Circular RNA (circRNA) has attracted extensive attention in studies related to the malignant progression of cancer, including hepatocellular carcinoma (HCC). Therefore, its molecular mechanism in HCC needs to be further explored.
Materials and methodsThe expression levels of circ_0008285, microRNA (miR)-384 and ribonucleotide reductase subunit M2 (RRM2) mRNA were detected by quantitative real-time polymerase chain reaction (qRT-PCR). Cell proliferation was analyzed using cell counting kit-8 assay and 5-ethynyl-2’-deoxyuridine assay, cell apoptosis was analyzed by flow cytometry, and cell migration and invasion were detected by transwell assay. Protein level was detected by western blot. The relationships between miR-384 and circ_0008285 or RRM2 were predicted by bioinformatics software and validated by dual luciferase reporter assay and RNA immunoprecipitation (RIP) assay.
ResultsCirc_0008285 expression is elevated to HCC tissues and cell lines. Silencing of circ_0008285 inhibited the proliferation, migration and invasion of HCC cells but accelerated cell apoptosis in vitro and impeded HCC tumorigenesis in vivo. Mechanistically, circ_0008285 directly interacted with miR-384, and miR-384 silencing attenuated the effects of circ_0008285 interference on cell proliferation, migration, invasion, and apoptosis. RRM2 was a direct target of miR-384, and RRM2 overexpression reversed the effects of miR-384 overexpression on cell proliferation, migration, invasion, and apoptosis. In addition, circ_0008285 regulated RRM2 expression by sponging miR-384.
ConclusionIn this study, circ_0008285 could promote the malignant biological behaviors of HCC cells through miR-384/RRM2 axis and has the potential to become a therapeutic target for HCC, providing a new idea for targeted therapy of HCC.
Hepatocellular carcinoma (HCC) is a malignant tumor originating from hepatocytes and hepatic bile duct cells. HCC is the most common form of primary liver cancer and is the second-leading cause of cancer-associated death [1]. Multiple risk factors for the development of HCC have been identified, including alcohol, smoking, and hepatitis virus [2]. Most HCC is detected at an advanced stage, but treatment options are lacking at this point. HCC patients at an early stage can be treated with local ablation, surgical resection, or liver transplantation [3]. Therefore, early detection and prevention of HCC development are the most influential strategy to improve patient outcomes [4]. The development of HCC is a multi-step transformational process involving many genes and epigenetics [5]. The specific molecular mechanisms regarding the occurrence and development of HCC have not been fully elucidated. Therefore, it is particularly important to explore new biomarkers and oncogenic molecules for HCC.
Among the family of non-coding RNAs, circRNA is a new RNA molecule that can exist stably on cells due to the absence of 3’-tail and 5’-cap structures, thus avoiding RNase R digestion [6, 7]. CircRNAs were found to be specifically expressed for disease stages and selectively enriched in different types of tissues [8]. A growing number of studies have signified a robust link between abnormal circRNA expression and the development and occurrence of cancers, including HCC [9–11]. CircRNA can affect gene expression mainly by sponging miRNA [12]. For example, circ_001680 can mediate the growth and migration of colorectal cancer tumors, while circ_001680 can reinforce the cancer stem cell (CSC) population in colorectal cancer by regulating miR-340/BMI1 axis [13]. Circ_0067934 targeted miR-545-3p/SLC7A11 axis to weaken ferroptosis in thyroid cancer [14]. Circ_0014130 acted as a sponge for miR-132-3p and promoted tumorigenesis and metastasis in bladder cancer by regulating the expression of KCNJ12 [15]. Circ_0008285 is derived from the CDYL gene and located at chr6:4891946-4892613, with a length of 667 bp. Circ_0008285 was significantly enhanced in cervical cancer and promoted cervical cancer progression by acting as a ceRNA for miR-211-5p to release SOX4 expression [16]. In addition, circ_0008285 is upregulated in HCC and is involved in apoptosis, proliferation and stemness of HCC cells [17]. However, the molecular mechanism of circ_0008285 in hepatocellular carcinogenesis was still not fully understood.
Our study revealed that circ_0008285 silencing hindered the proliferation, migration and invasion but triggered apoptosis in HCC cells. Furthermore, circ_0008285affected the development of HCC through miR-384/ribonucleotide reductase subunit M2 (RRM2) axis.
2Materials and methods2.1SubjectsCancer tissues and paired paracancerous tissues (>2 cm from the edge of cancerous tissues) of 55 HCC patients were surgically resected from HCC specimens and collected at The Affiliated Zhuzhou Hospital Xiangya Medical College CSU from 2019 to 2020. All tissues were frozen in liquid nitrogen urgently after resection and transferred to a -80°C refrigerator for storage. All patients were not treated with antitumor therapy such as radiotherapy, chemotherapy, or targeted drug therapy before surgery, and HCC tissues were postoperative histopathology confirmed.
2.2Cell culture and transfectionNormal hepatocyte cell lines (THLE-3) and HCC cell lines (Huh 7 and HCCLM3) were purchased from the ATCC (Manassas, VA, USA) and cultured in DMEM medium (including 10% fetal bovine serum (FBS) + 100 U/ml penicillin + 100 μg/ ml streptomycin) in an incubator with 5% CO2, 95 % relative humidity at 37°C.
Small interfering RNA targeting circ_0008285 (si-circ_0008285), miR-384 mimics (miR-384), miR-384 inhibitors (anti-miR-384), overexpression plasmids of RRM2 (pcDNA-RRM2) and negative controls (si-NC, miR-NC, anti-NC and pcDNA) were provided by GenePharma (Shanghai, China). Huh7 and HCCLM3 cells were inoculated in 6-well plates (5 × 105/well) and grown to 50% confluence, and the cultured cells were transfected using Lipofectamine™ 6000 (Beyotime, Shanghai, China).
2.3RNA extraction and qRT-PCRRNA was extracted from total cells and tissues using the Trizol (Beyotime) reagent. The extracted total RNA was reversely transcribed into cDNA according to the instruction manual of miScript RT Kit (TaKaRa, Dalian, China). cDNA was used as a template for qRT-PCR by using SYBR Premix Ex Taq II (TaKaRa). U6 (for miRNA or nucleus) or GAPDH (for circRNA, mRNA, or cytoplasm) was used as an internal control. Relative RNA levels were calculated by using 2−ΔΔCT method. The specific primer sequences are shown in Table 1.
Primers sequences used for PCR.
Cytoplasmic and nuclear RNA from HCC cells were isolated according to the instructions of the Cytoplasmic and Nuclear RNA Purification Kit (Beyotime), and the expression of circ_0008285 was detected by qRT-PCR.
2.5Cell counting kit-8 (CCK-8) assayThe transfected cells were planted on 96-well plates. 10 μL CCK-8 solution (Beyotime) was added after 0 h, 24 h, 48 h and 72 h of incubation, and the optical density values at 450 nm were tested by a microplate reader.
2.65-ethynyl-2’-deoxyuridine (Edu) stainingEdU Staining Proliferation Kit (Beyotime) was employed for the analysis of cell proliferation. The transfected cells were inoculated into 96-well plates, and each well was incubated with 50 μmol/L Edu solution for 2 h. The nuclear was then incubated with DAPI reaction solution for 30 min. The cells were observed under a fluorescence microscope.
2.7Cell apoptosis assayAnnexin V-FITC/PI apoptosis detection kit (Solarbio, Beijing, China) was utilized to analyze cell apoptosis. The cells were resuspended with Annexin V binding buffer and incubated with Annexin V-FITC and PI solution in the dark. Cell apoptosis was examined by a flow cytometer.
2.8Transwell assayTranswell chambers (Corning, Tewksbury, MA, USA) that precoated with (for invasion assay) or without (for migration assay) Matrigel matrix (Corning) were employed to detect cell migration and invasion. In brief, cells in non-serum medium were inoculated into the upper chamber of the transwell, and 10 % fetal bovine serum was added to the lower chamber and incubated for 24 h. Cells that migrated or invaded through the membranes were stained with 0.1% crystal violet solution for 30 minutes, and the results were analyzed by counting the number of migrated or invaded cells under the microscope.
2.9Western blottingTissues and cells were fully lysed using RIPA lysis solution (Beyotime) and total proteins were extracted. The total proteins were subjected to SDS-PAGE gel electrophoresis, and then the separated proteins were electro-transferred onto the PVDF membrane (Millipore, Billerica, MA, USA), and the membrane was blocked with 10% skim milk powder for 2 h. The membranes were incubated with primary antibodies including: anti-Bax (ab53154, 1:1000), anti-Bcl-2 (ab196495, 1:1000), anti-E-cadherin (ab133597, 1:2000), anti-Vimentin (ab137321, 1:2000), anti-N-cadherin (ab211126, 1:2000), anti-RRM2 (ab154964, 1:1000) and anti-β-actin (ab8227, 1:2000), overnight at 4°C. Then, the membranes were incubated with Goat Anti-Rabbit IgG H&L (HRP) second antibody (ab205718, 1:10000). Finally, the membranes were developed using ECL reagent (Millipore) and analyzed semi-quantitatively using ImageJ software. The antibodies used in this experiment were obtained from Abcam (Cambridge, MA, USA).
2.10Luciferase reporter assayBioinformatics was employed for the prediction of binding targets of circ_0008285 and RRM2. The wild-type (WT) sequences of circ_0008285 or RRM2 3′UTR containing miR-384 binding sequences were cloned into the pmirGLO vector (Promega, Madison, WI, USA), respectively, and the recombinant vectors were named circ_0008285-WT, RRM2 3′UTR-WT, circ_0008285-MUT and RRM2 3′UTR-MUT. The constructed plasmids and miR-384 mimics or miR-NC were co-transfected into HCC cells. The luciferase activity was measured at 48 h after transfection.
2.11RNA immunoprecipitation (RIP) assayMagna RNA immunoprecipitation kit (Millipore) was employed for RIP assay. HCC cells were lysed in RIP lysis buffer; then, cell lysates were incubated with magnetic beads labeled with anti-IgG (ab190475, Abcam) and anti-Ago2 (ab186733, Abcam). Then, qRT-PCR was applied to test the relative expression of circ_0008285, miR-384 and RRM2 in the precipitates.
2.12Xenograft mice modelShort hairpin RNA targeting circ_0008285 (sh-circ_0008285) or negative control (sh-NC) were packaged by lentivirus packaging to construct a stable transduced Huh7 cell line. The transfected Huh7 cells were adjusted to a concentration of 5 × 106/mL, inoculated into the right side of the back side of nude mice and then divided into 2 groups (6 nude mice for each group). The mice were placed in a specific pathogen-free room, and the general condition, time of graft tumor formation and size of the nude mice were recorded and observed. The longest diameter (L) and shortest diameter (W) of the xenograft tumors were measured every 7 days for 35 days with a vernier caliper, and their tumor volume (V) was calculated by using the formula: V = (L × W2)/2. The growth curve of the transplanted tumor was generated. 35 days later, the nude mice were killed, the tumor tissues were removed, tumor weights were measured, and subsequent experiments were performed. All animal experiments were approved by the Animal Care and Use Committee of The Affiliated Zhuzhou Hospital Xiangya Medical College CSU NO. ZX19617 and complied with the guidelines of the National Institutes of Health
2.13Statistical analysisThe experimental data of 3 replicates were statistically analyzed and statistical images were plotted using GraphPad Prism 7.0 software. The measurement data were presented as mean ±standard deviation. The linear relationship between the expression levels of circ_0008285, miR-384 and RRM2 in cancer tissues was assessed by Spearman's correlation test. Student t-test or one-way analysis of variance (ANOVA) was used to compare the differences between the groups. Statistical significance was deemed when P<0.05.
2.14Ethical statementsWritten informed consent was obtained from all participants and this study was permitted by the Ethics Committee of The Affiliated Zhuzhou Hospital Xiangya Medical College (CSU. NO. ZX19617).
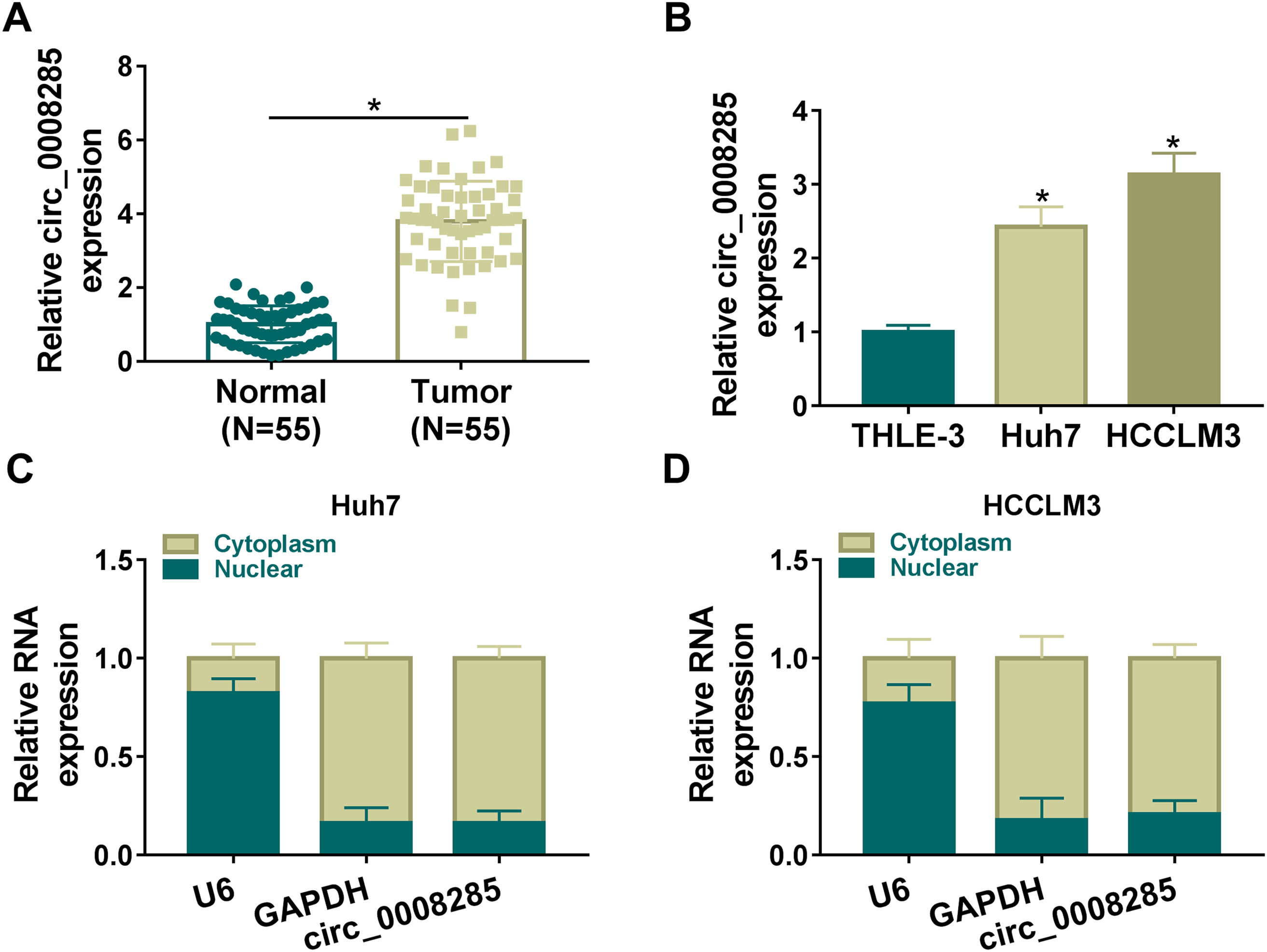
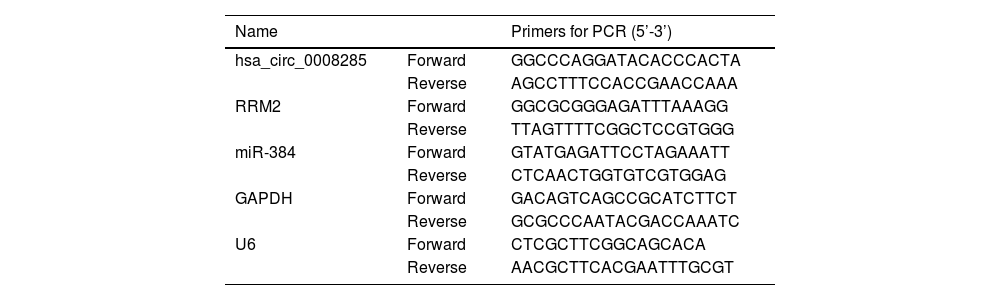
3Results3.1Circ_0008285 was highly expressed in the tissues and cells of HCCFirst, 55 pairs of HCC tissues and non-cancerous tissues were analyzed by qRT-PCR analysis. Results showed that circ_0008285 expression was significantly increased in HCC tissues compared to paraneoplastic tissues (Fig. 1A). Similarly, circ_0008285 expression was significantly upregulated in HCC cells (Huh7 and HCCLM3) compared to normal hepatocytes (THLE-3) (Fig. 1B). Cytoplasmic and nuclear RNA analysis showed that circ_00008285 was mainly presented in the cytoplasm (Fig. 1C-D). These results suggested that the aberrant expression of circ_0008285 in HCC tissues and cell lines may be closely associated with the progression of HCC.
Circ_0008285 was highly expressed in HCC tissues and cells
(A) qRT-PCR analysis of the expression level of circ_0008285 in the tissues of 55 patients with HCC and paracancerous tissues. (B) qRT-PCR assay was employed to detect the expression level of circ_0008285 in HCC cell lines (Huh7 and HCCLM3) and non-cancerous cell lines (THLE-3). (C-D) Subcellular fractionation location assay was used to analyze the distribution of circ_0008285 in the nucleus or cytoplasm. *P < 0.05.
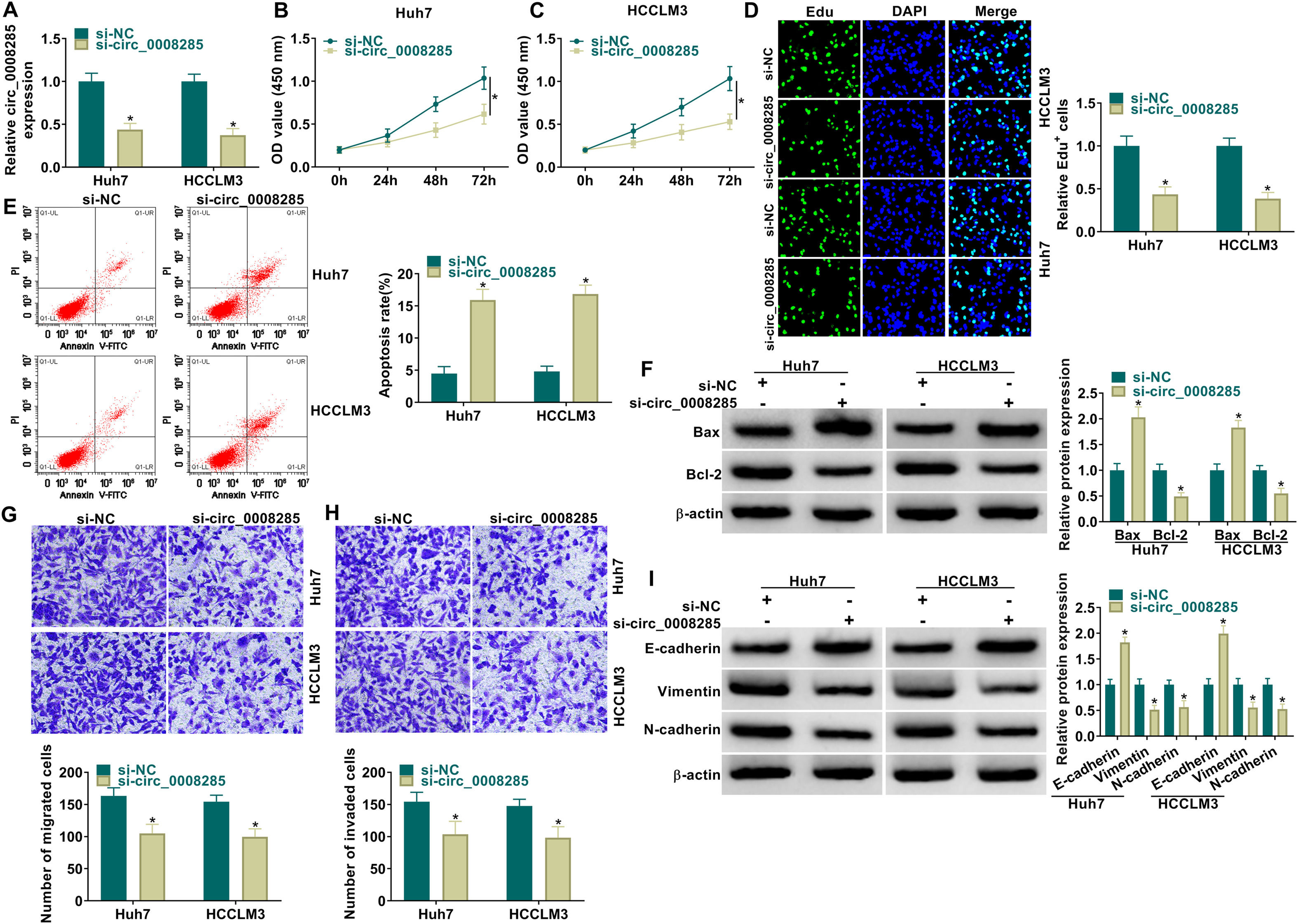
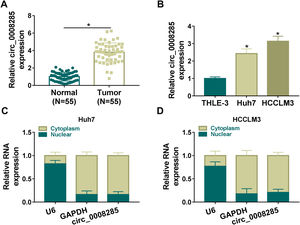
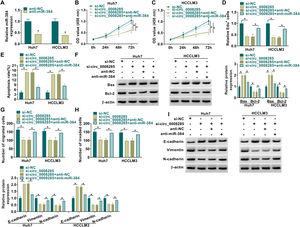
Next, to further explore the biological function of circ_0008285 in HCC, the expression of circ_0008285 was silenced in Huh7 and HCCLM3 cells. First, the cells were transfected with si-NC (as control) or si-circ_0008285. qRT-PCR was applied to analyze the transfection efficiency, and the results showed that transfection of si-circ_0008285 significantly decreased the expression of circ_0008285 in the cells (Fig. 2A). CCK8 and Edu assays showed that circ_0008285 silencing significantly inhibited the proliferative ability of cells (Fig. 2B-D). Flow cytometry analysis showed that knockdown of circ_0008285 significantly increased the apoptosis rate of Huh7 and HCCLM3 cells (Fig. 2E). The expression levels of pro-apoptotic protein Bax and anti-apoptotic protein Bcl-2 in HCC cells were detected by Western blot. In HCC cells with silencing of circ_0008285, the expression level of Bax was significantly upregulated and Bcl-2 expression level was significantly down-regulated in cells, clarifying that circ_0008285 interference promoted apoptosis in HCC cells (Fig. 2F). In addition, knockdown of circ_0008285 significantly inhibited cell migration and invasion (Fig. 2G-H). Epithelial-mesenchymal transition (EMT), which represents the metastatic tumor process, was examined by Western blot. The results showed that silencing of circ_0008285 significantly increased E-cadherin protein levels and decreased Vimentin and N-cadherin protein levels (Fig. 2I). In summary, we concluded that circ_0008285 silencing inhibited the proliferation, migration, invasion and EMT, but promoted apoptosis in HCC cells.
Circ_0008285 knockdown impeded cell proliferation, migration and invasion, while induced cell apoptosis in HCC cells
Huh7 and HCCLM3 cells were transfected with si-circ_0008285 or si-NC. (A) qRT-PCR was performed to detect the expression of circ_0008285 in Huh7 and HCCLM3 cells. (B-D) CCK-8 and EdU assays were employed to detect the proliferation ability of transfected cells. (Magnification, 200 ×). (E) The Annexin-V FITC/PI staining was used to assess cell apoptotic rate. (F) Western blotting assay was used to assess the protein levels of Bax and Bcl-2 in transfected cells. (G-H) Transwell assay was used to determine cell migration and invasion abilities. (Magnification, 100 ×). (I) Western blotting assay was employed to assess the protein levels of E-cadherin, Vimentin and N-cadherin in transfected cells. *P < 0.05.
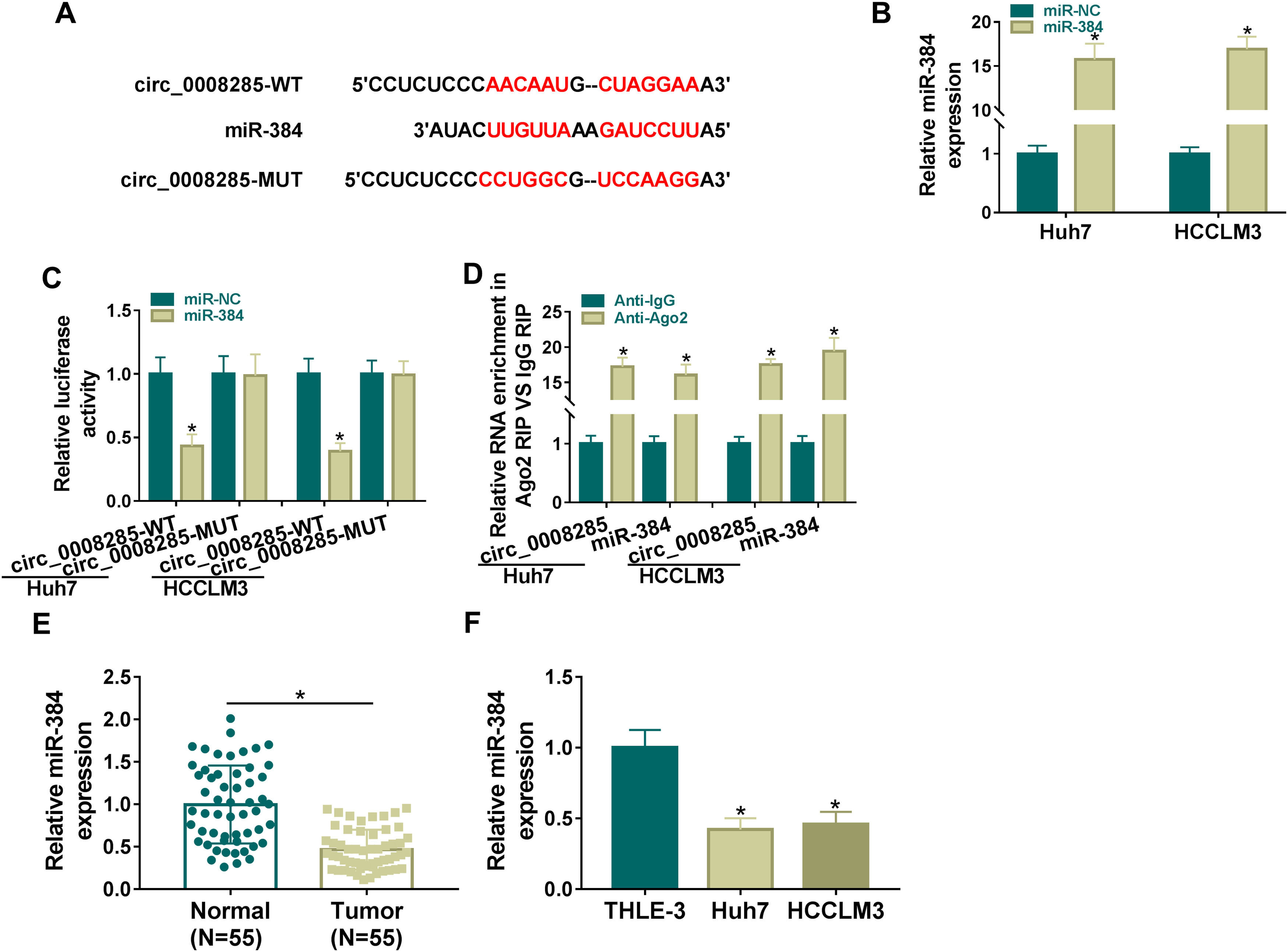
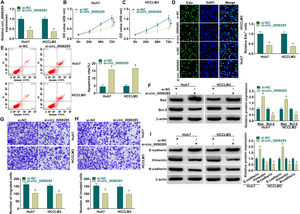
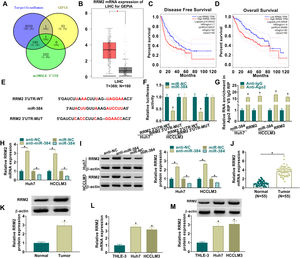
To further explore the regulatory mechanism of circ_0008285 in HCC, bioinformatics software was used to predict the potential miRNAs interacted with circ_0008285. Circinteractome database predicted the existence of complementary binding sites between circ_0008285 and miR-384 (Fig. 3A). To confirm the targeting relationship between circ_0008285 and miR-384, the overexpression efficiency of miR-384 mimics was first examined. qRT-PCR results showed that miR-384 expression was dramatically increased in HCC cells after transfection with miR-384 mimics (Fig. 3B). Subsequently, dual luciferase reporter assay showed that miR-384 overexpression significantly inhibited the luciferase activity of circ_0008285-WT group, while it had no significant effect on the luciferase activity of circ_0008285-MUT group (Fig. 3C). RIP analysis showed that compared with the anti-IgG group, circ_0008285 and miR-384 were significantly enriched in the immunoprecipitation of the anti-Ago2 group (Fig. 3D). It was found that miR-384 expression was significantly downregulated in HCC tissues and cell lines compared with adjacent normal tissues and normal hepatocytes (Fig. 3E-F). The above results suggested that miR-384 was targeted by circ_0008285.
Circ_0008285 directly targeted miR-384
(A) The wild type or mutant type complementary binding sites between circ_0008285 and miR-384 were presented. (B) qRT-PCR was performed to detect the expression of miR-384 in Huh7 and HCCLM3 cells transfected with miR-NC or miR-384 mimics. (C) Dual luciferase reporter assay was used to verify the binding relationship between circ_0008285 and miR-384. (D) RIP assays were used to detect the enrichments of circ_0008285 and miR-384 in anti-Ago2 groups. (E) qRT-PCR was used to detect miR-384 level in tumor tissues and paracancerous tissues. (F) qRT-PCR assay was used to detect the expression level of miR-384 in THLE-3, Huh7 and HCCLM3 cells. *P < 0.05.
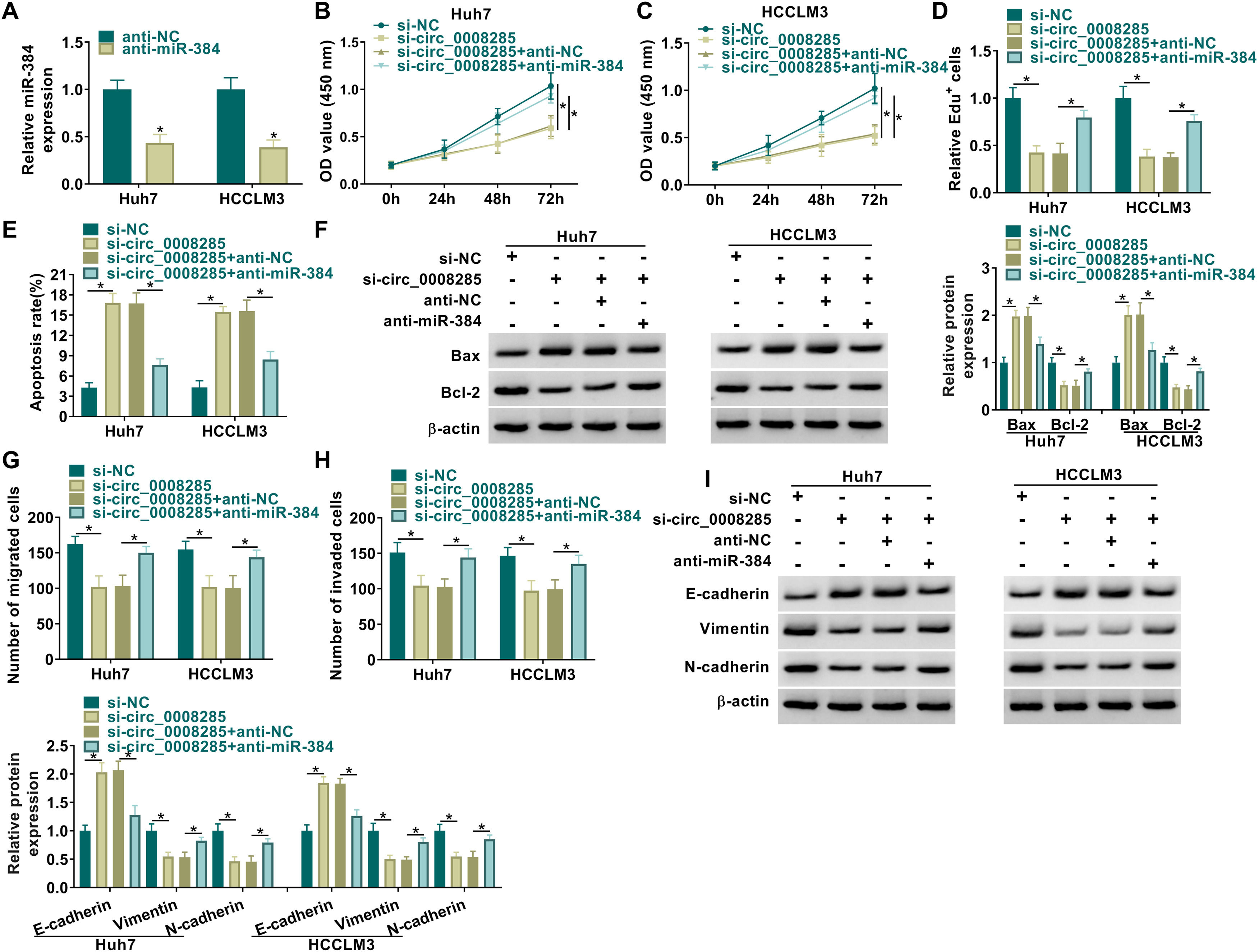
CircRNAs are extremely rich in binding sites for miRNAs and therefore act like sponges by absorbing miRNAs [18]. The relationship between circ_0008285 and miR-384 has been demonstrated; however, the biological behavior of HCC that they regulate still needs to be determined. We transfected si-NC, si-circ_0008285, si-circ_0008285+anti-NC, or si-circ_0008285+anti-miR-384 in Huh7 and HCCLM3 cells and then performed a series of rescue experiments. First, the effectiveness of miR-384 inhibitor was verified by qRT-PCR (Fig. 4A). CCK8 and Edu assays showed that miR-384 inhibitor partially attenuated the inhibition effect of circ_0008285 interference on cell proliferation (Fig. 4B-D). In addition, miR-384 knockdown attenuated promoting effect of circ_0008285 downregulation on cell apoptosis (Fig. 4E). Western blot results showed that miR-384 deletion reversed the effect of circ_0008285 silencing on Bax and Bcl-2 protein levels (Fig. 4F). Transwell assay results showed that silencing of circ_0008285 resulted in a significant inhibition in cell migration and invasion, while the addition of anti-miR-384 resulted in a significant increase in cell migration and invasion (Fig. 4G-H). Downregulation of miR-384 reversed the effects of si-circ_0008285 on E-cadherin, Vimentin and N-cadherin levels (Fig. 4I). Overall, circ_0008285 silencing suppressed the malignant phenotype of HCC cells by targeting miR-384.
MiR-384 downregulation attenuated the influence of circ_0008285 interference on cell proliferation, migration, invasion, and apoptosis in HCC cells
(A) The silencing efficiency of miR-384 inhibitor was detected by qRT-PCR. (B-I) Huh7 and HCCLM3 cells were transfected with si-NC, si-circ_0008285, si-circ_0008285+anti-NC or si-circ_0008285+anti-miR-384. (B-D) CCK-8 and EdU assays were employed to detect cell proliferation ability. (E) The Annexin-V FITC/PI staining was used to assess apoptotic rates. (F) Western blotting assay was utilized to assess the protein levels of Bax and Bcl-2. (G-H) Transwell assays were used to determine cell migration and invasion ability. (I) Western blotting was used to assess the protein levels of E-cadherin, Vimentin and N-cadherin. *P < 0.05.
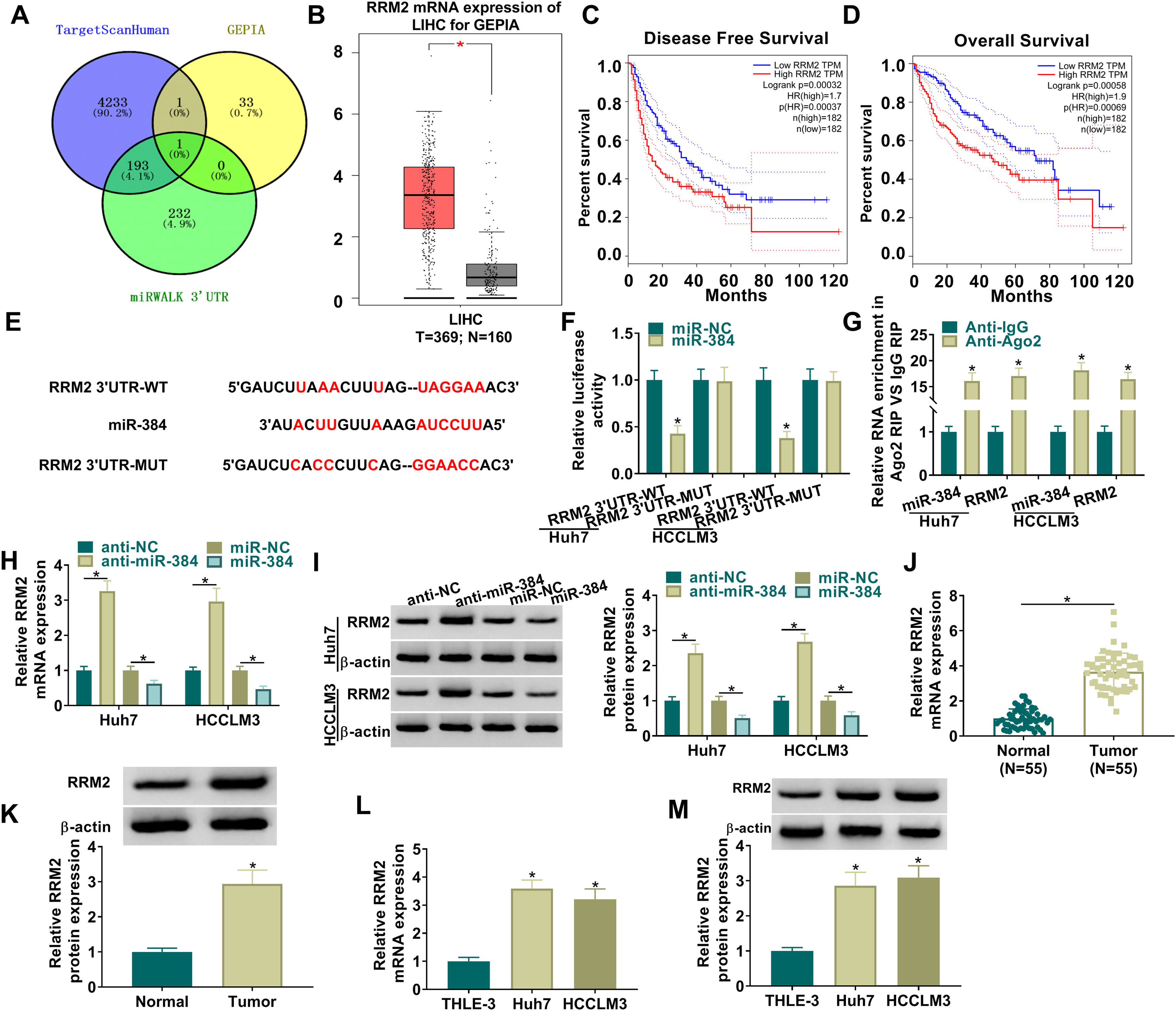
To further clarify the mechanism of miR-384, the target genes of miR-384 were predicted by online bioinformatics databases. We predicted the target gene RRM2 by TargetScanHuman, miRWALK and GEPIA simultaneously (Fig. 5A). RRM2 was found to be highly expressed in HCC tissues in the GEPIA database (Fig. 5B). Besides, according to the disease-free survival and overall survival data of the GEPIA database, we found that high RRM2 expression is associated with poorer disease-free survival and overall survival (Fig. 5C-D). MiR-384 and RRM2 were shown to contain complementary binding sites in the TargetScanHuman online database (Fig. 5E). Dual luciferase reporter assay showed that miR-384 overexpression significantly inhibited the luciferase activity of RRM2 3’UTR-WT group, while it had no significant effect on the luciferase activity of RRM2 3’UTR-MUT group (Fig. 5F). It has been widely known that miRNAs modulate the expression of target gene by binding to Ago2, the key component of RNA-induced silencing complex (RISC). Then RIP analysis was conducted, and the results showed that compared to the anti-IgG group, miR-384 and RRM2 were significantly enriched in the anti-Ago2 immunoprecipitation group (Fig. 5G). In addition, RRM2 mRNA and protein levels were significantly elevated by miR-384 inhibitor and were significantly down-regulated by miR-384 mimics (Fig. 5H-I). As expected, RRM2 mRNA and protein levels were significantly upregulated in HCC tissues and cell lines compared to adjacent normal tissues and normal hepatocytes (Fig. 5J-M). These results suggested that miR-384 could target RRM2.
MiR-384 targeted RRM2
(A) The binding sites between miR-384 and RRM2 were predicted by online prediction tools (TargetScanHuman, miRWALK, and GEPIA). (B) Expression of RRM2 in HCC tissues was analyzed according to the GEPIA database. (C-D) The overall survival and disease-free survival analysis of RRM2 gene of interest can be presented. (F) Dual luciferase reporter assay to verify the binding relationship between miR-384 and RRM2. (G) RIP assays were used to detect the enrichment of miR-384 and RRM2 in anti-Ago2 groups. (H-I) qRT-PCR and Western blot analysis were performed to determine the effects of miR-384 mimics and inhibitors on RRM2 mRNA and protein levels. (J-K) qRT-PCR and Western blot was used to detect RRM2 mRNA and protein levels in tumor tissues and paracancerous tissues. (L-M) qRT-PCR and Western blot analysis was performed to detect RRM2 mRNA and protein levels in THLE-3, Huh7 and HCCLM3 cells. *P < 0.05.
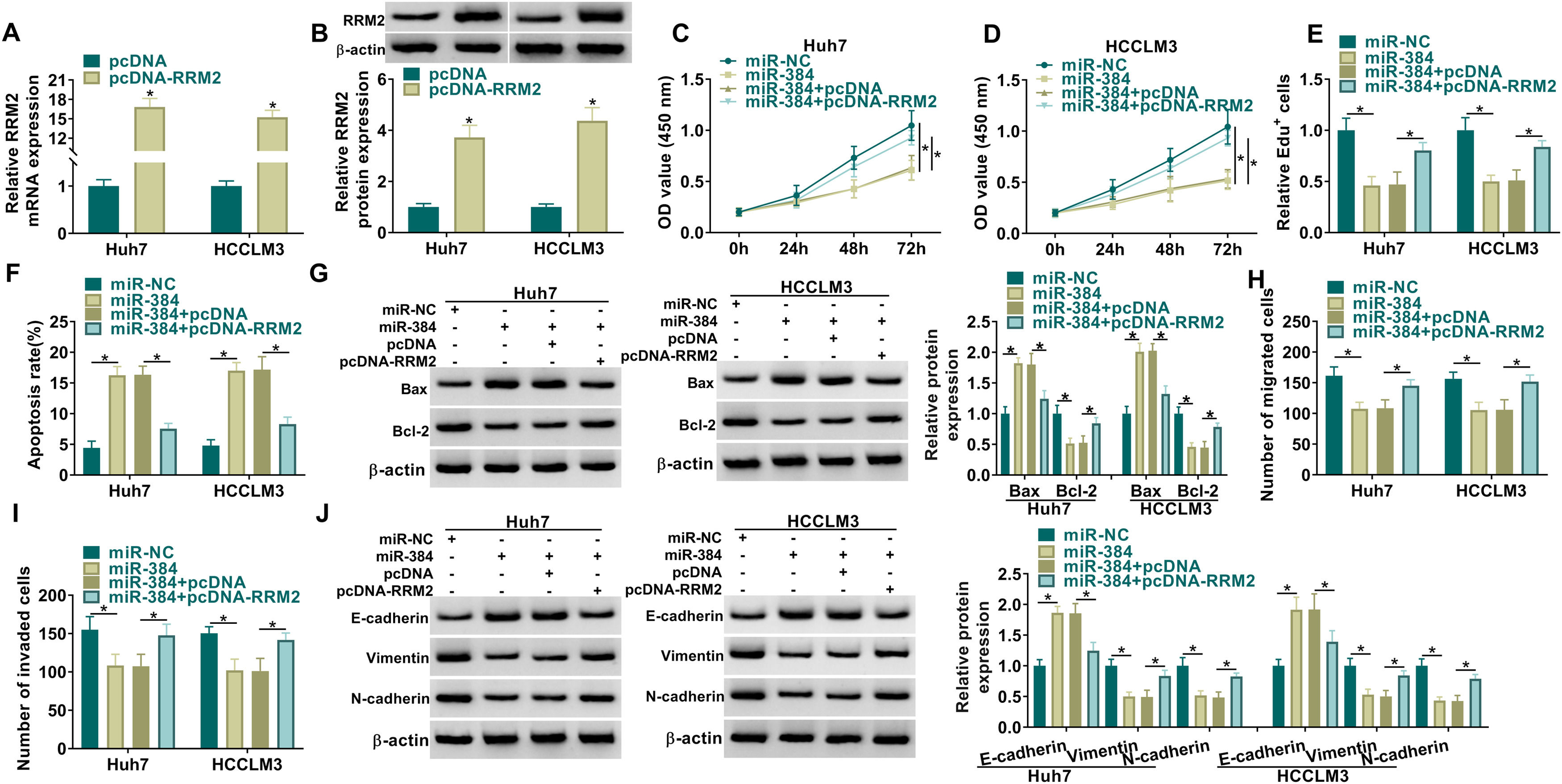
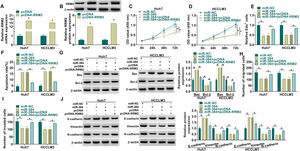
MiRNAs could regulate the expression of their target genes through post-transcriptional regulation [18, 19]. To determine whether RRM2 is involved in miR-384-mediated biological functions, we transfected miR-NC, miR-384, miR-384+pcDNA, or miR-384+pcDNA-RRM2 in Huh7 and HCCLM3 cells, and then performed a series of rescue experiments. First, qRT-PCR and Western blot results showed that RRM2 mRNA and protein levels were significantly elevated in the pcDNA-RRM2 group compared to the pcDNA group (Fig. 6A-B). In addition, overexpression of miR-384 significantly inhibited cell proliferation and promoted apoptosis, which were reversed by overexpression of RRM2 (Fig. 6B-F). Overexpression of miR-384 expression elevated the expression level of Bax and decreased the expression level of Bcl-2, which could be restored by RRM2 overexpression (Fig. 6G). Cell migration and invasion were inhibited by miR-384 mimics, and RRM2 overexpression impeded these effects (Fig. 6H-I). Furthermore, overexpression of RRM2 reversed the effects of miR-384 mimics on E-cadherin, Vimentin and N-cadherin protein levels (Fig. 6J). Taken together, miR-384 overexpression inhibited the malignant behaviors of HCC cells by targeting RRM2.
RRM2 overexpression impaired the effect of miR-384 overexpression on proliferation, migration, invasion, and apoptosis of HCC cells
(A-B) qRT-PCR and Western blot analysis were used to detect the overexpression efficiency of RRM2. (C-J) Huh7 and HCCLM3 cells were transfected with miR-NC, miR-384, miR-384+pcDNA, or miR-384+pcDNA-RRM2. (C-E) CCK-8 and EdU assays were utilized to detect cell proliferation ability. (F) The Annexin-V FITC/PI staining was used to assess apoptotic rates. (G) Western blotting assay was employed to assess the protein levels of Bax and Bcl-2. (H-I) Transwell assays were used to determine cell migration and invasion ability. (J) Western blotting to assess the protein levels of E-cadherin, Vimentin and N-cadherin. *P < 0.05.
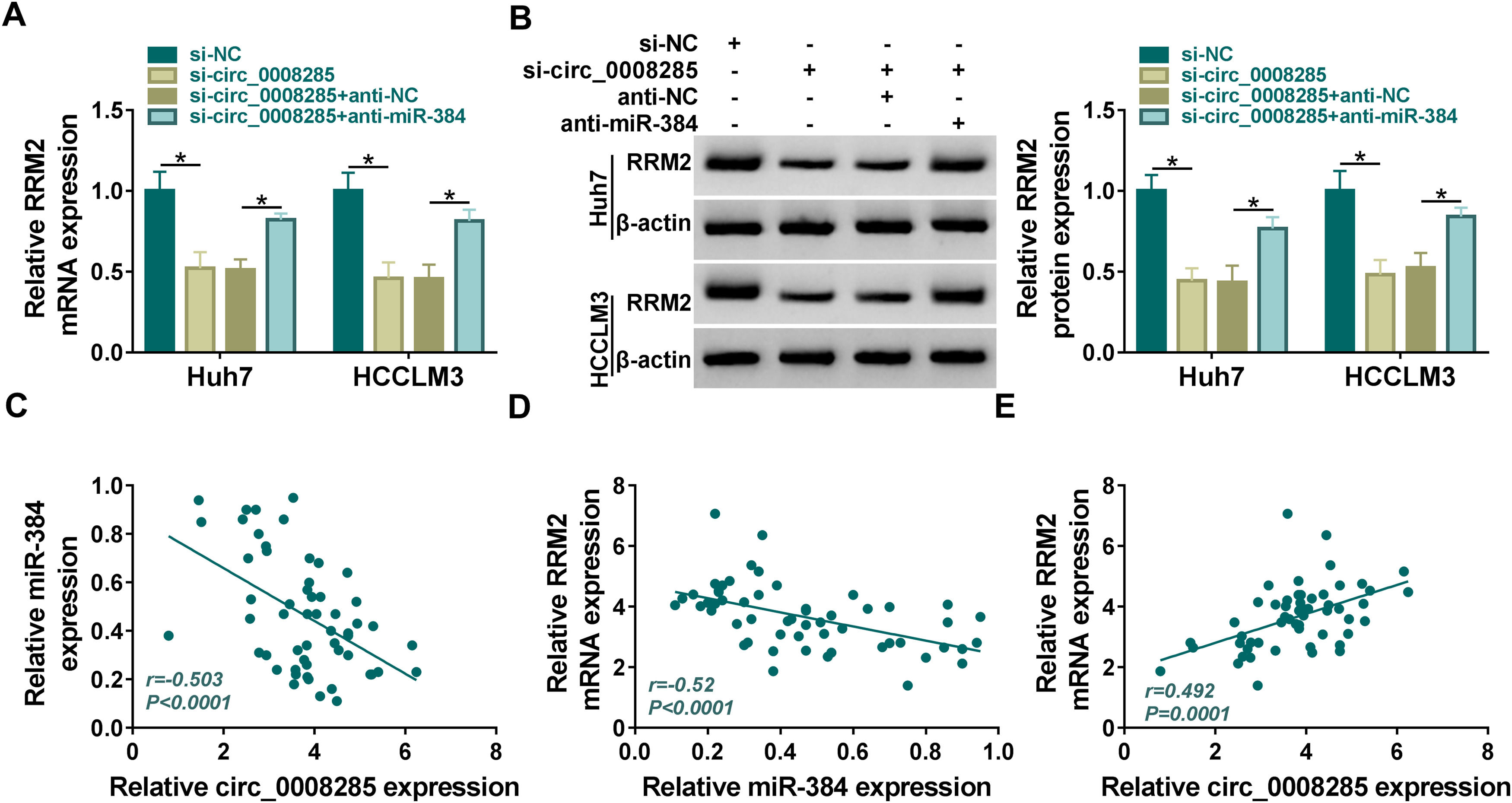
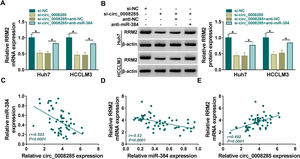
Next, we further explored whether circ_0008285 could act as a miR-384 sponge to regulate RRM2 expression. The results showed that in Huh7 and HCCLM3 cells, circ_0008285 silencing caused a great reduction in RRM2 mRNA and protein levels, and miR-384 inhibitor attenuated these effects (Fig. 7A-B). Furthermore, in HCC tissues, miR-384 expression was negatively correlated with circ_0008285 and RRM2 expression levels, while circ_0008285 expression was positively correlated with RRM2 expression (Fig. 7C-E). These results implied that circ_0008285 can regulate the expression of RRM2 by sponging miR-384.
Circ_0008285 regulated RRM2 expression by targeting miR-384
(A-C) Huh7 and HCCLM3 cells were transfected with si-NC, si-circ_0008285, si-circ_0008285+anti-NC or si-circ_0008285+anti-miR-384. And RRM2 mRNA and protein levels were detected by qRT-PCR and Western blot analysis. (C-E) Spearman correlation analysis for the correlation among circ_0008285, miR-384, and RRM2 mRNA in HCC tissues. *P < 0.05.
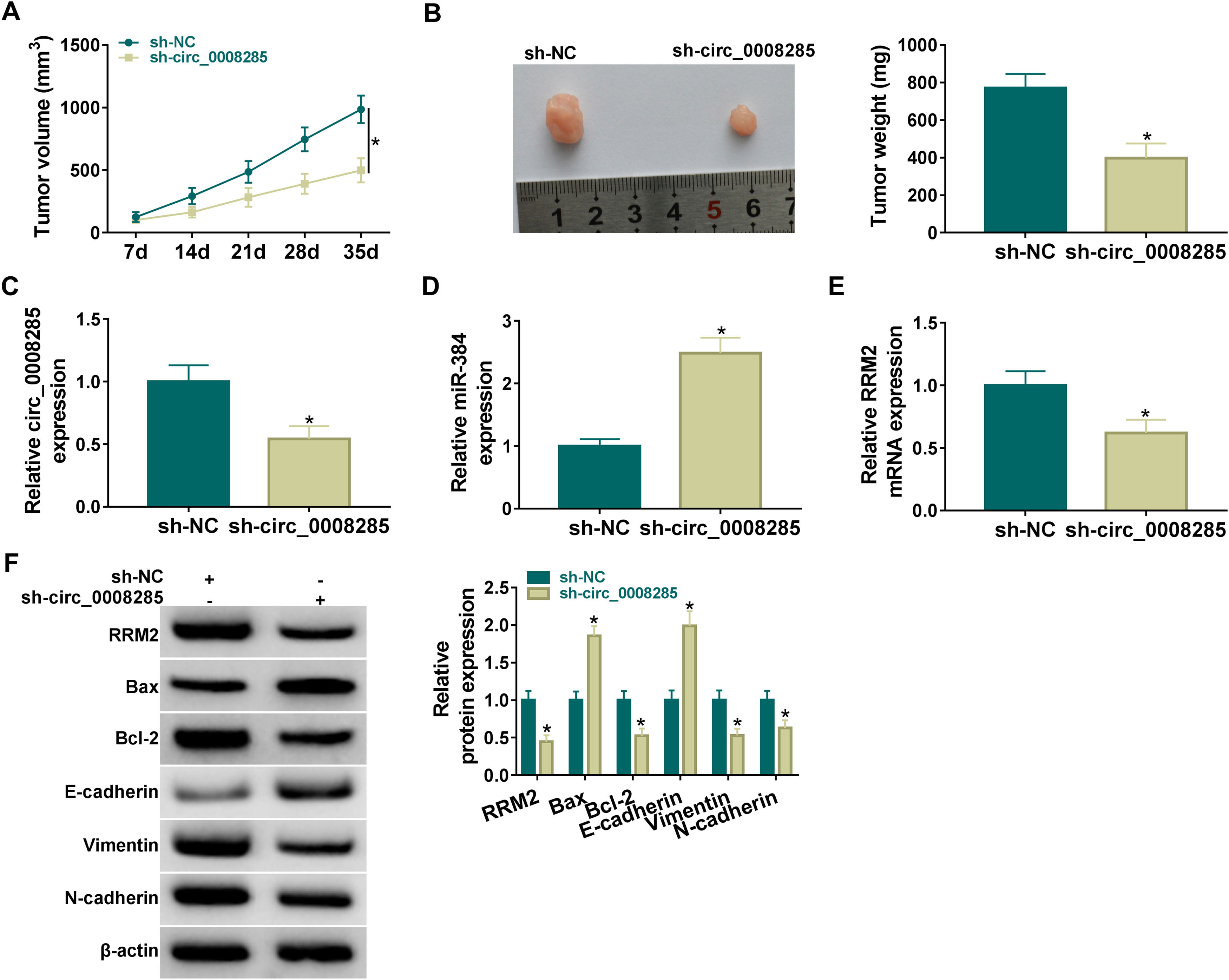
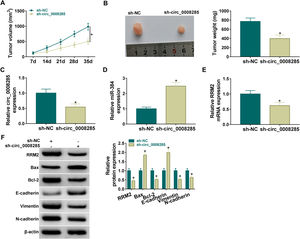
Finally, the effect of circ_0008285 silencing on tumor growth was investigated by in vivo tumorigenesis assay. The results showed that tumor volume and weight were suppressed in the sh-circ_0008285 group compared to the sh-NC group (Fig. 8A-B). qRT-PCR results showed that the levels of circ_0008285 and RRM2 mRNA were significantly reduced, and miR-140-3p level was significantly increased in xenograft tumor tissues with circ_0008285 knockdown (Fig. 8C-E). In addition, silencing of circ_0008285 significantly suppressed the expression levels of RRM2, Bcl-2, Vimentin and N-cadherin but elevated the expression levels of Bax and E-cadherin in xenograft tumor tissues (Fig. 8F). Altogether, circ_008285 silence inhibited tumor growth by miR-384/RRM2 axis.
Silencing of hsa_circ_0008285 inhibited tumor growth in vivo
(A-B) Effect of circ_0008285 silencing on tumor volume and weight. (C-E) The effects of circ_0008285 silencing on circ_0008285, miR-384 and RRM2 mRNA expression levels in xenograft tumors were examined by qRT-PCR. (F) The protein levels of RRM2, Bax, Bcl-2, E-cadherin, vimentin, and N-cadherin were evaluated by western blot in xenograft tumors. *P < 0.05.
In the past few years, an accumulating body of research has established that circRNAs can be used as diagnostic or predictive biomarkers for certain diseases, particularly cancer. CircRNAs are aberrantly expressed in cancer and play an integral oncogenic or anti-oncogene effects in the progression of tumors [20]. In addition, circRNAs can significantly regulate the biological phenotype of cells, which might be new tumor markers [21, 22]. For example, a low expression level of circ_0035445 in gastric cancer was associated with distant metastasis and tumor size; functionally, upregulation of circ_0035445 blocked cell migration, boosted apoptosis, and suppressed cell cycle progression [23]. In colorectal cancer, circ_001659 might be a useful serum biomarker for the detection and prognosis of colorectal cancer; in vitro experiments showed that circ_001659 could expedite cell invasion and migration [24]. In addition, circ_001569 boosted cell proliferation, migration and invasion of pancreatic cancer but curbed apoptosis [25]. A previous study has shown that circ-CDYL (circ_0008285) promoted the tumor-initiating properties and chemoresistance of HCC cells [17]. However, large-scale identifications of circ_0008285 expression in HCC tumorigenesis were not yet reported. In this study, we demonstrated that circ_0008285 was abnormally unregulated in HCC, and silencing of circ_0008285 significantly constrained proliferation, migration, invasion and EMT but impelled apoptosis in HCC cells. In vivo studies showed that silencing of circ_0008285 inhibited tumor growth in vivo. These studies revealed the oncogenic role of circ_0008285 in HCC, suggesting that it could be an important marker for the therapy of HCC.
Studies show that miRNAs are engaging in a wide range of biological processes and have a significant correlation between cancer diagnosis and prognosis [26]. MiRNAs can directly bind to downstream target genes to induce mRNA degradation or inhibit translation, thereby participating in various physiological and pathological processes [27, 28]. For example, miR-145 was found to be downregulated in prostate cancer and to be a putative tumor suppressor by reducing cell growth and increasing cell death [29]. In breast cancer cells, downregulation of miR-105-3p retarded cell proliferation, suppressed cell migration, invasion, and reinforced apoptosis [30]. It was found that miR-384 induced autophagy and apoptosis through negative control of COL10A1 in non-small cell lung cancer cells [31]. In this study, miR-384 expression was declined in HCC, and overexpression of miR-384 confined cell proliferation, migration, invasion and EMT but reinforced apoptosis in vitro.
Notably, circRNAs can serve as miRNA sponges, thereby modulating gene expression [32]. Circ_0000517 could sponge miR-326 to retard its expression, and inhibition of miR-326 abolished the effect of circ_0000517 knockdown on the malignant behaviors of breast cancer cells [33]. Circ_0001017 acted as a sponge for miR-197 and inhibited the development of gastric cancer [34]. Circ_0007385 suppressed the malignant behaviors by targeting miR-519d-3p in non-small cell lung cancer cells [35]. We predicted the binding sites between circ_0008285 and miR-384, and also verified that circ_0008285 could adsorb miR-384. Circ_0008285 was negatively correlated with miR-384 expression in HCC tissues. In addition, anti-miR-384 reversed the effects of circ_0000218 silencing on the proliferation, migration, invasion and apoptosis of HCC cells. Besides, circ_0008285 reinforced HCC cell progression through sponging miR-384.
It has been revealed that circRNA can adsorb miRNA to regulate mRNA, and the circRNA-miRNA-mRNA regulatory network has an important regulatory role in cancer. For example, Circ_0007841 accelerates ovarian cancer development by intensifying the expression of MEX3C through the inhibition of miR-151-3p activity [36]. RRM2 acts as a pro-oncogene in HCC, and high expression of RRM2 may be a helpful marker for predicting early recurrence and may be a marker of poor prognosis after curative resection in HCC [37]. In this study, RRM2 was identified and confirmed as a potential target for miR-384. And miR-384 negatively moderated RRM2 expression; furthermore, RRM2 mRNA and protein levels were significantly elevated in HCC. Overexpression of RRM2 was able to reverse the suppression effects of miR-384 mimics on the malignant phenotypes of HCC cells. These data suggested that miR-384 regulated the development of HCC by targeting RRM2. Furthermore, circ_0008285 expression was positively correlated with RRM2 in HCC. Circ_0008285 silencing decreased RRM2 expression, while miR-384 inhibitor attenuated this effect. This finding implied that circ_0008285 could regulate RRM2 expression by sponging miR-384. Besides that, previous studies also showed the downstream target mRNAs or pathways of RRM2 in multiple diseases. For instance, RRM2 binds to ANXA1 to activate AKT signaling in renal cancer, thus promoting tumor growth and sunitinib resistance [38]. Overexpression of RRM2 can improve the damage of cardiac structure and function by activating Hippo-YAP Pathway [39]. However, the lack of target gene detection of RRM2 is a certain limitation in our work, which will be studied in future research.
5ConclusionCirc_0008285 could perform its vital functions in the progression of HCC by miR-384/RRM2 axis, which provided a new experimental basis for using circ_0008285 as a marker for the diagnosis and treatment of HCC.
FundingThis study was supported by: the Natural Science Foundation of Hunan Province [2021JJ70076]
Author's contributionsAll persons who meet authorship criteria are listed as authors, and all authors certify that they have participated sufficiently in work to take public responsibility for the content, including participation in the concept, design, analysis, writing, or revision of the manuscript. All the authors gave their final approval of the version to be published and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the article are appropriately investigated and resolved. Shuang Peng designed the research and wrote the manuscript. Lai Yi, Yiling Bin and Lingzhi Liao collected and analyzed the data. Weiming Qu and Hongsai Hu reviewed and edited the manuscript. All authors read and approved the final manuscript.
None.