group were notably reduced compared with the sh-NC group (Figure 7B and 7C). These data suggested the pivotal role of circ_0000291 in driving tumor growth in vivo. The expression levels of circ_0000291 and UBE2T were reduced in the sh-circ_0000291 group compared with the sh-NC group, while miR-1322 exhibited the increased expression (Figure 7D and 7E). IHC assays showed that the staining intensity of the proliferation indicator PCNA was reduced by circ_0000291 knockdown in xenograft tumor tissues (Figure 7F). These data suggested that circ_0000291 might play an oncogenic role at least partly by targeting the miR-1322/UBE2T axis in vivo.
Circ_0000291 knockdown reduces the tumorigenic potential in vivo. (A) Tumor growth of mice injected with Huh-7 cells stably expressing sh-NC or sh-circ_0000291 was recorded every week. Tumor volume was calculated as length × width2 × 0.5. (B) Representative images of xenograft tumors in the sh-NC group and sh-circ_0000291 group were shown. (C) The average final tumor weight in the sh-NC group and sh-circ_0000291 group was recorded. (D) RT-qPCR was conducted to examine the expression of circ_0000291, miR-1322, and UBE2T mRNA in xenograft tumor tissues. (E) Western blot assay was conducted to determine the protein level of UBE2T in xenograft tumor tissues. (F) Immunohistochemical staining was conducted to measure the expression of PCNA in xenograft tumor tissues. *P<0.05.
HCC is a common liver malignancy worldwide [2]. Despite advances in clinical intervention strategies, the high recurrence and metastasis hinder the therapeutic effect and lead to poor outcomes [3,4]. Hence, understanding the mechanism underlying HCC tumorigenesis is urgently needed for the early diagnosis and treatment of HCC.
Increasing studies identify circRNAs as potential biomarkers for multiple malignancies [26,27]. For example, circ_0000745, circ_0044516, circ_0007534, and circ_0104824 are identified as the potential biomarkers for gastric cancer [28], prostate cancer [29], colorectal cancer [30], and breast cancer [31], respectively. As for HCC, circ_0000517 [32], circ_104075 [33], and circ_0067934 [34] have been identified as potential biomarkers. Previous studies have demonstrated that circ_0000291 can act as an oncogene in gastric cancer [18] and breast cancer [19]. However, in HCC, no studies proved whether circ_0000291 dysfunction is causally involved in its tumorigenesis. The data of the three GEO datasets (GSE97332, GSE94508, and GSE164803) showed that circ_0000291 is enhanced in HCC tissue specimens compared with adjacent healthy tissue specimens. Similar to the previous studies [35,36], we observed that circ_0000291 abundance was enhanced in human HCC. HCC patients with high circ_0000291 abundance displayed a poor survival rate, suggesting that circ_0000291 might be a novel prognostic marker for HCC patients. Through loss-of-function experiments, we found that circ_0000291 exerted an oncogenic activity in HCC cells by facilitating cell proliferation, motility, and stemness and suppressing cell apoptosis.
CircRNAs can absorb miRNAs to reduced the activity of miRNAs and up-regulate downstream gene expression [37,38]. We found that circ_0000291 could directly bind to miR-1322 and negatively regulate miR-1322 expression in HCC cells. Previous studies demonstrated that miR-1322 suppressed the progression of lung adenocarcinoma [39] and HCC [40]. In HCC, circ-HOMER1 is reported to facilitate the growth and aggressiveness of HCC cells by sponging miR-1322 [40]. Through rescue assays, we observed that miR-1322 knockdown largely reversed anti-tumor effects of circ_0000291 interference in HCC cells, suggesting that circ_0000291 silencing restrained HCC progression largely by up-regulating miR-1322.
UBE2T was further confirmed as the downstream gene of miR-1322. UBE2T is a ubiquitin-conjugating enzyme that plays a vital role in cell proliferation [41]. UBE2T has been identified as an oncogene in multiple cancers [42–44]. In HCC, UBE2T is reported to promote the radio-resistance and proliferation of HCC cells [43,45-47]. Our results showed that UBE2T expression exhibited an opposite correlation to miR-1322 level and a positive correlation to circ_0000291 expression in HCC tissues. UBE2T overexpression largely overturned the anti-tumor impacts mediated by circ_0000291 knockdown in HCC cells, indicating that circ_0000291 interference restrained HCC progression largely by down-regulating UBE2T. More importantly, we first established that circ_0000291 positively modulated UBE2T expression by absorbing miR-1322. Additionally, xenograft tumor assays demonstrated that circ_0000291 knockdown significantly suppressed the tumorigenic potential in vivo, and its function might be partly dependent on the modulation of the miR-1322/UBE2T cascade. The direct evidence about the novel regulatory network in vivo is lacking at present, which will be further elucidated in further work. Moreover, due to the limited sample size, a larger cohort of patients with HCC should be recruited to determine the diagnostic and prognostic value of circ_0000291 in HCC. Additionally, the two HCC cell lines are also limited for studying the function of circ_0000291 in HCC progression.
Based on the findings in this study, inhibition of circ_0000291 appears to represent a potential anti-HCC strategy that does not hinder the malignant behaviors of HCC cells but enhances apoptosis. We envision that the circ_0000291 shRNA may be a starting point for the development of circRNA-based molecular therapies against HCC. Future work should be to determine the long-term efficacy and safety of such agents in multiple experimental models. Although inhibition of circ_0000291 may represent a novel therapeutic opportunity for HCC, its dichotomous functions warrant careful evaluation of the potential benefit and adverse side effects when this treatment is applied at different stages of this disease. With the intense investigation of circRNA functions in cancer, we propose that circRNA-based molecular therapies have potential values for clinical use in future personalized medicine.
5ConclusionsIn summary, we proved that circ_0000291 contributed to cell proliferation, migration, invasion, and stemness and hampered cell apoptosis in HCC cells through the miR-1322/UBE2T axis. The circ_0000291/miR-1322/UBE2T regulation cascade may be a potential target for HCC therapy.
FundingThis work was supported by 2017 Zhejiang Medical Association Clinical Research Fund Project Fibrinogen Research Special Project2017ZYC-A57
Declaration of interestNone
Author contributionsFang Wang conception and design of the study, Shanshan Zhong acquisition of data, Fang Wang drafting the article or revising it critically for important intellectual content. All authors final approval of the version to be submitted.
None