To study the effect of eukaryotic initiation factor 3B (EIF3B) on the invasion and migration of hepatocellular carcinoma (HCC) and its potential mechanism.
Materials and MethodsThe clinical significance of EIF3B expression was studied with The Cancer Genome Atlas (TCGA) and Gene Expression Profiling Interaction Analysis datasets. Immunohistochemical staining and western blotting were used to examine EIF3B expression in cell lines and tissues from HCC patients. The scratch assay and transwell assay were used to measure the invasion and metastasis of different HCC cell lines in vitro. The molecular mechanism of EIF3B was determined using RNA-seq and identification of dysregulated signaling pathways. Western blotting was used to verify the alterations of EIF3B signaling functioned in the promotion of HCC progression.
ResultsElevated expression of EIF3B in HCC correlated significantly with aggressive clinicopathologic characteristics, including advanced tumor grade and poor prognosis. Studies with cultured cells indicated that EIF3B knockdown inhibited HCC cell invasion and metastasis by depressing the epithelial-mesenchymal transition (EMT). EIF3B also activated the TGFBI/MAPK/ERK signaling pathway by increasing the levels of pMEK and pERK.
ConclusionsOur results indicate that EIF3B functions as an oncogene in HCC that accelerates cell invasion, metastasis, and the EMT by stimulation of the TGFBI/MAPK/ERK signaling pathway. EIF3B is a potential target for the treatment of HCC.
Primary hepatocellular carcinoma (HCC) is responsible for up to 90% of all primary liver cancers. Among all cancers, primary liver cancer has among the highest global rates of incidence and mortality, and nearly half of all global liver cancer cases are in China [1-2]. Currently, surgical resection is often the best option for patients with early liver cancer. Unfortunately, owing to the typically subtle nature of onset, most of these patients receive diagnosis with intermediate or advanced-stage cancer, and surgery is not typically a treatment option. The high rates of recurrence and metastasis are the main reasons these patients have a high risk for mortality and poor prognosis [3]. The exact mechanisms underlying HCC invasion and metastasis are still unclear. Therefore, it is imperative to examine the molecular and physiological basis of HCC progression so that drugs can be developed against new targets. Recent research that examined protein translation, particularly the onset of translation, demonstrated that numerous eukaryotic initiation factors (EIFs) function in the dysregulation of protein synthesis that is characteristic of many tumors [4].
EIFs are essential protein complexes that mediate the translation of mRNAs. EIF3 has 13 subunits and is the largest and most important EIF. EIF3B, one of its subunits, is generally considered the main scaffolding protein because it promotes the binding of other components with different affinities [5]. Recent research indicated that EIF3B participates in cell proliferation, tumor invasion, and metastasis, and that elevated expression of EIF3B was closely connected to the initiation and development of malignant tumors. In particular, there is evidence of abnormal patterns of EIF3B expression in prostate cancer, ovarian tumors, and non-small cell lung cancer (NSCLC), and that its overexpression correlates with poor cancer prognosis. [6-8]. Furthermore, the down-regulation of EIF3B inhibits β-catenin signaling and represses the hyperplasia, invasion, and metastasis of endometrial tumor cells [9]. Other studies reported abnormal expression of EIF3B in osteosarcoma, esophageal squamous cell cancer, and renal cell tumors, and that functions in the invasion and metastasis of these cancers [10-12]. EIF3B hastens the progression of gastric cancer by upregulating the PI3K/AKT/mTOR signaling pathway, and activation of this pathwayis related to worse outcome in these patients [13].
Studies of HCC based on The Cancer Genome Atlas (TCGA) implied that overexpression of EIF3B mRNA was related with shorter overall and recurrence-free survival. Yue et al. also provided evidence that elevated EIF3B mRNA expression was an independent prognostic biomarker for liver cancer. These findings also suggested that EIF3B may function in the onset and promotion of HCC [14].
However, the molecular mechanism by which EIF3B regulates HCC invasion and metastasis remains to be elucidated. Therefore, we evaluated EIF3B expression in HCC tissues, examined its association with the clinicopathological features of patients, and analyzed its potential biological function in HCC cells, with a focus on the mechanisms of invasion and metastasis.
2Materials and Methods2.1Cell lines, antibodies and tissue samplesFetal calf serum, Dulbecco's modified Eagle medium (DMEM), trypsin, and puromycin were acquired from Gibco (USA). The normal human hepatic cell (L-02) and HCC cell lines (HepG2, Huh7, Sk-Hep-1, and MHCC-97H) were all purchased from the National Collection of Authenticated Cell Cultures (Shanghai, China). Antibodies against the following proteins were used for analyses of protein expression: EIF3B (ab124778, Abcam), β-actin (3700S,CST), E-cadherin (ab1416, Abcam), N-cadherin (ab76011, Abcam), vimentin (ab188499, Abcam), Snail (ab85936, Abcam), TGFBI (ab170874, Abcam), p-ERK (4370T, CST), t-ERK (4695T, CST), p-MEK (41G9,CST), and t-MEK (47E6,CST). All chemicals were analytical-grade reagents and water was double distilled before use. In 2021, pairs samples of liver cancer tissues and matched normal liver tissue were received from the Department of Hepatobiliary Surgery, Fujian Cancer Hospital (Fuzhou, China).These collected tissue samples were stored in liquid nitrogen immediately after dissection. None of these patients received any other treatment before the operation. Fifty-nine paraffin-embedded specimens collected from HCC patients undergoing radical hepatectomy from January 2015 to January 2019 were obtained from the Pathology Department, Fujian Cancer Hospital. This research was approved by the Ethics Committee of Fujian Cancer Hospital (Reference NO.SQ2021-015-01). All patients signed informed consent documents.
2.2Cell cultureAll cells were cultured in DMEM supplemented with 10% (v/v) fetal calf serum (FCS) in an incubator at 37°C with 5% CO2. Cells were grown to approximately 70% confluence prior to the wound scratch assay and the migration assay. Cells were stained with crystal violet for counting.
2.3Analysis of public databasesThe RNA-seq data and clinical information from HCC patients were from TCGA (http://gdc.cancer.gov/). Gene Expression Profiling Interaction Analysis was applied to characterize the overall survival and disease-free survival of patients with high or low EIF3B expression.
2.4RNA extraction, Reverse transcription, and qRT-PCRExtraction of complete RNA from frozen tissues or exponentially growing cells (1 × 106) was performed by grinding in liquid nitrogen and use of the Trizol reagent (Invitrogen Life Technologies, USA). cDNA was reverse-transcribed from 1μg RNA using a reverse transcription kit (Promega, USA). qRT-PCR was performed according to Roche Light-Cycler 480 software program(Roche, USA) and the relative expression of EIF3B was calculated using the 2−ΔΔCt method. The primer sequences of EIF3B were: 5′-CGGTGCCTTAGCGTTTGTG-3′(forward) and 5′-CGGTCCTTGTTGTTCTTCTGC-3′(reverse); the internal reference primer sequences of GAPDH were 5′-TGACTTCAACAGCGACACCCA-3′(forward) and 5′-CACCCTGTTGCTGTAGCCAAA-3′(reverse). The amplification conditions consisted of a hot start (95°Cfor 10 min) followed by 40 cycles of amplification (95°Cfor15 s, 60°Cfor 10 s, and 72°Cfor 1 s), and then maintenance at 4°C.
2.5Western blot assayWhen the cells reached 70% confluence, they were lysed with a solution that contained phosphatase and protease inhibitors. The BCA reagent kit (Beyotime, China) was used to quantify protein concentration. Then 25μg of protein was added to each lane, 8%-12% SDS-PAGE was performed to separate the proteins, and the bands were transferred onto PVDF membranes. After use of 5% skimmed milk powder for blocking, the first antibody (1:1000–1:5000) was added at 4°C overnight. Then, the membrane was incubated with anti-rabbit or anti- mouse horseradish peroxidase (HRP)-conjugated secondary antibody (1:3000). Finally, the membranes were visualized using the Bio-Rad gel imaging system (ECL, Millipore).
2.6Lentivirus and plasmid transfectionMHCC-97H and Huh7 cells were transfected with EIF3B shRNA lentivirus and negative control lentivirus, which were constructed by Genechem Co., Ltd. (China), for 12h with DMEM medium plus with polybrene. The transfection medium was changed to 10% DMEM with FBS. HCC cells with stable EIF3B knockdown were screened with puromycin (1.5μg/mL) for 14 days, and EIF3B transfection efficiency was assessed by western blotting. Vectors that overexpressed the transforming growth factor beta-induced gene (TGFBI) and empty vectors; TGFBI short hairpin RNA(shRNA) and EIF3B overexpressing vector (Genechem Biotechnology; Shanghai, China), were transfected into MHCC-97H shEIF3B cells using plasmid transfection technology. Western blotting was used to assess the transfection efficiency at 72 h after transfection.
2.7Scratch assayThe density of Huh7-shEIF3B, Huh7-con, MHCC-97H-shEIF3B, and MHCC-97H-con cells were adjusted to 5 × 105 and inoculated into 6-well plates until 70% confluence occurred. The cell scratching experiments were performed the next day. The suspended cells were discarded and the remaining adherent cells were cultured for another 48 h. Wound healing of the cell monolayer was evaluated by light microscopy before scratching (0 h) and 48 h after scratching.
2.8Transwell assayFor the cell migration assay, 10μL of fibronectin (1μg/μL) was spread evenly on the membrane of a Transwell lower chamber and placed in a 37 °C incubator for 4h. Serum-free DMEM was used to adjust cell density to 5 × 102/μL. Then, 100μL of a cell suspension was seeded into the upper wells, and 600 μL of complete medium containing 20% FBS was added into the bottom wells. After incubation at 37 °Cfor 48h, cells that did not migrate were wiped away with a cotton swab, and cells that migrated to the lower insert was stained with 0.1% crystal violet following fixation with methanol. Subsequently, the total number of cells in the five different fields were measured by optical microscopy (× 200).
For cell invasion experiments, the invasive insert was precoated with diluted matrigel (100μL). The remaining steps were the same as the migration assay.
2.9Cell RNA extraction and sequencingTotal RNA was extracted from transfected Huh7 and MHCC-97H cells (shEIF3B and sh-NC) using the Trizol reagent (Invitrogen Life Technologies, USA). A Nanodrop 2000 instrument (Thermo Fisher Scientific, USA) was employed to determine RNA concentration and purity. The quality and quantities of RNA samples were estimated with the QIAxcel RNA QC Kit version 2.0. For library construction, 1 µg RNA (concentration > 200 ng/μL) was used for sequencing, reading quality check, alignment, clustering, expression evaluation, Gene Ontology (GO) enrichment analysis, and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis by the Wuhan SeqHealth Co., Ltd. (China). The CollibriTM Stranded RNA Library Prep Kit for Illumina (Thermo Fisher), HiSeq 2000 Sequencing System (Illumina), TopHat version2.1.1, HTSeq version0.12.4, DEGseq2, and Hypergeometric test were used for these analyses.
2.10ImmunohistochemistryParaffin-embedded tumor and adjacent tissue samples were subjected to routine dewaxing and hydration. Then, antigen retrieval was performed and the tissue sections were subsequently incubated with an anti-EIF3B antibody at 4°C overnight. After washing, sections were incubated with horseradish peroxidase (HRP)-conjugated anti-IgG (Abcam, USA) as the secondary antibody. The sections were developed using aDAB kit (GeneTech, Shanghai, China), followed by counterstaining with hematoxylin. The evaluation method is as follows: Positive reactions were defined as those showing brown signals in the cell cytoplasm. The intensity was scored as follows: O, negative; 1, weak; 2, moderate; and 3, strong. The frequency of positive cells was defined as follows: O, less than 5%; 1, 5% to 25%; 2, 26% to 50%; 3, 51%to 75%; and 4, greater than 75%. The intensity score is multiplied by the frequency score to get the IHC score. Each section was independently evaluated by 2 pathologists who were blinded to patient data.
2.11ELISAFor analysis of TGFBI expression, cells with transient interference of EIF3B and control cells were compared by collection of culture supernatants and detection with an ELISA kit following the manufacturer's protocol (ab55426, Abcam, USA).
2.12Statistical analysisAll data were reported as means ± standard deviations (SDs) from three independent replicates. Comparisons of different groups were conducted using Student's t-test and results were presented using GraphPad Prism version 7.0 (USA). A p-value below 0.05 was defined as statistically significant.
2.13Ethical statementWritten informed consent was obtained from each patient included in the study and the study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the Ethics Committee of Fujian Cancer Hospital (NO.SQ2021-015-01).
3Results3.1HCC cells have upregulation of EIF3BWe first analyzed EIF3B expression in the dataset for HCC in TCGA.The results indicated greater EIF3B expression in HCC samples than in normal control samples (Fig. 1A). We then analyzed the relationship of EIF3B expression with multiple demographic and pathologic parameters of HCC in TCGA. The results indicated that EIF3B expression was unrelated to patient age, gender, or lymph node metastasis, but was related with tumor grade (Fig. 1B). Thus, EIF3B was highly expressed in human HCC and positively correlated with tumor malignancy.
High EIF3B expression in HCC indicates poor prognosis. A, Expression of EIF3B in carcinomas and adjacent carcinomas according to HCC data from TCGA. B, Relationship of EIF3B expression with tumor grade according to HCC data from TCGA. C–D, Association of EIF3B expression with OS and DFS. E, Expression of EIF3B (qPCR) in HCC tissues and control tissues. F, Immunohistochemistry of EIF3B in HCC and corresponding normal tissues. G, Quantitative analysis of EIF3B in F. H, Statistical analysis of the correlation between EIF3B expression and OS.
Data are mean ± standard deviation (SD); *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
We then examined the prognostic significance of EIF3B expression using Gene Expression Profiling Interactive Analysis (GEIPA). Kaplan-Meier plots showed that the overall survival (OS) and disease-free survival (DFS) were significant shorter in patients with higher expression of EIF3B (Fig. 1C and 1D).
We then validated this bioinformatics analysis by analyzing mRNA expression in 13 pairs of fresh primary HCC tumors and corresponding normal tissues. The results demonstrated that HCC tissues had significantly higher expression of EIF3B (Fig. 1E). For confirmation, we used IHC to detect EIF3B in 59 HCC tissues and adjacent tissue samples. In agreement, the results revealed that EIF3B expression was higher in tumor tissues than in adjacent tissues (Fig. 1F and 1G). In addition, we also conducted statistical analysis on the clinical data of these HCC patients. The results showed that the expression of EIF3B is significantly correlated with patient 's stage and survival, but not with age, gender or lymph node metastasis (Fig. 1H and Table 1). In conclusion, these results demonstrated that high expression of EIF3B in HCC may be related to poor prognosis.
Correlation analysis between EIF3B expression and pathologic parameters of patients.
| Case number | IHC Score | P value | |
|---|---|---|---|
| <6 | ≥6 | ||
| Age | |||
| ≤50 | 13 | 16 | 0.7864 |
| >50 | 11 | 19 | |
| Gender | |||
| Male | 10 | 18 | 0.8233 |
| Female | 14 | 17 | |
| Lymph node metastasis | |||
| Yes | 12 | 17 | 0.9153 |
| No | 12 | 18 | |
| Clinical stage | |||
| Ⅰ | 4 | 4 | 0.0328 * |
| Ⅱ | 6 | 8 | |
| Ⅲ | 7 | 10 | |
| Ⅳ | 7 | 13 | |
We then used qRT-PCR and western blotting to measure the expression of EIF3B mRNA and protein in a line of normal human hepatocellular epithelial cells(L-02) and four lines of HCC cells (HepG2, Huh7, MHCC-97H, and SK-Hep-1). Compared to the other cell lines, the expression of EIF3B mRNA and protein were significantly higher in Huh7 and MHCC-97H cells (Fig. 2A and 2B). To study the effects of EIF3B in liver cells, we performed transduction of Huh7 and MHCC-97H cells using a lentivirus-expressing shRNA to knockdown EIF3B. At 72h after transduction, more than 90% of the cells expressed the GFP marker, confirming successful transduction (Fig. 2C). We confirmed the reduced expression of EIF3B in transduced cells using qRT-PCR and western blotting (Fig. 2D and 2E).
EIF3B knockdown suppresses invasion and metastasis of HCC cells in vitro. A–B, qRT-PCR and western blotting of EIF3B in four human HCC cell lines and one normal liver cell line. C, Transduction efficiency (GFP fluorescence, × 100) of EIF3B shRNA lentivirus in MHCC-97H and Huh7 cells after 72 h. D, EIF3B expression (qRT-PCR) in lentivirus-infected MHCC-97H and Huh7 cells. E, EIF3B expression (western blotting) in lentivirus-infected Huh7 and MHCC-97H cells. F, Effect of EIF3B on HCC cell migration (scratch assay). G, Effects of EIF3B on HCC cell invasion and metastasis (transwell assay). H–I, Quantitation of results from the migration and invasion assays. Tests were performed in triplicate and values were indicated as means ± SDs; **P<0.01, ***P <0.001.
We then performed a scratch assay to assess the impact of EIF3B on HCC cell migration. The results showed that cells with EIF3B depletion had significantly attenuated wound healing compared to control HCC cells (Fig. 2F). Similarly, compared with the control group, the cell migration and invasion of the two cell lines transfected with the shEIF3B vector were significantly decreased (Fig. 2G, 2H, and 2I). Overall, these results indicated that EIF3B led to promotion of cell migration and invasion.
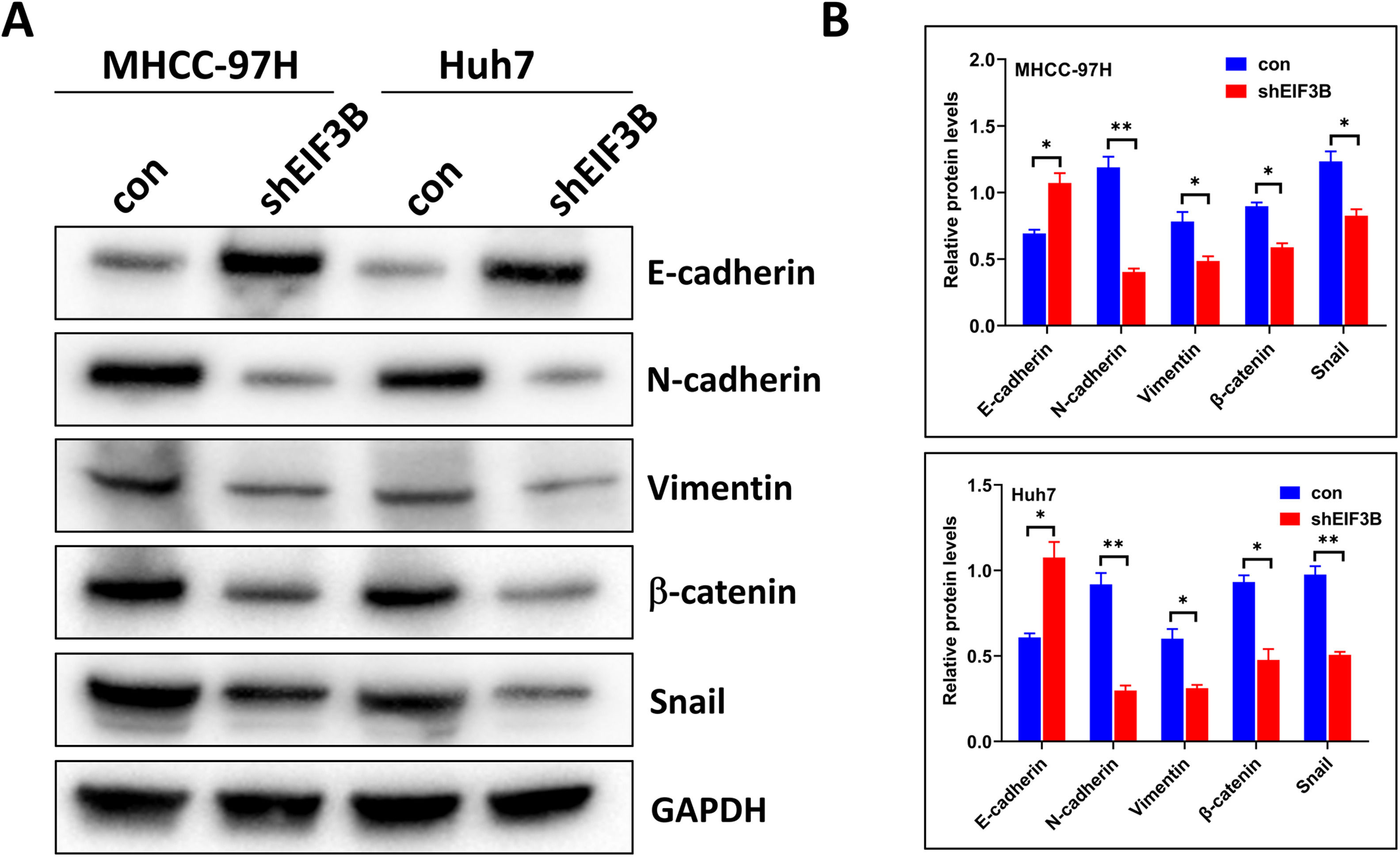
3.3EIF3B knockdown impairs the epithelial-to-mesenchymal transitionof HCC cellsThe epithelial-mesenchymal transition (EMT) plays a key role in the metastasis of various types of tumors. We therefore examined the expression of multiple EMT-associated molecules in MHCC-97H and Huh7 cells that had EIF3B knockdown. Western blotting showed that four mesenchymal markers (N-cadherin, vimentin, β-catenin, and Snail) had significantly lower expression in cells with EIF3B knockdown than control cells. In contrast, the expression of an epithelial marker (E-cadherin) was significantly greater in the MHCC-97H-shEIF3B and Huh7-shEIF3B cells relative to control cells (Fig. 3A and 3B). These results suggest that EIF3B expression promoted the EMT of MHCC-97H and Huh7 cells.
3.4EIF3B knockdown inhibits the expression of TGFBIWe then performed RNA-seq of cells with EIF3B knockdown and control cells to identify the downstream proteins regulated by EIF3B. In this analysis, we used a change in expression of ‘more than 2.0-fold'or ‘less than 2.0-fold’ (based on p<0.05) as the cut-off thresholds, and presented data for genes with the greatest differences in expression as a heat map (Fig. 4A). We then compared the differentially expressed genes (DEGs). Transcriptome analysis showed that relative to control cells, MHCC-97H-shEIF3B cells had 1912 DEGs, Huh7-shEIF3B cells had 567 DEGs, and 160 of the same DEGs were in both cell lines (Fig. 4B). In addition, the RNA-seq analysis indicated that TGFBI was significantly downregulated in MHCC-97H and Huh7 cells following EIF3B knockdown (Fig. 4C).
EIF3B regulates several downstream proteins with potential functions in HCC. A, Heatmap of 2479 mRNAs that were significantly up-regulated (red, +2.0-fold or more change) or down-regulated (blue, −2.0-fold or more change) by EIF3B knockdown in two lines of HCC cells. B, Number of DEGs that were unique to each cell type, and that were common to both cell types. C, Heat map and hierarchical clustering of the top 20 DEGs identified by RNA-seq. D, Western blotting of TGFBI in two lines of HCC cells. E, ELISA of TGFBI in culture supernatants from two lines of HCC cells. Data are mean ± SD; **P<0.01.
We then performed western blotting and ELISA to validate these changes in expression at the protein level. The findings indicated that expression of TGFBI was significantly lower in HCC cells with EIF3B knockdown than control cells (Fig. 4D) and the ELISA results indicated that secretion of TGFBI was significantly less in cells with EIF3B knockdown than control cells (Fig. 4E).
3.5TGFBI functions in EIF3B-regulated HCC invasion and metastasisThere is evidence that TGFBI plays an essential role in the invasion and metastasis of urothelial carcinoma [15]. Therefore, we assumed that EIF3B stimulated the invasion and metastasis of HCC cells by activating TGFBI. To validate this effect in MHCC-97H cells, we restored the expression of TGFBI in MHCC-97H-shEIF3B cells using a TGFBI-overexpressing vector (genetic rescue). The western blotting results demonstrated the overexpression of TGFBI in MHCC-97H cells (Fig. 5A). We then performed chamber migration experiments and modified matrigel invasion assays for 48 h with replenishment of TGFBI expression to compare the extent of cell invasion and metastasis. The results suggested that high-expression of TGFBI obviously increased the invasion and metastasis that were suppressed by EIF3B deletion (Fig. 5B). In addition, the scratch assay experiments confirmed that the restoration of TGFBI expression reversed the impact of EIF3B knockdown on cell migration (Fig. 5C). In summary, these results show that EIF3B increased the invasion and metastasis of HCC cells by increasing the level of TGFBI.
TGFBI overexpression increases cell migration and invasion of MHCC-97H cells. A, Western blotting of TGFBI in control cells, cells with EIF3B knockdown, and cells with EIF3B knockdown and genetic rescue of TGFBI. B, Transwell assays, with or without Matrigel coating, after treatment for 48h, and quantitation of these results. C, Scratch assay of cell migration after treatment for 48h. Data are shown as means ± SDs from three separate experiments. **P<0.01, ***P<0.001, ns: P>0.05.
We then conducted Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of the DEGs that were down-regulated in MHCC-97H-shEIF3B and Huh7-shEIF3B cells. This analysis led to the identification of the top 20 essential GO terms (biological processes, cell components, and molecular functions; Fig. 6A) and the 20 most enriched KEGG pathways (Fig. 6B). These results showed enrichment of the mitogen-activated protein kinase (MAPK) pathway in MHCC-97H-shEIF3B and Huh7-shEIF3B cells.
EIF3B regulates pathways that contribute to HCC progression. A, GO analysis of the molecular functions and biological processes of mRNAs regulated by EIF3B in HCC cells. B, KEGG pathway analysis of key pathways related to EIF3B in HCC. C, Effect of TGFBI expression on the phosphorylation of ERK and MEK in two lines of HCC cells. D, Western blotting was performed to evaluate the expression of shTGFBI and oeEIF3B on the phosphorylation of ERK and MEK in the MHCC-97H cells. E, Migration and Invasion assay of HCC cells after treatment. F, Wound healing assay of HCC cells after treatment. Results are expressed as means ± SDs of three independent experiments. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, ns: P>0.05.
TGFBI may provide a downstream signal to MAPK signaling, a well-established signaling pathway that functions in cell proliferation and apoptosis [16]. We therefore used western blotting to investigate changes in the MAPK signal transduction pathway. The results indicated downregulation of p-MEK and p-ERK in MHCC-97H-shEIF3B and Huh7-shEIF3B cells relative to control cells, but there were no differences in the total levels of MEK and ERK (phosphorylated and non-phosphorylated forms)(Fig. 6C). To further verify whether EIF3B enhances the migration and invasion abilities of hepatocellular carcinoma (HCC) cells through TGFBI, MHCC-97H cells were transfected with TGFBI shRNA and oeEIF3B plasmids for 48 hours. The Western blotting, Wound healing and Transwell migration/invasion assays revealed that upregulation of EIF3B significantly increased the expression of pMEK/ERK and enhanced the migration and invasion potential of HCC cells, whereas downregulation of TGFBI remarkedly attenuated the expression of pMEK/ERK as well as the migration and invasion capacities of HCC cells (Fig. 6D-F). Furthermore, due to the knockdown of TGFBI, the effect of EIF3B on pMEK/ERK expression and its influence on the metastasis of HCC cells were both apparently suppressed (Fig. 6D-F). Therefore, these results indicate that EIF3B activates the MAPK/ERK pathway in HCC cells, primarily regulating the metastasis of HCC via TGFBI rather than through a bystander effect.
4DiscussionPrevious researchers suggested that EIF3B functions as the central scaffolding subunit for the EIF3 complex, which plays an important role in the initiation and promotion of protein translation. We therefore examined the role of EIF3B in HCC, a common malignancy that has high rate of morbidity and mortality worldwide. We focused on the effect of EIF3B by performing in vitro experiments and bioinformatics analysis to determine its effects and potential mechanism.
The TCGA data showed that EIF3B was upregulated in HCC patients and significantly related to tumor grade. Moreover, a high level of EIF3B was associated with shorter OS and DFS, suggesting that EIF3B expression may be a prognostic risk for this cancer. In accordance with the bioinformatics results, our in vitro studies demonstrated that EIF3B expression was obviously elevated in HCC tissues and HCC cell lines relative to matched normal tissues and normal hepatocytes. In addition, we investigated the biological role of EIF3B by determining the influence of EIF3B suppression on cell migration, invasion, and the EMT of HCC and its effect on the MAPK/ERK signaling pathway. Our results suggest that EIF3B has promise for use as a molecular marker for targeted therapy and prognostic evaluation of HCC. Consistent with our results, Ma et al. demonstrated that reduction of EIF3B significantly reduced hyperplasia and metastasis of gastric carcinoma cells in vitro and in vivo [17]. Other researchers reported that EIF3B expression was higher in breast carcinoma and pancreatic adenocarcinoma than corresponding normal tissues, and was also related to poorer patient outcome and tumor progression [18-19].
An important finding in the present study is that EIF3B knockdown suppressed the EMT-like phenotype in human hepatoma cells. The EMT is a pivotal process in the initiation of cancer progression, because it facilitates the spread of tumor cells from the original site to adjacent areas, and contributes to tissue infiltration and the development of metastatic foci [20-21]. Similar to our findings, previous research reported activation of the EMT in gastric cancer after EIF3B overexpression, and that EIF3B knockdown led to decreased cell invasion and migration in endometrial cancer [13,22].
To date, there is little known about how EIF3B promotes HCC invasion and metastasis. We therefore performed RNA-seq to identify the molecular signaling pathways activated by EIF3B in HCC cells. The bioinformatics and in vitro results demonstrated that EIF3B affected tumor invasion and metastasis via the TGFBI/MAPK/ERK signaling pathway. The MAPK/ERK signal pathway is more active during tumor metastasis, and evidence indicates it promotes the proliferation and migration of cancer cells [23]. MAPK signaling is stimulated in various cancers: hepatocellular tumors, ovarian cancer, pancreatic cancer, and breast cancer [24-27]. Abnormal activation of MAPK/ERK signaling also occurs in more than 50% of humans with HCC [28]. Likewise, Chen et al. showed that exosomes that originated from the promotion of MAPK signaling accelerated HCC progression and recurrence by inducing the EMT [29]. However, very few studies examined the effects of EIF3B on MAPK signaling in HCC. We found that HCC cells with EIF3B knockdown had reduced levels of pERK and pMEK, and that this suppressed the malignant characteristics of these cells. These results support the hypothesis that EIF3B promotes the EMT and the aggressive characteristics of hepatic tumor cells by activating the TGFBI/MAPK/ERK pathway.
5ConclusionsOur results showed that EIF3B was dramatically up-regulated in HCC cells and tissues, and that in vitro knockdown of EIF3B repressed the tumorigenicity of HCC cells by impairing cell migration and invasion due to repression of the TGFBI/MAPK/ERK signaling pathway. Thus, our results suggest that EIF3B has promise for use as a therapeutic target in patients with HCC.
Authors’ ContributionsConception and design were by Y.Y and L.W.; investigation by L.W. and C.H.; development of methodology by Z.Z., J.L., and M.C.; acquisition of data were by J.L. and L.Z.; analysis and interpretation of data were done by W.L; writing, review, and revision of article were taken care of by Y.Y., L.W. and C.H.; all coauthors have reviewed and approved of the article before submission.
Funding InformationThis study was supported by the Startup Fund for scientific research, Fujian Medical University (Grand number: 2020QH1234), Technology of Fujian Province (Grant number: 2020J011108), Joint Funds for the Innovation of Science and Technology of Fujian province (Grant number: 2021Y9194, 2022J011048), and the Fujian Provincial Health Technology Project (Grant number: 2020CXA013).