Liver cancer, predominantly hepatocellular carcinoma (HCC), is a highly lethal disease that caused about 830,000 deaths worldwide in 2020 [1]. This poor outcome has been attributed to the lack of timely diagnosis and the frequent presence of intrahepatic metastasis. Therefore, understanding the mechanisms underlying hepatocarcinogenesis and metastasis is essential for developing better diagnostic approaches and therapeutic options for HCC patients. The occurrence of HCC is multi-step and evolves from a precancerous lesion to advanced HCC. During HCC development, primary tumor masses spawn pioneer cells that move out and undergo intravasation and survival in the circulatory system, followed by extravasation and traveling to other sites where they can succeed in founding new colonies [2,3]. Cancer cells that can accomplish these steps have rapid growth and metastatic capacity due to the accumulation of genetic and epigenetic alterations, including changes in circular RNA (circRNA) expression [4].
CircRNAs are a class of endogenous non-coding RNAs that are conserved and stable, and numerous circRNAs appear to be specifically expressed in certain cell types or developmental stages. The cell type- or stage-specific expression patterns indicate that circRNAs may play essential roles in numerous physiological and pathophysiological processes [5]. With expression profiling technologies being applied to circRNAs, accumulating evidence has shown that circRNAs contribute to the initiation and progression of HCC [5–9]. However, only a few studies have examined circRNA expression profiles in more than ten samples [8,10,11,12], leading to highly inconsistent results. Furthermore, the development of HCC is a multi-step process that progresses from chronic hepatitis, cirrhosis, and primary HCC to metastatic HCC. However, most studies have only compared the differences in circRNA expression profiles between HCC tissue and adjacent normal tissue [10,13,14], resulting in only a partial understanding of the circRNA expression changes during hepatocarcinogenesis and metastasis.
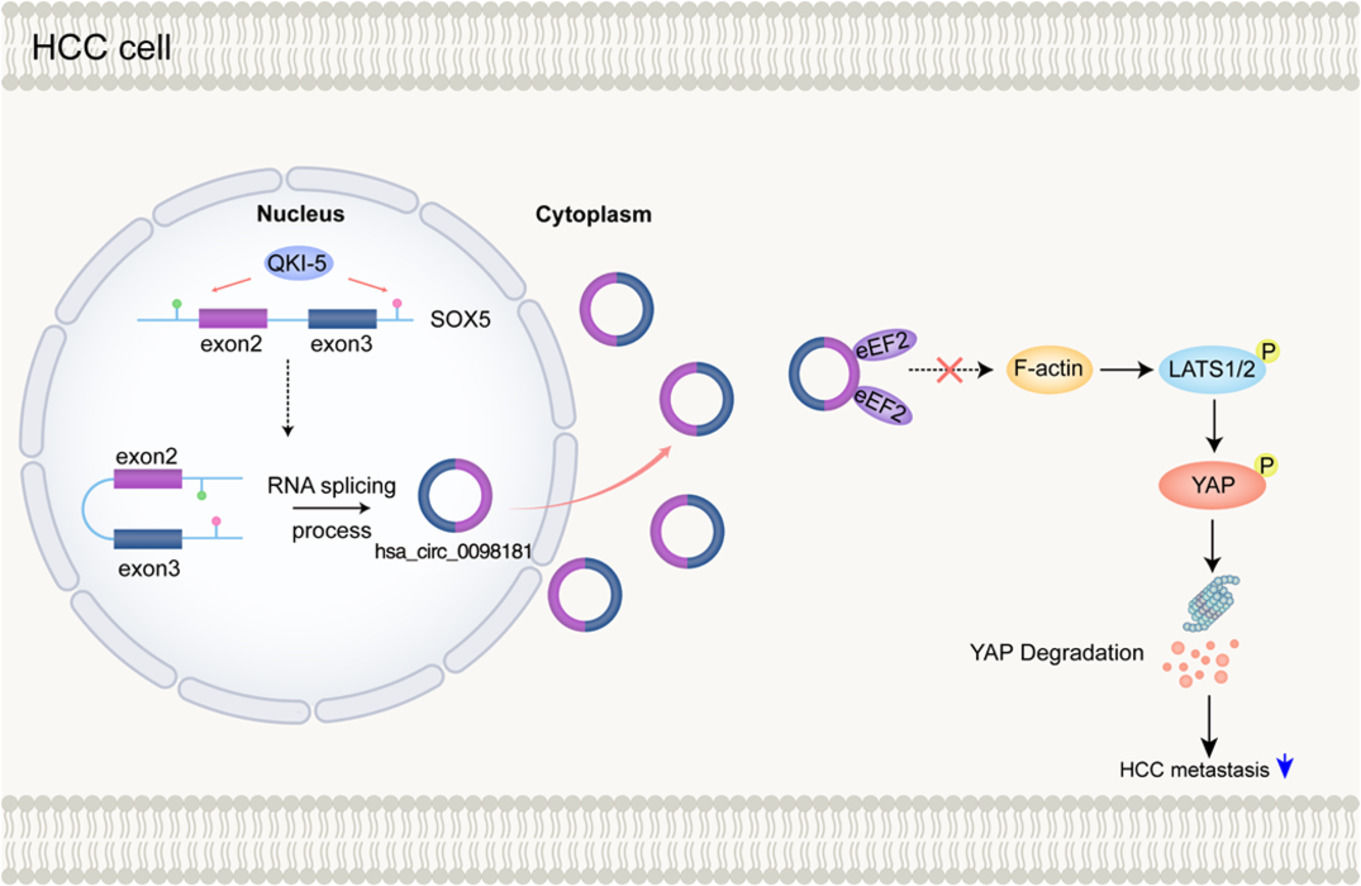
In the present study, we analyzed the circRNA expression profiles of 30 samples, including HCC tissues and adjacent non-tumorous tissues from ten patients without venous metastases, and HCC tissues from ten patients with venous metastases, making our results more robust. We also examined the expression of candidate circRNAs in HCC cell lines and HCC samples. Finally, we focused on one particular circRNA, hsa_circ_0098181, which is derived from exons 2 and 3 of the SOX5 gene. A previous study found that hsa_circ_0098181 suppressed HCC by sponging miR-18a-3p and targeting peroxisome proliferator-activated receptor-α [15]. In contrast, our research focused on the effect of hsa_circ_0098181 on HCC metastasis explored its protein partners that may regulate specific signaling pathways and examined the biogenesis of hsa_circ_0098181 in HCC.
2Materials and Methods2.1Patients and samplesTumor tissues and matched adjacent non-tumorous tissues were collected from 143 patients who underwent HCC resection between 2009 and 2015 at the Eastern Hepatobiliary Surgery Hospital (Shanghai, China). The inclusion criteria of primary HCC patients are described in the Supplementary Materials and Methods. The clinical characteristics of the 143 patients are summarized in Supplementary Table S1.
2.2CircRNA microarray experimentsMicroarray experiments were conducted using the Arraystar Human circRNA Array V2 (Arraystar Inc., Rockville, MD, USA) consisting of probes for 13,617 circRNAs. Sample preparation and microarray hybridization were performed according to Arraystar's standard protocols. Agilent Feature Extraction software (version 11.0.1.1, Agilent Technologies, Santa Clara, CA, USA) was used to analyze the acquired array images. The microarray data were deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (accession number: GSE121714).
2.3Cell linesHuh-7, HepG2, and SK-Hep-1 were purchased from GeneChem Corporation (Shanghai, China). QGY-7703 was purchased from the cell bank of Central South University (Changsha, China). QGY-7703 cells were cultured in RPMI 1640 medium (Corning, NY, USA) with 10% fetal bovine serum (FBS, Biological Industries, Beit Haemek, Israel). Huh-7 and HepG2 were maintained in DMEM high-glucose medium (Corning) with 10% FBS. SK-Hep-1 cells were cultured in MEM medium (Corning) with 10% FBS. All cells were incubated at 37°C in a humidified atmosphere containing 5% CO2. The cells were validated to be mycoplasma-free.
2.4Construction of stable cell lines with hsa_circ_0098181 overexpressionThe hsa_circ_0098181 plasmid and empty plasmid were transfected into Huh-7 and HepG2 cells with LipofectamineTM 3000 reagent (Invitrogen, Carlsbad, CA, USA), and then the cells were treated with puromycin (1 µg/ml, Cayman Chemical Company, Ann Arbor, MI, USA) for four weeks to generate stably hsa_circ_0098181 overexpression cell lines. The clones expressing puromycin resistance were obtained, expanded, and the expression of hsa_circ_0098181 was identified using quantitative real-time PCR (qRT-PCR). For functional studies, two Huh-7 clones (clones 5 and 6) and two HepG2 clones (clones 3 and 7) were selected.
2.5In vivo metastasis assayFor the in vivo metastasis assay, 5 × 106 hsa_circ_0098181-overexpressing cells (Huh7 clone 6) or negative control (NC) cells were injected via the tail vein into male BALB/c nude mice, which were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China) (n=8 per group). After eight weeks, the mice were euthanized, and their lungs and livers were harvested and fixed with phosphate-buffered neutral formalin, followed by hematoxylin-eosin staining. The numbers of lung and liver metastases were counted in a blinded manner.
2.6Biotin-labeled RNA pull-downBiotin-labeled hsa_circ_0098181 (sense) and control (antisense) probes were designed and synthesized by RiboBio (Guangzhou, China). RNA pull-down assay was performed as previously described [5,16]. Briefly, 1 × 107 cells were fixed with 1% formaldehyde for 10 min, lysed, and sonicated. After centrifugation, 50 μl of the supernatant was retained as the input, and the remaining portion was incubated in a probes-streptavidin dynabeads (M-280, Invitrogen) mixture overnight at 30°C. The next day, the dynabeads-probes-circRNAs mixture was captured using magnets (Invitrogen) and washed four times with wash buffer. The protein was eluted with a cocktail of 100 μg/ml RNase A (Solarbio, Beijing, China), 0.1 U/μl RNase H (Thermo Scientific, Wilmington, DE, USA), and 100 U/ml DNase I (Thermo Scientific) at 37°C for 30 min [17].
2.7Silver staining and mass spectrometry analysisHsa_circ_0098181 binding proteins were boiled in the loading buffer and subjected to SDS-PAGE. The proteins were detected by silver staining using the PAGE Gel Silver Staining Kit (Leagene Biotechnology, Beijing, China). The specific bands were excised and analyzed by mass spectrometry at Shanghai Applied Protein Technology Co. Ltd (Shanghai, China).
2.8RNA-binding protein immunoprecipitationRNA-binding protein immunoprecipitation (RIP) was performed according to the instructions of the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Burlington, MA, USA). The co-precipitated RNA was extracted using TRIzol reagent (Invitrogen). Then, the purified RNAs were detected by qRT-PCR using specific primers (Supplementary Table S2).
2.9Statistical analysisStatistical analyses were performed using SPSS version 22.0 (IBM Corporation, Armonk, NY, USA). The optimal cut-off value of the relative expression of hsa_circ_0098181 in HCC was determined by the K-adaptive partitioning statistical algorithm in R software [5]. Disease-free survival (DFS) and overall survival (OS) were calculated by the Kaplan-Meier method, and the differences between the survival curves were compared using the log-rank test. Differences between the experimental groups were compared by the Student's t-test, Wilcoxon rank-sum test, and Wilcoxon signed-rank test, as appropriate. P-values < 0.05 were considered statistically significant.
2.10Other materials and methodsThe detailed procedures of other methods are described in the Supplementary Materials and Methods.
2.11Ethical StatementWritten informed consent was obtained from each patient included in the study, and the study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the Committee for the Conduct of Human Research of the Eastern Hepatobiliary Surgery Hospital (EHBHKY2019-01-001).
All animal experiments complied with the ARRIVE guidelines and were carried out in accordance with the U.K. Animals (Scientific Procedures) Act, 1986 and associated guidelines.
3Results3.1CircRNA expression profiles in adjacent non-tumorous tissues, primary HCCs without venous metastases, and primary HCCs with venous metastasesThe circRNA expression profiles of 30 samples were determined in this study. Among these patients, ten samples accompanied by venous metastases established at surgery were classified as the metastatic group (M), ten patients whose tumors were not accompanied by metastases and recurrence at 5-year follow-up were classified as the non-metastatic group (T), and ten adjacent non-tumorous tissues of the T group were classified as the normal group (N). Patients with and without venous metastases were matched in terms of age, sex, etiology and cirrhosis. The circRNA expression profiles were compared among these three groups.
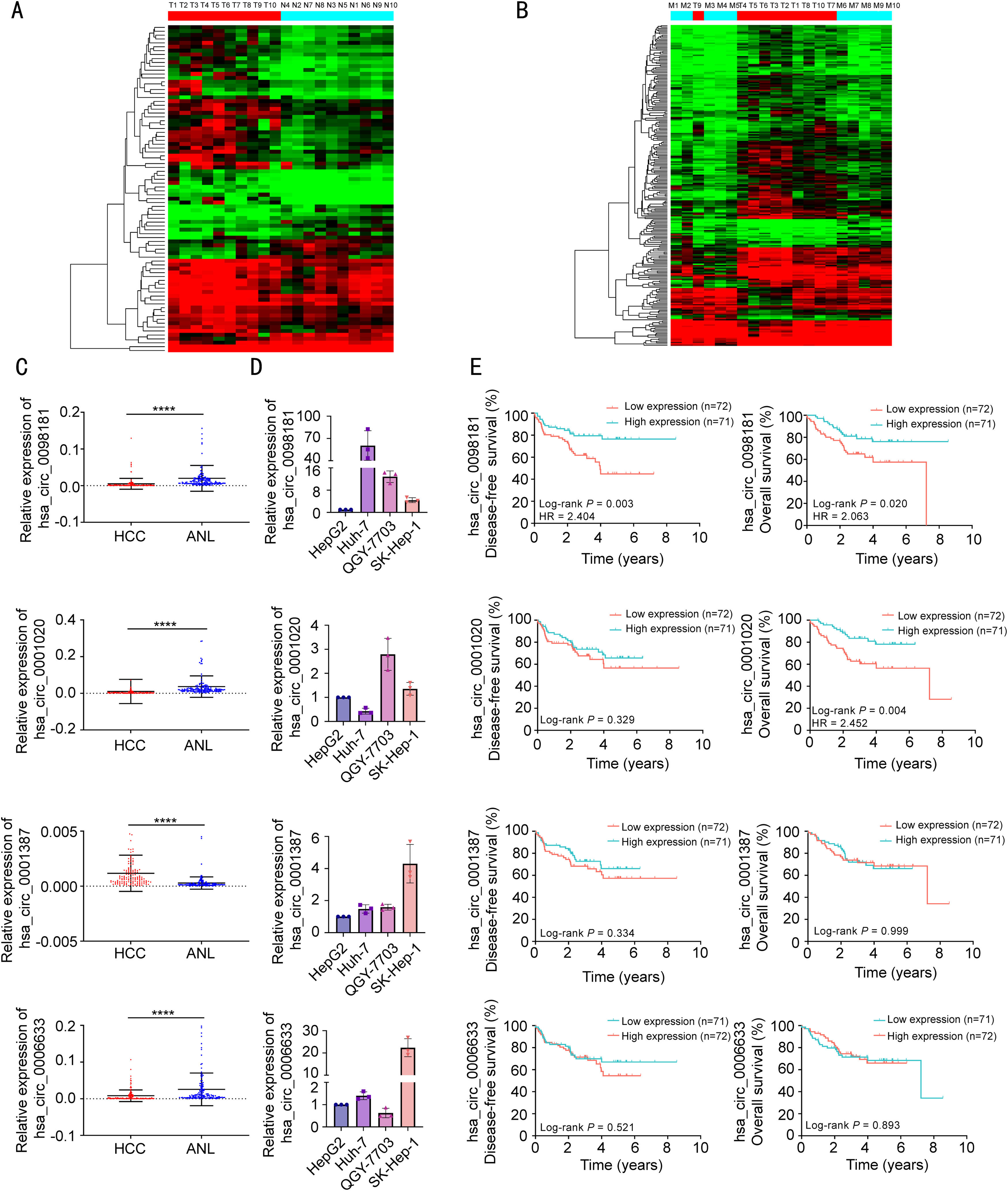
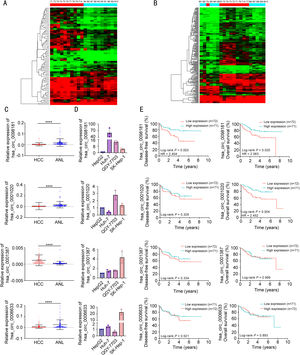
CircRNA deregulation in primary HCCs. A comparison of the ten pairs of T and N samples showed significant deregulation in 84 circRNAs. Among these, 66 were significantly up-regulated in primary HCC samples, and 18 were down-regulated (Figs. 1A, S1A).
Identification of circRNAs during hepatocarcinogenesis and metastasis using circRNA microarrays. (A) Hierarchical clustering of 10 HCC samples without venous metastases (red bars at the top left) and 10 adjacent non-tumorous liver sections (blue bars at the top right). (B) Hierarchical clustering of 10 HCC samples with venous metastases (blue bars) and 10 HCC samples without venous metastasis (red bars). (C) The expression levels of four circRNAs were evaluated in 143 pairs of HCC tissues and matched non-tumorous tissues by qRT-PCR. ANL, adjacent non-tumorous liver tissues. ⁎⁎⁎⁎P<0.0001. (D) The expression levels of four circRNAs were evaluated in four HCC cell lines by qRT-PCR. (E) Kaplan-Meier analysis showed the correlations between the four circRNA expression levels and DFS or OS.
CircRNA deregulation in HCCs with venous metastases. To determine whether circRNA deregulation contributes to HCC metastasis, we compared the circRNA expression levels between ten M and ten T samples. Overall, 238 circRNAs were found to be differentially expressed between these two groups (Figs. 1B, S1B). Among these, 58 circRNAs were up-regulated, and 180 were down-regulated in HCCs with venous metastasis.
CircRNA deregulation during HCC metastatic growth. By comparing the M and N groups, a set of significantly deregulated circRNA was identified. In total, 400 circRNAs were deregulated in the M group. Among these, 119 circRNAs were up-regulated and 281 were down-regulated in this comparison (Fig. S1C-D). Interestingly, the deregulated circRNA set included some of the deregulated circRNAs identified in primary HCCs (32/84, 38%). Furthermore, the set covered more than half of the differentially expressed circRNAs identified in HCCs with venous metastases (137/238, 58%). These findings indicated that part of the pattern of deregulation of circRNAs was likely already established during primary HCC development, and a substantial qualitative change in circRNA expression is required for HCC metastasis.
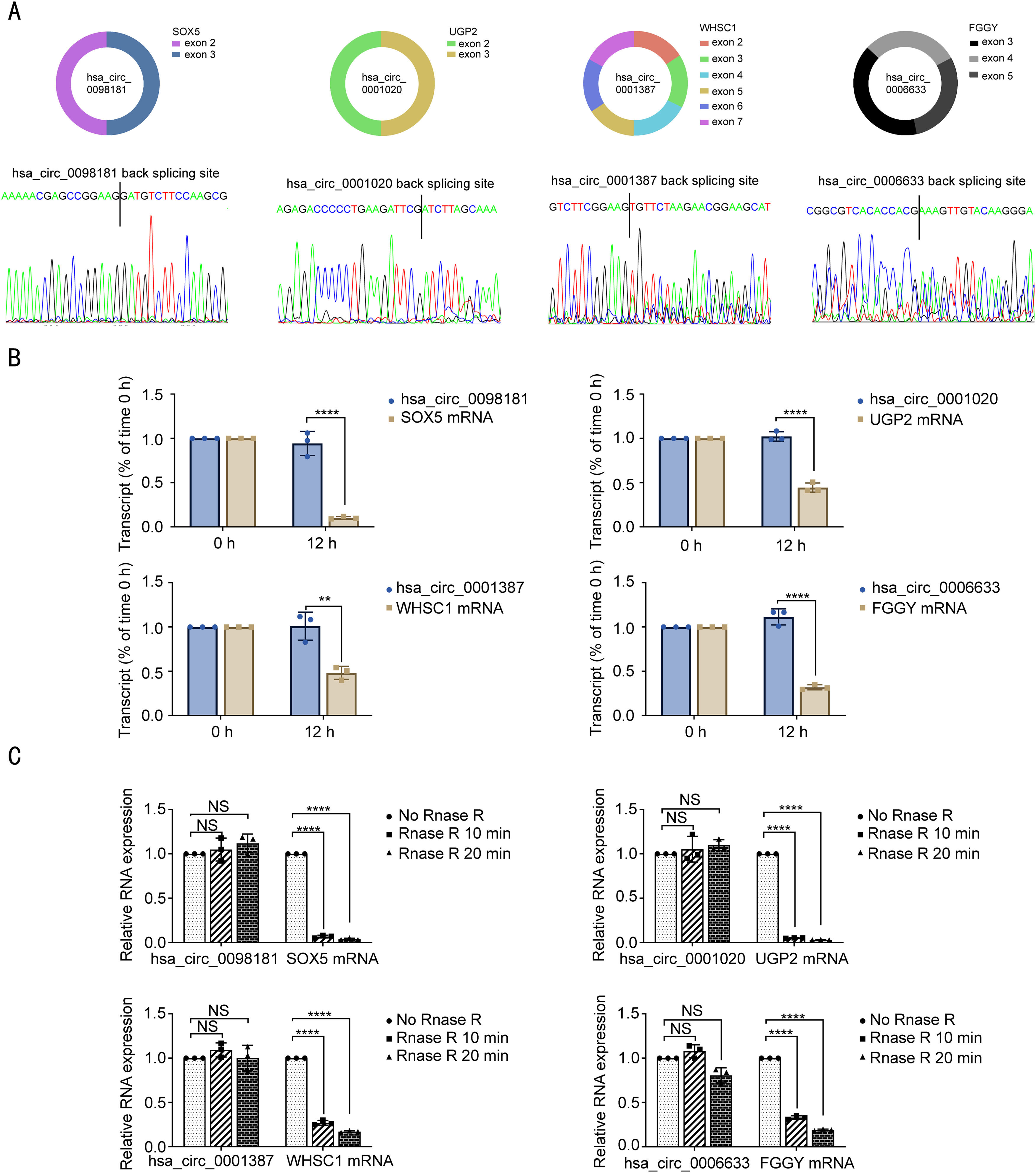
To further explore the changes in circRNA expression throughout the process from chronic hepatitis to HCC with venous metastases, MetaboAnalyst software (www.metaboanalyst.ca) was used to identify the circRNAs with an increasing or decreasing trend in disease progression. A total of 73 circRNAs had a correlation coefficient ≥ 0.5, among which 14 were up-regulated and 59 were down-regulated (Supplementary Table S3). To increase the robustness of the analysis and reduce the high number of differentially expressed circRNAs, we selected 13 circRNAs that were above the cut-off point of a 3-fold change with a correlation coefficient ≥ 0.5 to be further validated by qRT-PCR (Supplementary Table S4). Among these, one circRNA (hsa_circ_0001387 derived from exons 2-7 of the WHSC1 gene) was significantly increased and three circRNAs (hsa_circ_0098181 derived from exons 2 and 3 of the SOX5 gene, hsa_circ_0001020 derived from exons 2 and 3 of the UGP2 gene, and hsa_circRNA_0006633 derived from exons 3-5 of the FGGY gene) were significantly decreased in 143 HCC tissues compared to the adjacent non-tumorous tissues (Fig. 1C). We also examined the expression of these four circRNAs in HCC cell lines (Fig. 1D). Furthermore, the Kaplan-Meier survival curves showed that patients with lower hsa_circ_0098181 expression had worse DFS and OS using the median value of hsa_circ_0098181 expression as the cut-off. Patients with lower hsa_circ_0001020 expression also had shorter OS. However, the expressions of hsa_circ_0001387 and hsa_circRNA_0006633 were not correlated with the DFS or OS of patients (Fig. 1E).
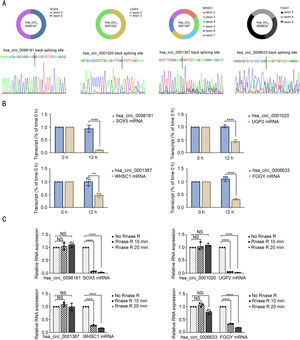
Next, we performed a series of experiments to determine the existence and circular characteristics of these four circRNAs. Specific back-splicing sites were examined by Sanger sequencing (Fig. 2A). Furthermore, actinomycin D and RNase R treatment validated that these four circRNAs were more stable and relatively resistant to RNase R than their linear messenger RNAs (Fig. 2B-C). These data suggested that they were circular RNAs generated from their messenger RNAs by back-splicing.
Identification of hsa_circ_0098181, hsa_circ_0001020, hsa_circ_0001387 and hsa_circ_0006633 as circRNAs. (A) Sketch map for the four circRNAs. (B-C) The expression levels of the four circRNAs and their corresponding mRNAs were detected by qRT-PCR after treatment with actinomycin D for 12 h or with RNase R for 10 min and 20 min. ⁎⁎P<0.01, ⁎⁎⁎⁎P<0.0001; NS, no significance.
We also conducted preliminary experiments to examine the roles of these four circRNAs. Only hsa_circ_0098181 overexpression had a significant effect in inhibiting HCC cell migration. Therefore, we focused on the effect of hsa_circ_0098181 on HCC metastasis.
3.2Decreased hsa_circ_0098181 expression is associated with metastasis-associated clinical characteristics and poor prognosis in HCC patientsTo assess the clinical significance of hsa_circ_0098181 in HCC, we divided patients into two groups according to their clinical characteristics. As shown in Supplementary Table S5, hsa_circ_0098181 was significantly correlated with unfavorable metastatic features, including incomplete tumor encapsulation and tumor embolus, indicating that hsa_circ_0098181 was associated with HCC metastasis.
The K-adaptive partitioning statistical algorithm was used to calculate the most efficient cut-off value for hsa_circ_0098181 expression that could most significantly distinguish between the outcomes of HCC patients. Kaplan-Meier survival analysis indicated that patients with lower hsa_circ_0098181 expression (n=34) had shorter DFS and OS than those with higher hsa_circ_0098181 levels (n=109; Fig. S2). Multivariate Cox proportional analysis further indicated that the expression level of hsa_circ_0098181 was an independent prognostic factor for DFS and OS (Supplementary Tables S6 and S7).
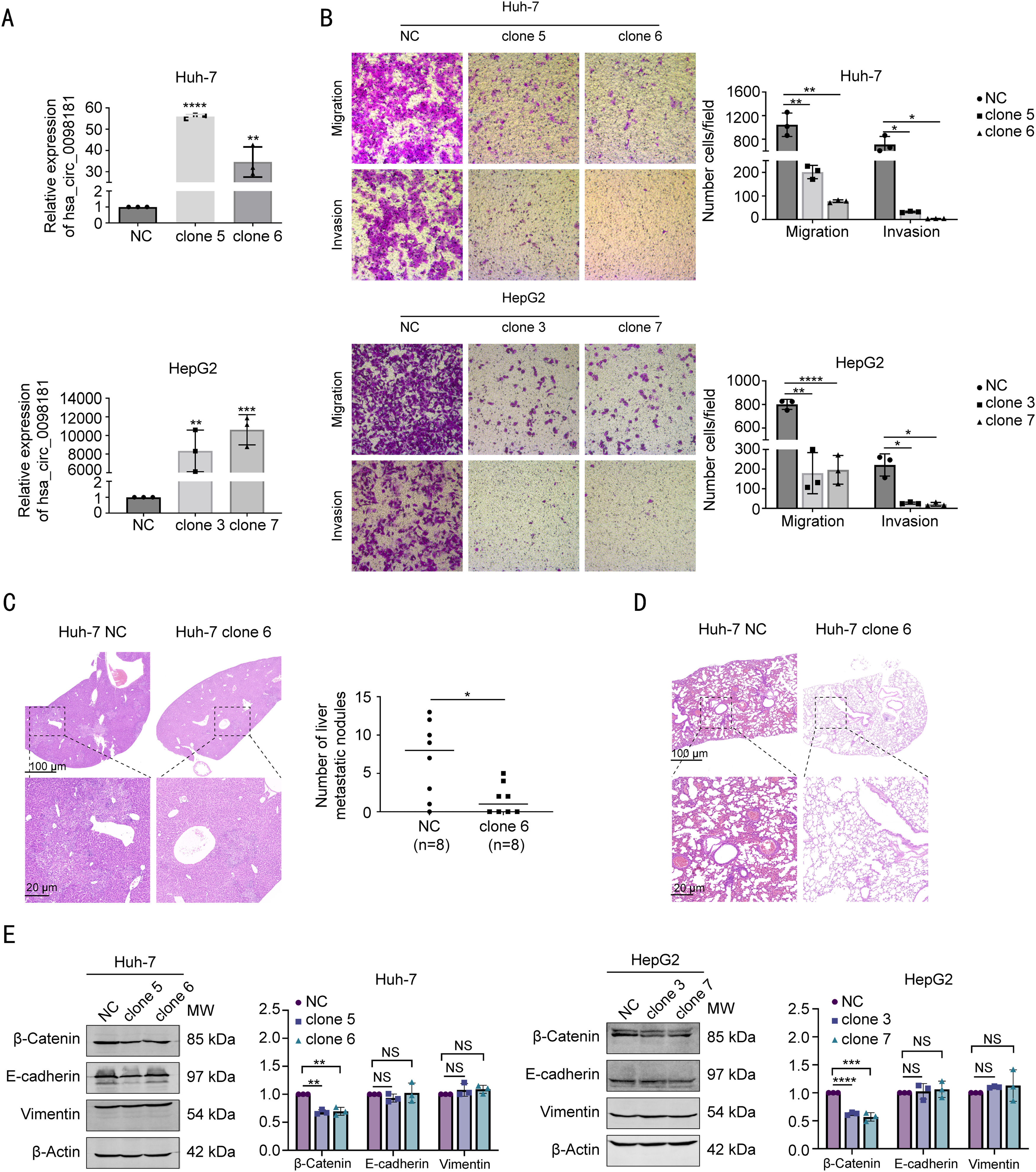
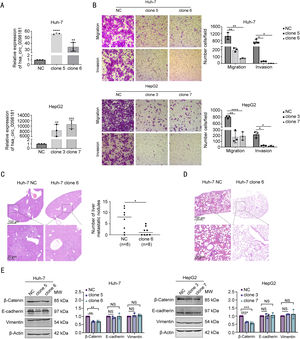
3.3Hsa_circ_0098181 impedes HCC metastasis in vitro and in vivoThe results of qRT-PCR revealed that the endogenous expression of hsa_circ_0098181 was low in different HCC cell lines (Cthsa_circ_0098181−CtACTB > 15, Supplementary Table S8). We selected Huh-7 and HepG2 cells to establish stable overexpression cell lines. For functional studies, we selected two Huh-7 clones (clones 5 and 6) and two HepG2 clones (clones 3 and 7), all of which effectively up-regulated the expression of hsa_circ_0098181 and had no effect on linear mSOX5 (Figs. 3A, S3). Elevated hsa_circ_0098181 expression impeded the migratory and invasive capacity of the Huh-7 and HepG2 cell lines, as demonstrated by the Transwell migration and invasion assays (Fig. 3B).
Hsa_circ_0098181 impedes HCC metastasis in vitro and in vivo. (A) The expression levels of hsa_circ_0098181 in Huh-7 and HepG2 cells after overexpressing hsa_circ_0098181 were detected by qRT-PCR. (B) The migratory and invasive capacities of Huh-7 and HepG2 cells after overexpressing hsa_circ_0098181 were evaluated by Transwell migration and invasion assays, respectively. (C) (Left) Representative images of hematoxylin-eosin staining of liver tissues from nude mice. Scale bar, 100 μm in the upper panel; 20 μm in the lower panel. (Right) The number of metastatic liver nodules in nude mice was compared between the two groups. (D) Representative images of hematoxylin-eosin staining of lung tissues from nude mice. Scale bar, 100 μm in the upper panel; 20 μm in the lower panel. (E) The expression levels of EMT-related proteins β-catenin, E-cadherin and Vimentin after overexpressing hsa_circ_0098181 were determined by western blot analysis. β-Actin was used as an internal control. Data were shown as mean ± standard deviation of three independent experiments. *P<0.05, ⁎⁎P<0.01, ⁎⁎⁎P<0.001, ⁎⁎⁎⁎P<0.0001; NS, no significance.
To explore whether hsa_circ_0098181 suppresses metastasis in vivo, an in vivo metastasis model was established to compare the metastatic nodules formed in the lungs and livers of nude mice after injection with Huh-7 NC or Huh-7 clone 6 cells through the tail vein. Nude mice were monitored over eight weeks, and the experiment ended when mice injected with NC cells displayed decreased physical and mental activity. The metastatic nodules that formed on the surfaces of the liver and lungs were counted. As shown in Fig. 3C, fewer metastatic nodules were observed on the liver surfaces of mice injected with clone 6 cells. Metastatic nodules were absent in the lungs of all animals tested; however, five of the eight nude mice injected with NC cells presented with inflammatory cell infiltration into the lung tissues (Fig. 3D).
Epithelial-mesenchymal transition (EMT) is associated with cancer metastasis. We investigated whether hsa_circ_0098181 could influence the EMT of HCC cells. The expressions of EMT-related proteins E-cadherin, β-catenin, and Vimentin were examined. Western blot analysis showed that elevated hsa_circ_0098181 decreased the expression of β-catenin in Huh-7 and HepG2 cell lines but did not affect E-cadherin or Vimentin protein levels (Fig. 3E). Taken together, these data suggested that hsa_circ_0098181 impedes HCC metastasis in vitro and in vivo.
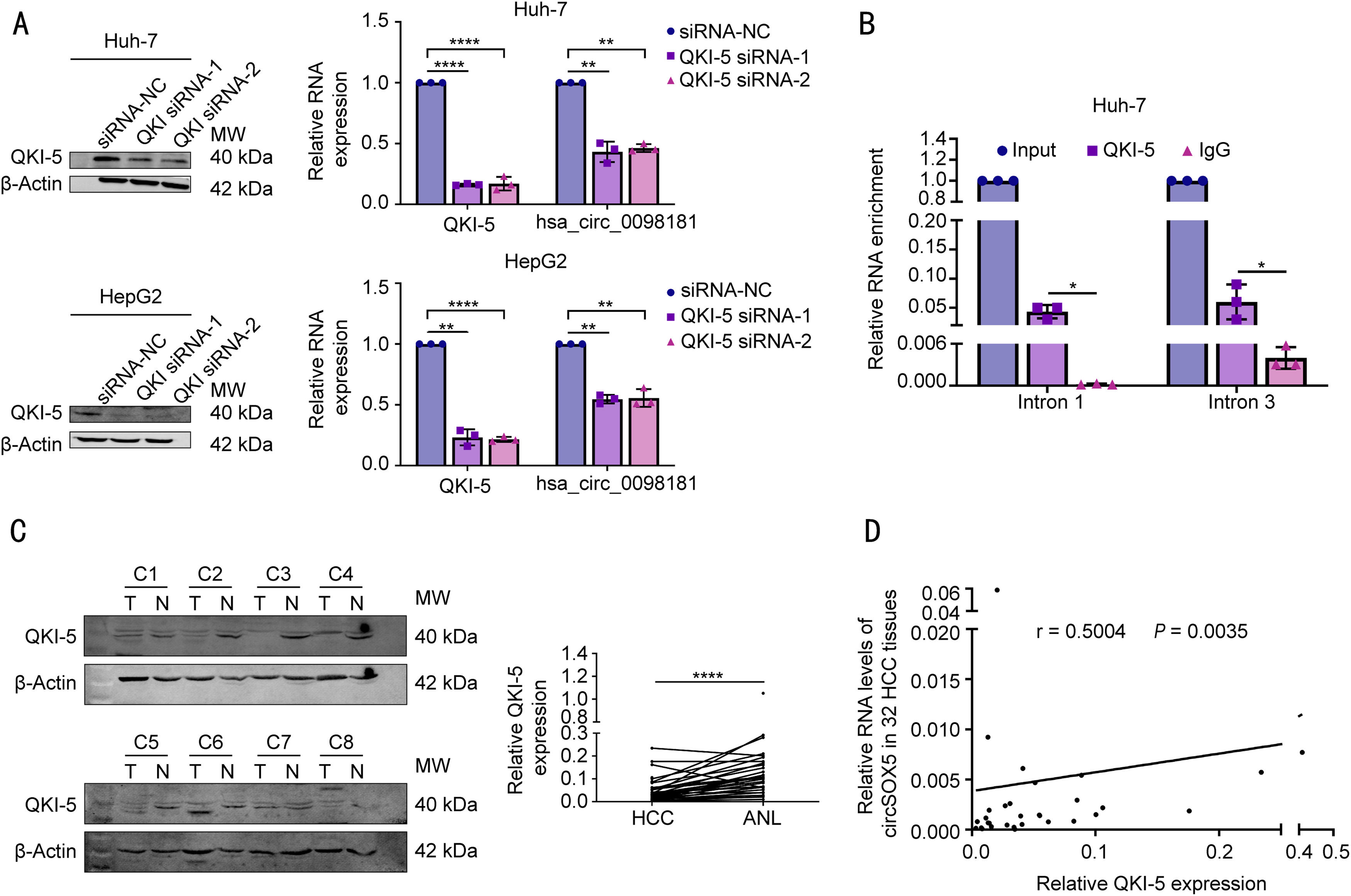
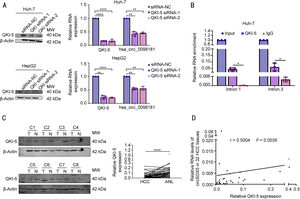
3.4QKI-5 modulates the biogenesis of hsa_circ_0098181 in HCCNext, we explored why hsa_circ_0098181 was repressed in HCC. RNA splicing-related RNA-binding proteins (RBPs) have been associated with circRNA biogenesis in many previous studies. Herein, we focused on five RBPs that reportedly regulate the biogenesis of circRNAs in HCC (QKI-5, DExH-box helicase 9) or have been predicted to bind to flanking regions of hsa_circ_0098181 (epithelial splicing regulatory protein 2, RNA binding motif protein 5, and RNA binding motif single stranded interacting protein 3). The qRT-PCR results indicated that after QKI-5 knockdown in Huh-7 cells, hsa_circ_0098181 was down-regulated, but knockdown of the other four RBPs did not alter the expression of hsa_circ_0098181 (Figs. 4A, S4). In addition, QKI-5 knockdown in HepG2 cells produced similar results (Fig. 4A).
QKI-5 modulates the biogenesis of hsa_circ_0098181 in HCC. (A) (Left) The expression levels of QKI-5 protein were detected by western blot analysis upon QKI-5 depletion using RNA interference in Huh-7 and HepG2 cells. (Right) The expression levels of QKI-5 mRNA and hsa_circ_0098181 were detected by qRT-PCR upon QKI-5 depletion using RNA interference in Huh-7 and HepG2 cells. (B) Anti-QKI-5 RIP experiments were performed to detect the enrichment of intron 1 and intron 3 of SOX5 pre-mRNA. (C) The expression levels of QKI-5 protein were evaluated in 32 pairs of HCC tissues and matched non-tumorous tissues by western blot analysis. ANL, adjacent non-tumorous liver tissues. (D) The correlation between the expression of QKI-5 protein and the expression of hsa_circ_0098181 in 32 HCC tissues was measured by Spearman's rank correlation analysis. *P<0.05, ⁎⁎P<0.01, ⁎⁎⁎⁎P<0.0001.
The effect of QKI-5 on circRNA formation can be achieved by direct binding of QKI-5 to the pre-mRNA [18]. Therefore, we investigated whether QKI-binding sequences (rich in ACUAA) on the flanking introns were required for hsa_circ_0098181 biogenesis. Several putative QKI-5 binding sites are present in the flanking introns. We identified one putative site located upstream (named intron 1) and one putative site located downstream (named intron 3), which are closest to the cyclization exons. Binding sites located in more remote regions play a minor role in circRNA biogenesis [18]. Anti-QKI-5 RIP assay revealed a significant enrichment of introns 1 and 3 (Fig. 4B). Furthermore, western blot analysis indicated that QKI-5 was down-regulated in HCC tissues compared to adjacent non-tumorous tissues (Fig. 4C), and the expression level of hsa_circ_0098181 was positively correlated with the protein level of QKI-5 (n=32, Fig. 4D).
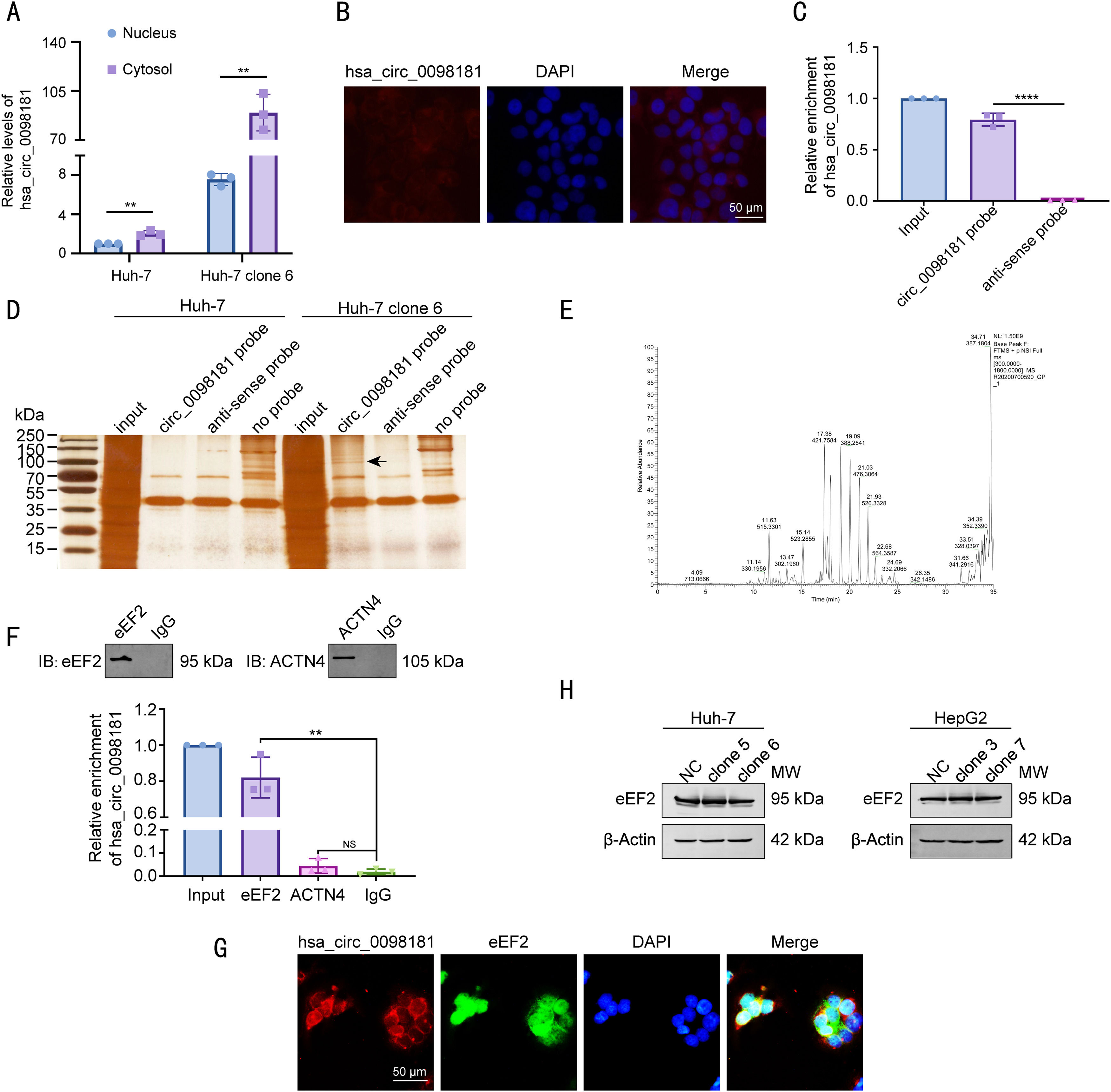
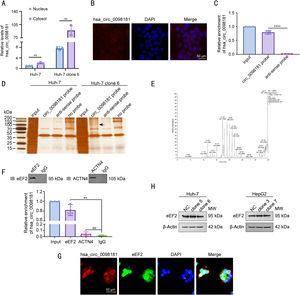
3.5Hsa_circ_0098181 is distributed in the cytosol and interacts with eEF2To dissect the underlying mechanism by which hsa_circ_0098181 impedes HCC metastasis, the localization and protein partners of hsa_circ_0098181 within HCC cells were investigated. We detected the expression of hsa_circ_0098181 in the cytosol and nuclear fractions of Huh-7 and Huh-7 clone 6 cells using qRT-PCR. The results showed an abundant expression of hsa_circ_0098181 in the cytoplasm (Fig. 5A), which was verified by in situ hybridization (Fig. 5B). RNA pull-down assay using a biotinylated hsa_circ_0098181 probe was conducted after confirming the specificity and effectiveness of the probe for hsa_circ_0098181 (Fig. 5C). A variety of potential binding proteins were enriched, and a specific protein band was found at ཞ100 kD after silver staining (Fig. 5D). Mass spectrometric analysis results indicated that eEF2 and actinin alpha 4 could interact with hsa_circ_0098181 (Fig. 5E).
Hsa_circ_0098181 is distributed in the cytosol and interacts with eEF2. (A) The expression levels of hsa_circ_0098181 in the nuclear and cytosol fractions of Huh-7 and Huh-7 clone 6 cells were detected by qRT-PCR. (B) The localization of hsa_circ_0098181 was detected by RNA in situ hybridization in Huh-7 cells. Scale bar, 50 μm. (C) The enrichment ability of the hsa_circ_0098181 probe for hsa_circ_0098181 was detected by qRT-PCR. (D) The proteins that interacted with hsa_circ_0098181 were explored by RNA pull-down with lysates from Huh-7 and Huh-7 clone 6 cells. The arrow indicates the specific band precipitated by the hsa_circ_0098181 probe. (E) The proteins that interacted with hsa_circ_0098181 were identified by mass spectrometric analysis. (F) The proteins that interacted with hsa_circ_0098181 were validated by RNA-binding protein immunoprecipitation assay. NS, no significance. (G) The colocalization of hsa_circ_0098181 and eEF2 was confirmed by dual RNA-fluorescent in situ hybridization and immunofluorescence staining assays. Scale bar, 50 μm. (H) The expression levels of the eEF2 protein after overexpressing hsa_circ_0098181 were determined by western blot analysis. β-Actin was used as an internal control. ⁎⁎P<0.01, ⁎⁎⁎⁎P<0.0001.
Next, the RIP assay verified that an antibody against eEF2 but not actinin alpha 4 could enrich hsa_circ_0098181 (Fig. 5F). Furthermore, dual RNA-fluorescent in situ hybridization and immunofluorescence assays confirmed the colocalization of hsa_circ_0098181 and eEF2 in Huh-7 clone 6 cells (Fig. 5G). However, elevated hsa_circ_0098181 did not change the protein level of eEF2 in Huh-7 and HepG2 cells (Fig. 5H). Collectively, these data suggested that hsa_circ_0098181 interacts directly with eEF2 and sequesters it in HCC cells.
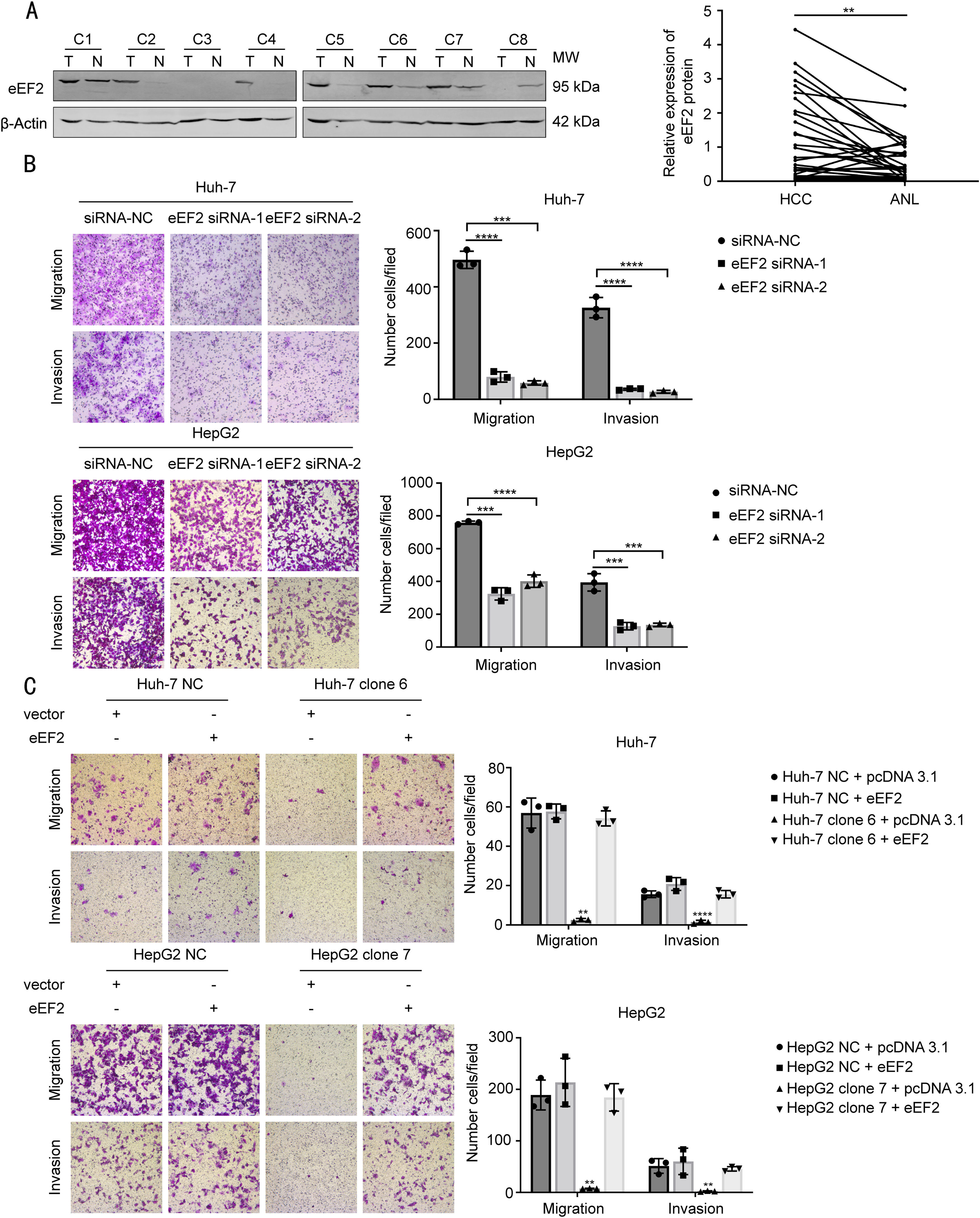
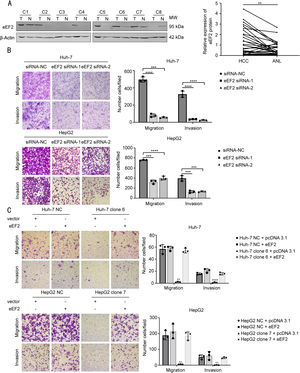
3.6Up-regulation of eEF2 attenuates metastatic suppression by hsa_circ_0098181 in HCC cellsPrevious studies have reported that eEF2 increases in HCC tissues [19]; however, no studies have investigated the effect of eEF2 on HCC metastasis. To study the roles of eEF2 in HCC metastasis, we first examined the amount of eEF2 in 32 pairs of HCC and adjacent non-tumorous tissues by western blot analysis, and found that eEF2 expression was higher in HCC tissues (Fig. 6A). Furthermore, compared to the NC, knockdown of the eEF2 gene significantly reduced the migratory and invasive capacity of HCC cell lines (Figs. 6B and S5).
Up-regulation of eEF2 attenuates the suppression of metastasis by hsa_circ_0098181 in HCC cells. (A) The expression levels of eEF2 protein were evaluated in 32 pairs of HCC tissues and matched non-tumorous tissues by western blot analysis. ANL, adjacent non-tumorous liver tissues. (B) The migratory and invasive capacities of Huh-7 and HepG2 cells after eEF2 knockdown using RNA interference were evaluated by Transwell migration and invasion assays, respectively. (C) The migratory and invasive capacities of eEF2 transiently transfected Huh-7 clone 6 and HepG2 clone 7 cells were evaluated by Transwell migration and invasion assays, respectively. ⁎⁎P<0.01, ⁎⁎⁎P<0.001, ⁎⁎⁎⁎P<0.0001.
The recombinant eEF2 plasmid was then transiently transfected into cells overexpressing hsa_circ_0098181 to detect whether the suppression of HCC cell metastasis by hsa_circ_0098181 could be reversed. Transwell migration and invasion assays revealed that the inhibitory effect of hsa_circ_0098181 was blocked by overexpressing eEF2 (Fig. 6C).
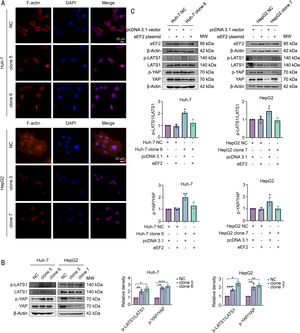
3.7Hsa_circ_0098181 prevents the formation of F-actin and activation of the Hippo signaling pathway by sequestering eEF2eEF family members play an important role in polypeptide chain elongation. Apart from its canonical function, eEF2 has been shown to co-localize with F-actin [20,21], and together with the eEF family member eEF1A1, has been reported to play a role in the formation of F-actin bundles [22]. Furthermore, F-actin was reported to bind to LATS1 and inhibit its phosphorylation, leading to the inactivation of the Hippo signaling pathway in HCC [23]. Therefore, we propose that hsa_circ_0098181 may prevent F-actin formation and then activate the Hippo signaling pathway by sequestering eEF2.
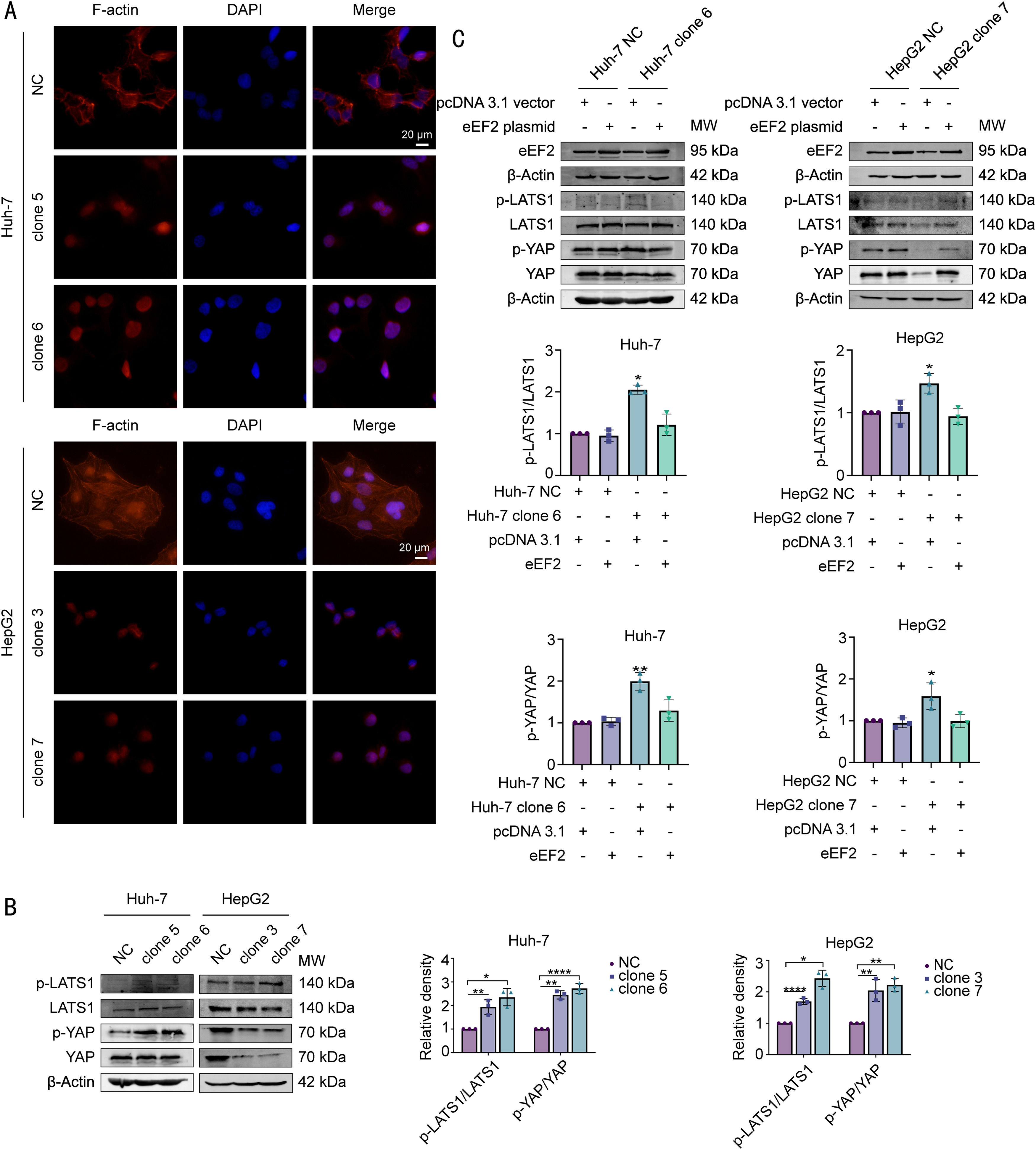
To test this hypothesis, we first performed FITC-phalloidin staining to observe changes in F-actin in cells overexpressing hsa_circ_0098181. The results indicated that overexpression of hsa_circ_0098181 markedly prevented the formation of F-actin (Fig. 7A). Next, western blot analysis was performed to detect the expression of the key components of the Hippo signaling pathway including phosphorylated LATS1 (p-LATS1, ser909), LATS1, phosphorylated YAP (p-YAP, ser127), and YAP in cells overexpressing hsa_circ_0098181. As expected, the results showed that elevated hsa_circ_0098181 increased the p-LATS1/LATS1 and p-YAP/YAP ratios in HCC cells, leading to the degradation of YAP, especially in HepG2 cells (Fig. 7B). Rescue experiments showed that overexpressing eEF2 abolished activation of the Hippo signaling pathway induced by elevated hsa_circ_0098181 (Fig. 7C). Together, these data suggested that hsa_circ_0098181 suppresses HCC metastasis through the Hippo signaling pathway.
Hsa_circ_0098181 prevents the formation of F-actin to activate the Hippo signaling pathway by sequestering eEF2. (A) Effects of elevated hsa_circ_0098181 on the formation of F-actin were determined by FITC-phalloidin staining. Scale bar, 20 μm. (B) The protein expression levels of Hippo signaling components (p-LATS1, LATS1, p-YAP, and YAP) after overexpressing hsa_circ_0098181 were determined by western blot analysis. (C) The protein expression levels of Hippo signaling components (p-LATS1, LATS1, p-YAP, and YAP) were measured by western blot analysis in eEF2 transiently transfected Huh-7 clone 6 and HepG2 clone 7 cells. *P<0.05, ⁎⁎P<0.01, ⁎⁎⁎⁎P<0.0001.
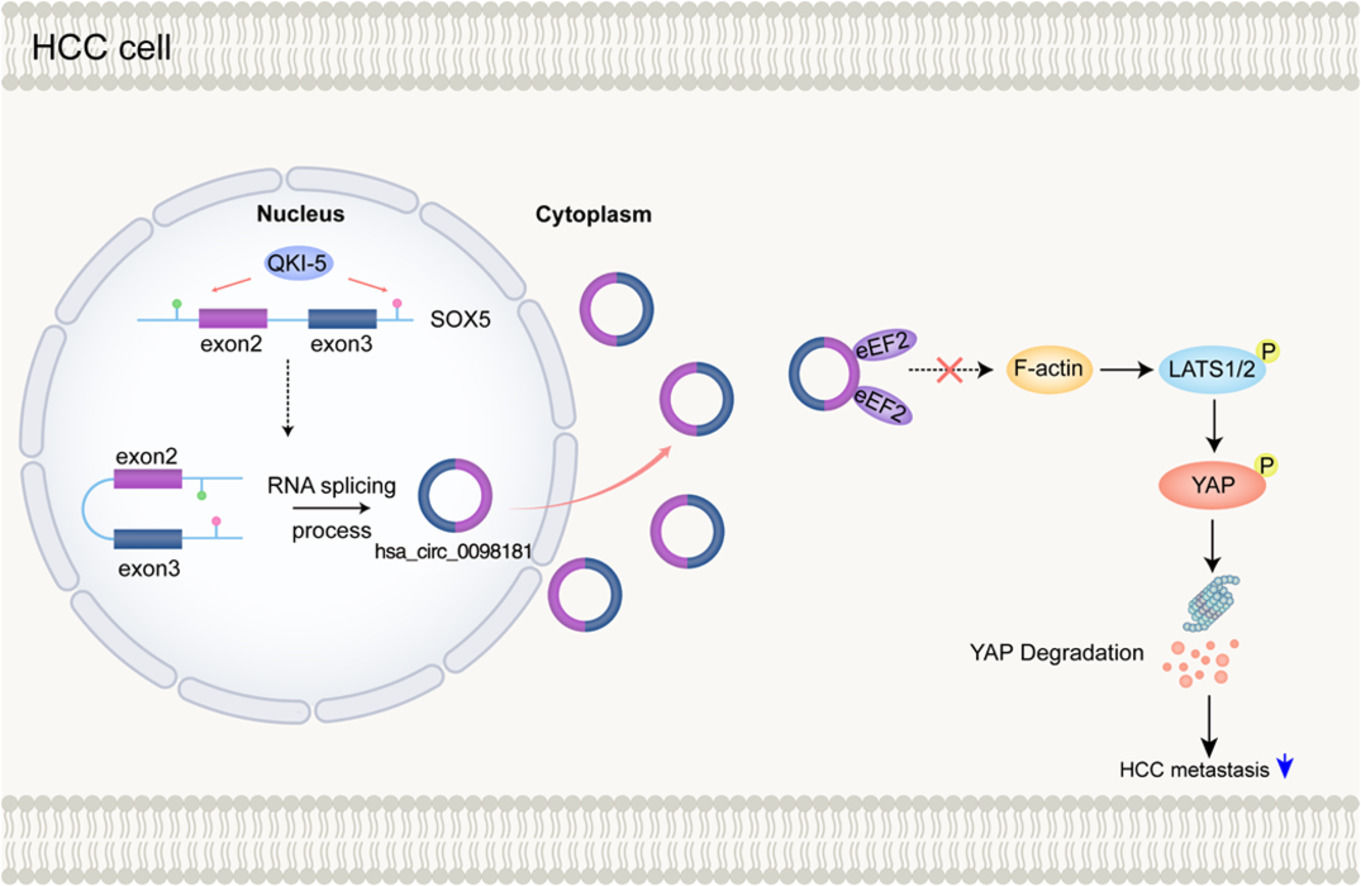
In this study, we demonstrated that the expression profiles of circRNA among adjacent chronic hepatitis tissues, primary tumors without venous metastases, and primary tumors with venous metastases were significantly different. We focused on the roles and mechanisms of hsa_circ_0098181 in HCC and showed that hsa_circ_0098181 was significantly repressed in HCC tissues. Also, for the first time to the best of our knowledge, we provided evidence that hsa_circ_0098181 can delay HCC metastasis by activating the Hippo signaling pathway via sequestration of eEF2. Furthermore, we found that the down-regulation of hsa_circ_0098181 was caused by the down-regulation of QKI-5 in HCC.
Currently, several studies have screened for differentially expressed circRNAs using high-throughput analysis in HCC. However, most studies examined circRNA expression profiles in only three pairs of HCC samples [11,12], leading to highly inconsistent results. Furthermore, hepatocarcinogenesis is a multi-step process that begins with background liver diseases and then proceeds towards early and advanced HCCs. Using a total of 30 samples, our study compared the changes in circRNA expression profiles beginning in chronic hepatitis, continuing to primary HCC without venous metastases, and extending to primary HCC with venous metastases. The circRNA profiles of these samples provide new insights into hepatocarcinogenesis. Part of the pattern of circRNA deregulation is likely already established during primary HCC development, and a substantial qualitative change in circRNA expression may be required for HCC metastasis.
EMT is regarded as an essential step in cancer metastasis [24,25]. In the present study, we verified that the elevated expression of hsa_circ_0098181 decreased β-catenin expression but did not influence the levels of E-cadherin or Vimentin. We propose two explanations for this result. First, the expression levels of EMT markers may not change simultaneously. In the study by Korpal et al., miR-200s promoted the metastatic colonization of cancer cells; however, EMT inducers such as Snail and Twist and mesenchymal markers such as N-cadherin and Vimentin were unaffected by the ectopic expression of miR-200s [26]. Another study reported that loss of Neogenin1 in colorectal carcinoma cells may contribute to metastasis by promoting partial EMT. Neogenin1 knockdown can disrupt the adherens junctions but does not influence the protein levels of E-cadherin [27]. Second and more importantly, EMT alone is insufficient to account for hsa_circ_0098181 phenotypes, suggesting that hsa_circ_0098181 probably influences other pathways during metastasis. Therefore, we used the RNA pull-down assay, mass spectrometric analysis and RIP assay to demonstrate how hsa_circ_0098181 interacts with and sequesters eEF2, leading to the disassociation of eEF2 from F-actin, thereby preventing F-actin formation and subsequently activating the Hippo signaling pathway. Indeed, compelling evidence has revealed the roles of the Hippo pathway in cancer metastasis, including HCC. Aberrant Hippo signaling promotes metastasis by specifically targeting cell migration/invasion [28,29], vascular invasion [30,31], resistance to anoikis [32], and cancer stem cells [33]. Furthermore, the Hippo, Wnt/β-catenin, and Notch pathways can form an interacting network to suppress HCC [34]. Phosphorylated YAP suppresses the nuclear translocation of β-catenin by binding directly to it in the cytoplasm [35], while the accumulation of β-catenin in the cytoplasm leads to its ubiquitination and degradation. Our results indeed confirmed that elevated hsa_circ_0098181 decreases the expression levels of YAP and β-catenin.
CircRNAs are universally recognized to participate in the promotion or suppression of HCC via different molecular mechanisms; for example, acting as an miRNA sponge, affecting transcriptional regulation or splicing, interacting with proteins, and translating into proteins [5,36,37,38]. Most studies have focused on the roles of circRNAs as competing endogenous RNAs [5,13]. In the past two years, an increasing number of studies have examined the interaction between circRNAs and proteins. CircRNAs can physically combine with specific proteins to facilitate or inhibit their function. For example, circBACH1 is significantly up-regulated in HCC tissues and combines directly with the human embryonic lethal abnormal vision-like protein HuR, facilitating its translocation from the nucleus to the cytoplasm, thereby downregulating p27 expression [39]. Another circRNA, circCPSF6, is also up-regulated in HCC tissues. Mechanistically, circCPSF6 competitively binds to the poly binding protein 2, interfering with its binding to YAP mRNA, thus sustaining the stability of YAP [40]. Our study confirmed that hsa_circ_0098181 can interact with eEF2 and inhibit its function. In addition, after silver staining, a protein band at ཞ70 kD was identified, which was more enriched than that of the control group (Fig. 5D), and mass spectrometric analysis was used to preliminarily identify this interacting protein. Whether this protein plays a vital role in the hsa_circ_0098181 suppression of metastasis will be further investigated.
Finally, we need to point out some shortcomings of this study. First, we only included HCC patients with a history of hepatitis B virus infection, and we will further verify whether the expression of hsa_circ_0098181 is reduced in HCC patients with other underlying liver diseases such as those related to hepatitis C and/or alcohol. Second, we only used one endogenous control β-actin for qRT-PCR. However, the expression of these reference genes may vary among different tissues and may change in response to experimental treatment. Therefore, it is important to choose more than one reference gene to improve qRT-PCR accuracy [41].
5ConclusionsOur study revealed differences in circRNA expression profiles during HCC development and metastasis, which offers new insights into hepatocarcinogenesis and metastasis. We identified a regulatory role for the QKI5-hsa_circ_0098181-eEF2-Hippo signaling pathway in HCC, which may provide new therapeutic strategies for HCC.
Data availability statementThe data generated in this study are publicly available in Gene Expression Omnibus at GSE121714 (https://www.ncbi.nlm.nih.gov/geo).
CRediT authorship contribution statementPing Gao: Conceptualization, Investigation, Formal analysis, Writing – original draft, Funding acquisition. Yuan Yang: Conceptualization, Resources, Writing – review & editing. Xiaowei Li: Investigation. Qi Zhao: Investigation. Yujin Liu: Investigation. Chunnan Dong: Formal analysis, Funding acquisition. Yanan Zhang: Investigation. Dianwu Liu: Conceptualization, Writing – review & editing, Supervision.