Warburg effect is attracting increasing attention as it is important for cancer progression. However, how cancer cells regulate glucose metabolism through glycolysis is still unknown. Here, we demonstrated the regulatory role of Ras related GTP binding D (RRAGD) in human hepatocellular carcinoma (HCC) cells.
Patients or Materials and MethodsKaplan-Meier’s analysis was used to analyze the correlation between RRAGD expression levels and prognosis of HCC patients from the Cancer Genome Atlas database. Two stable RRAGD knockdown HCC cell lines were created using shRNAs to investigate cancer progression and aerobic glycolysis. Western blot and quantitative reverse transcription polymerase chain reaction were performed to detect the expression levels of RRAGD and MYC.
ResultsRRAGD expression was elevated in HCC patients with poor prognosis. RRAGD knockdown could inhibit the proliferation, invasion and migration of Huh-7 and HepG2 cells. Interestingly, silence RRAGD was able to reduce the glucose uptake, lactate production and extracellular acidification rate of HCC. RRAGD expression level was up-regulated by oncogene MYC in HCC cells.
ConclusionThis study highlights RRAGD as an important cancer-promoting factor for cancer progression and aerobic glycolysis, and thereby it is a potential therapeutic target for HCC intervention.
Eighty to ninety percent of primary liver cancer cases are human hepatocellular carcinoma (HCC), which is one of the most malignant cancer worldwide and causes more than 700,000 deaths each year [1,2]. In the past two decades, the incidence of HCC has been increased one-fold in USA, moreover, 8% of population in the world are at high risk of new HCC development, which are induced by chronically infected with hepatitis viruses and obesity associated metabolic disorders, such as alcoholic and non-alcoholic steatohepatitis [2,3]. In general, HCC is chemotherapy resistant, and there are limited treatment options of surgical resection, local ablation and liver transplantation [1,4]. Multiple drugs have been developed for advanced HCC treatment, such as sorafenib, lenvatinib, regorafenib, and however, which might extend several months of survival time of patients [5–7]. The systematic failure in HCC treatment mainly due to limited understanding of the complicated molecular mechanisms in tumorigenesis of HCC.
Cancer cells can rewrite their metabolic program to promote survival, growth, proliferation, migration, and invasion through multiple pathways and metabolic processes [8–10]. Otto Warburg discovered that cancer cells tend to metabolize glucose through glycolysis, a less efficient process for generating ATP relative to oxidative phosphorylation, even in the presence of adequate levels of oxygen [11,12]. This process was named as Warburg effect (or aerobic glycolysis), which can promote tumor survival and growth with increased lactate production and glucose uptake [13,14]. Since Warburg effect is a major characteristic of cancer, more and more cancer therapeutic agents are being developed to target this pathway. Transcriptional factors play important regulatory role of Warburg effect, such as hypoxia-inducible factors that increase the expression levels of many glycolytic genes [15–17], oncogenic transcriptional factor MYC stimulates aerobic glycolysis and also activates glycolytic genes [18]. Conversely, tumor suppressor p53 can repress the translation of glycolytic genes and cause a reduction of aerobic glycolysis [19]. Although Warburg effect is a major characteristic of cancer cell metabolism, its causal relationship and downstream effectors is still unclear.
Ras-related GTP-binding protein D (RRAGD) is a monomeric GTP or GDP binding protein, which plays critical role in mediating amino acid stimulated mTOR signaling pathway, a key pathway in determination of cell growth and proliferation rate [20]. Recently, RRAGD has been reported involving the progression of ovarian cancer and colon cancer [21,22], however, the role of RRAGD in HCC is still unknown. In current study, we investigated the correlation between RRAGD expression levels and the prognosis of HCC patients, and found that high RRAGD expression closely associated with poor prognosis. The functional analyses demonstrated that RRAGD promotes cell proliferation, invasion and migration, importantly, it facilitates Warburg effect of HCC. Moreover, the RRAGD expression level was up-regulated by MYC, an oncogenic transcriptional factor, in human HCC cells.
2Materials and methods2.1Cell cultureHuh-7 and HepG2, two well-studied human HCC cell lines, were obtained from AcceGen (Fairfield, USA) and American Type Culture Collection (Manassas, USA), respectively. Both cells were cultured in Dulbecco's modified eagle medium (DMEM, Thermo Fisher Scientific, Waltham, USA) with adding 10% fetal bovine serum (FBS, Gibco), 2 mM L-glutamine, 100 μg/mL streptomycin, and 100 IU/mL penicillin in the humidified incubator at 37 °C with 5% CO2.
2.2Generation of stable RRAGD knockdown cell linesThe shRNA constructs of control, RRAGD, and MYC were ordered from GenePharma (Shanghai, China). In order to create stable RRAGD knockdown HCC lines, Viromer® GREEN (OriGene, Rockville, USA) was used to transfect control shRNA or RRAGD shRNAs (RRAGD-KD1 or RRAGD-KD2) into Huh-7 and HepG2 cells, and following the manufacturer’s instructions. Puromycin (Thermo Fisher Scientific, Waltham, USA) was applied to screen cell clones. The positive ones were expanded, and RRAGD expression was determined by Western blot. Stable MYC knockdown Huh-7 and HepG2 cells (MYC-KD1 and MYC-KD2) were created by using the above protocol.
2.3Cell proliferation assayA total of 1 × 103 HepG2 or Huh-7 cells was seeded in 96-well plate. After culturing for 72 h, cell proliferation of each well was measured by using Cell Counting Kit 8 (CCK8) (Beyotime, Shanghai, China) for day 1, 2, 3, and 4, following the manufacturer’s instructions. Data represent three independent experiments.
2.4Colony formation assayControl and RRAGD knockdown HepG2 or Huh-7 cells were resuspended and placed into a 6-well plate with 1 × 103 cells per well. Cells were cultured with normal culture medium for 10–14 days. When observable colonies reached the appropriate confluency, the cells were stained with crystal violet (0.2%)/formaline (10%), and crystal violet signal was quantified at 700 nm using GloMax® Discover Microplate Reader (Promega, Madison, USA).
2.5Transwell migration and invasion assaysInvasion and migration of the treated Huh-7 and HepG2 cells were measured by using 6.5 mm transwell chamber (8 μm pore, BD, San Jose, USA). In brief, serum-free medium was used to suspend HCC cells, which was cultured in the transwell chamber (upper) 5 × 104 cells/well. Complete DMEM media was added in the lower chamber. After 12- or 24 -hs culture, the migrated/invaded cells were fixed using 4% paraformaldehyde in the lower chamber, and then stained with crystal violet (0.1 mg/mL). The Corning matrigel matrix (Corning Inc., Corning, USA) was used for invasion assay. Nikon microscope were used to count cells. Three visual fields were counted randomly for each well, and the average was presented.
2.6Glucose uptake and lactate production measurementControl and RRAGD knockdown Huh-7 or HepG2 cells were measured by BioVision glucose uptake colorimetric assay kit (BioVision Inc., Milpitas, USA), and following the protocol provided by manufacturer. The new fluorescent analog of D-glucose, 2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-2-deoxyglucose (2-NBDG), was applied to glucose uptake determination. Briefly, 2 × 103 cells were placed in 96-well plate and starved with serum free culture medium for 12 h. Cells were then incubated with 100 μL Krebs–ringer–phosphate–HEPES (KRPH) buffer with 2% bovine serum albumin. Thirty minutes later, added 10 μL 2-NBDG (10 mM) to each well and incubated for 20 min. nicotinamide adenine dinucleotide phosphate generated by the treated cells was determined by enzymatic recycling amplification reaction. The absorbance at 412 nm was recorded by using GloMax® Discover Microplate Reader (Promega, Madison, USA) and normalized to protein concentration.
Treated cells were suspended in fresh DMEM medium without pyruvic acid and seeded in 96-well plate with the density of 2 × 103 cells/well. Six hours later, collected the culture medium and diluted by KRPH buffer with the ratio of 1:6. Lactate concentration was determined by lactate colorimetric assay kit (BioVision Inc., Milpitas, US), and following the manufacturer’s protocol. The absorbance at 450 nm was recorded by using GloMax® Discover Microplate Reader (Promega, Madison, USA) and normalized to protein concentration.
2.7Extracellular acidification rate (ECAR) assayECAR was detected by using the seahorse XF96 extracellular flux analyzer (Seahorse Bioscience, North Billerica, USA). Control and RRAGD knockdown Huh-7 or HepG2 cells were placed in an XF96 plate at a density of 1 × 104 cells/well and incubated for 12 h. One hour before measurement, the media were exchanged to XF media. XF Glycolysis Stress Test Kit (Seahorse Bioscience, North Billerica, USA) was used to detect the glycolytic capacity. The Glucose (10 mM), oligomycin (1 mM) and 2-deoxy glucose (2-DG, 50 mM) were diluted into XF media and loaded into the plate at 20-min, 50-min, and 80-min, respectively. ECAR was measured following the manufacturer’s instructions.
2.8RNA extraction and quantitative reverse transcription polymerase chain reaction (qRT-PCR)Total RNA of treated Huh-7 and HepG2 cells was extracted by using the TRIzol™ Reagent (Thermo Fisher Scientific, Waltham, USA). The cDNA was synthesized by using the M-MLV Reverse Transcriptase (Beyotime, Shanghai, China). The qRT-PCR was conducted using Real-time PCR System (Agilent, Santa Clara, USA). The expression levels of RRAGD was normalized by GAPDH, and calculated using the 2−ΔΔCT method. Primers: RRAGD, F: CTAGCGGACTACGGAGACG, R: ATGAGCAGGATTCTCGGCTTC; GAPDH, F: CCACATCGC TCA GAC ACC AT, R: ACCAGGCGC CCA ATA CG.
2.9Western blotTreated Huh-7 or HepG2 cells were lysed using the lysis buffer containing 150 mM NaCl, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate, 1% NP-40, 50 mM Tris (pH7.4), and protease inhibitor cocktails. Target proteins expression was determined by Western blot as previously described [23]. Primary antibody of MYC (ab32072, 1:1000 dilution), RRAGD (PA5-67003, 1:1000 dilution), and β-actin (#4967, 1:3000 dilution) were purchased from Abcam (Cambridge, UK), Thermo Fisher Scientific (Waltham, USA) and Cell Signaling Technology, Inc. (Danvers, USA), respectively. The Western blot results were quantified by using ImageJ software.
2.10Correlation and statistical analysisCorrelation and statistical analysis were performed by using Prism 7.0 software (https://www.graphpad.com/). Kaplan-Meier’s analysis was used to calculate the correlation between RRAGD expression and overall survival (OS) or recurrence-free survival (RSF) of HCC patients from The Cancer Genome Atlas (TCGA) database as previously described [24,25]. One-way and two-way analysis of variance (ANOVA) methods and Dunnett post hoc test were used to in this study. Data were showed as mean ± standard deviation (SD).
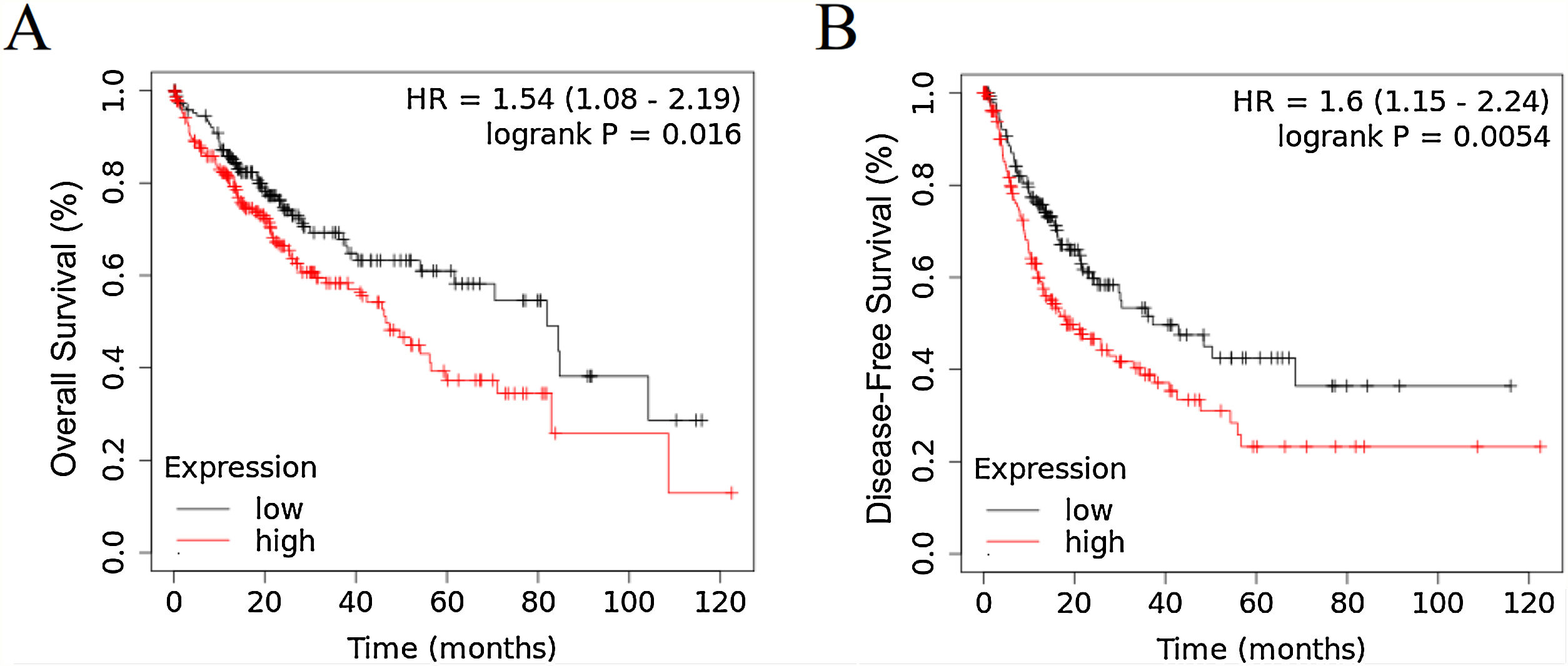
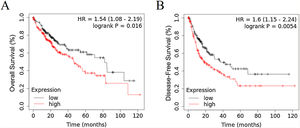
3Results3.1RRAGD expression is elevated in HCC patients with poor prognosisTo investigate the role of RRAGD in tumor prognosis of HCC patients, we analyzed the correlation between RRAGD expression and OS or RFS of HCC patients from TCGA database, a landmark cancer genomics program, by using Kaplan-Meier Plotter package (http://kmplot.com/analysis/) [24]. As shown in Fig. 1A and B, HCC patients with high RRAGD expression had significantly lower overall survival (logrank P = 0.016) and poorer recurrence-free survival (logrank P = 0.0054), respectively.
RRAGD expression level is elevated in HCC patients with poor prognosis. (A) Kaplan-Meier’s analysis of the correlation between RRAGD expression and overall survival (OS) of HCC patients from TCGA. (B) Kaplan-Meier’s analysis of the correlation between RRAGD expression and recurrence-free survival (RFS) of HCC patients from TCGA.
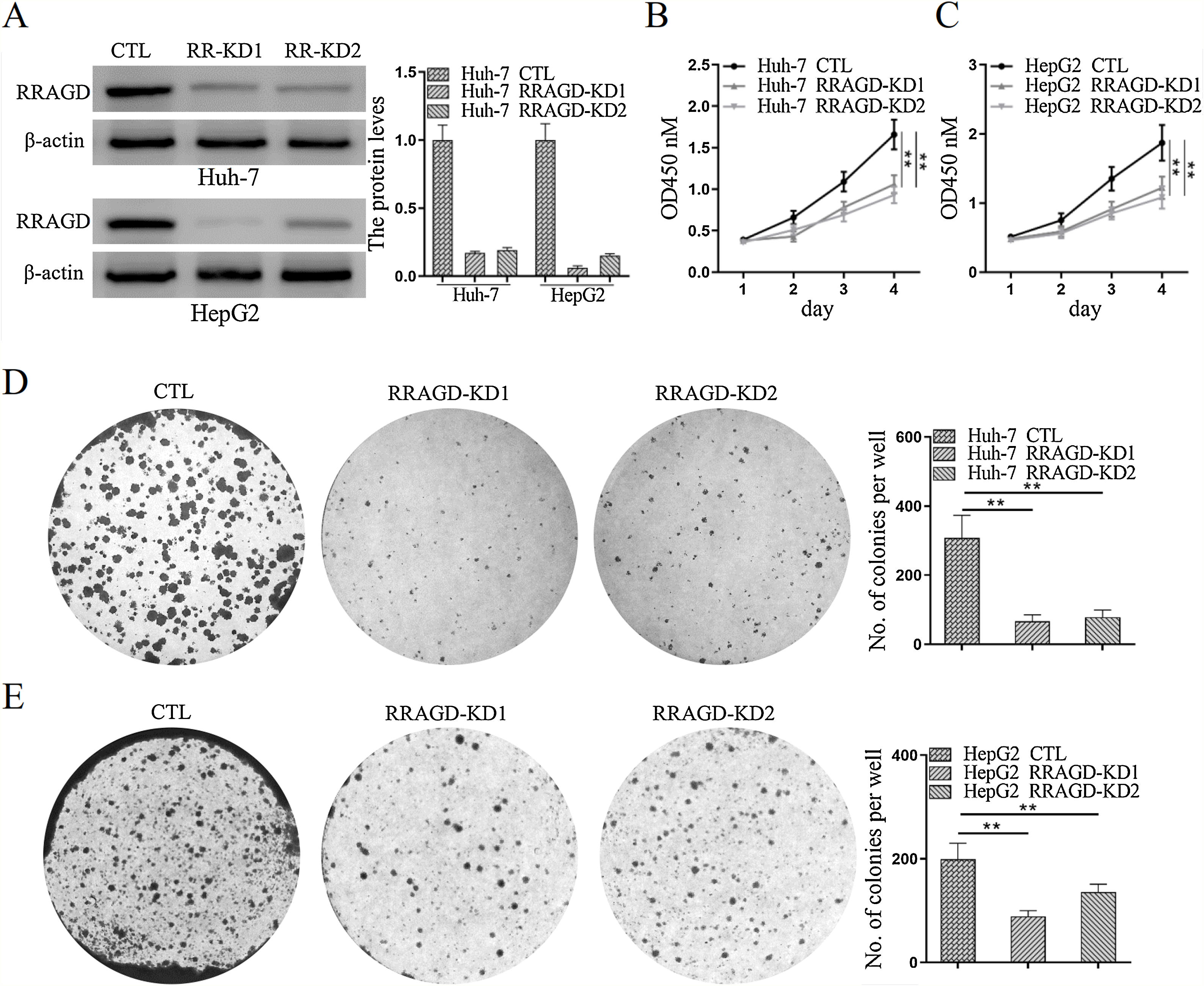
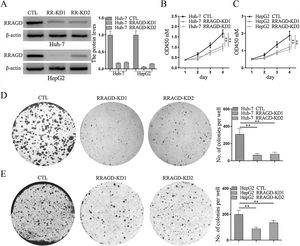
Two stable RRAGD knockdown lines, Huh-7 and HepG2, were created to further examine the functional role of RRAGD in HCC progression (Fig. 2A). CCK8 assay result showed that RRAGD knockdown was able to inhibit HCC cell proliferation significantly (Fig. 2B and C). Moreover, cell colony formation in RRAGD knockdown Huh-7 and HepG2 cells was dramatically decreased compared to that in control groups (Fig. 2D and E). These data suggested that silence of RRAGD could inhibit HCC cells proliferation in vitro.
RRAGD promotes HCC cells proliferation. (A) Western blot and quantification of RRAGD protein level in Huh-7 and HepG2 cells stably expressing Control (CTL) or shRRAGDs. β-actin served as loading controls. (B and C) Cell proliferation assay of Huh-7 and HepG2 cells were measured by CCK8. (D and E) Huh-7 and HepG2 cells were subjected to cell colony formation assay. Data were presented as mean ± S.D. of three independent experiments, ** P < 0.01.
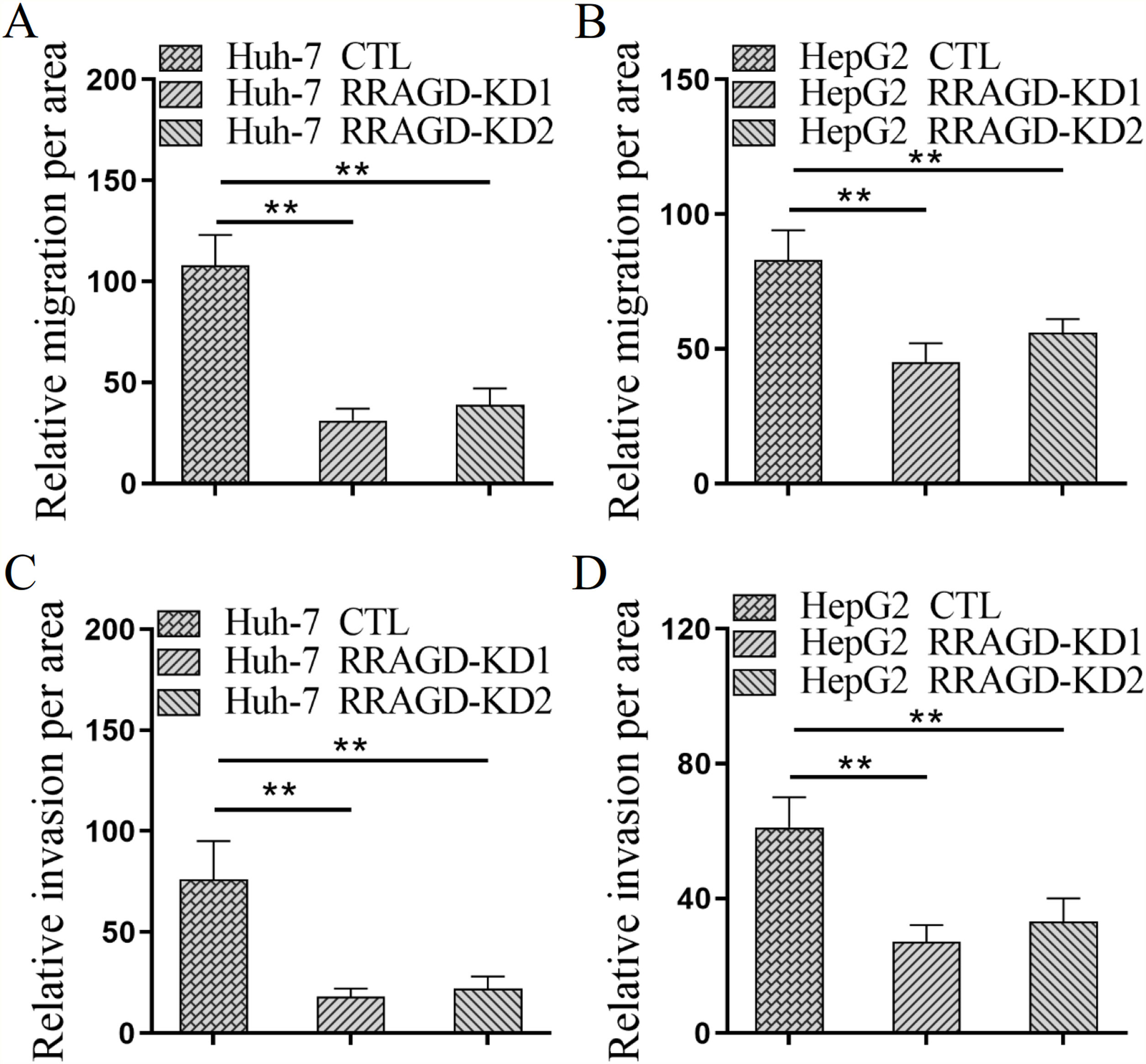
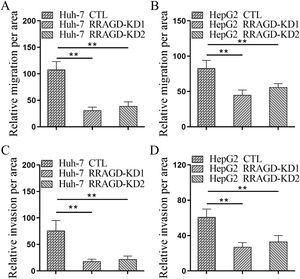
Next, we investigated the effect of RRAGD on HCC cells progression by using transwell migration and invasion assays. As shown in Fig. 3A, the migration ability of RRAGD knockdown Huh-7 cells was significantly reduced relative to control group. Transwell invasion assay showed that the invasion per area of RRAGD knockdown Huh-7 cells was dramatically smaller than that in control group (Fig. 3C). In line with the results of Huh-7 cells, RRAGD knockdown could inhibit the migration and invasion of HepG2 cells significantly (Fig. 3B and D). The above results indicated that RRAGD promotes while silence RRAGD suppresses HCC progression in vitro.
RRAGD promotes HCC cells migration and invasion. (A and B) Huh-7 and HepG2 cells were subjected to transwell migration assay. (C and D) Huh-7 and HepG2 cells were subjected to transwell invasion assay. Data were presented as mean ± S.D. of three independent experiments, ** P < 0.01.
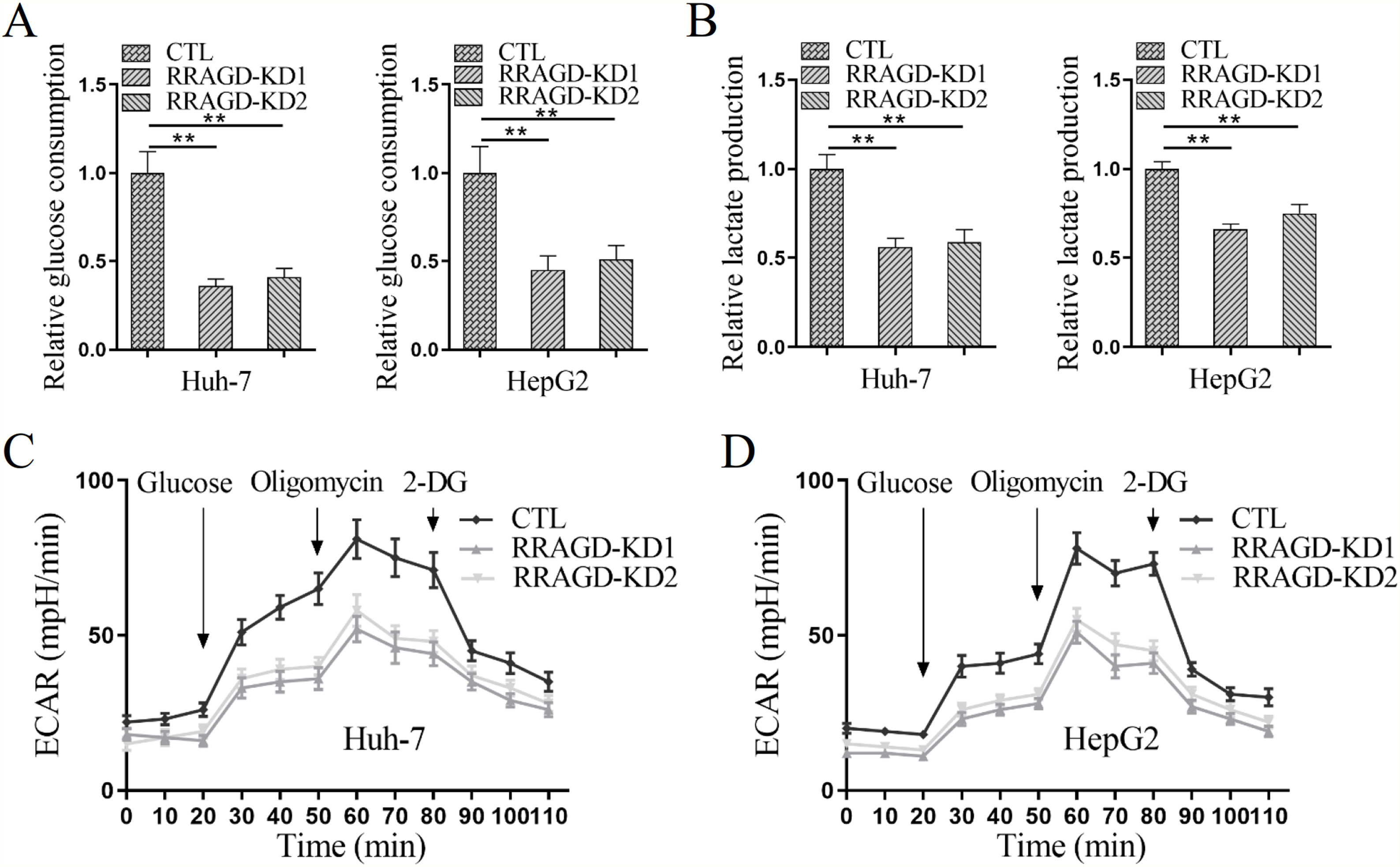
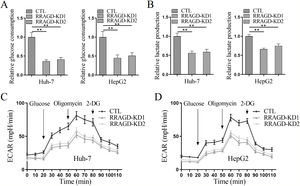
Since the Warburg effect of aerobic glycolysis is a key metabolic hallmark of cancer, we next examine the glucose consumption, lactate production, and extracellular acidification rate (ECAR) in HCC cells with or without RRAGD. Increased the ratio of glucose consumption and lactate production are two major features of tumor metabolism, in comparison with control groups, RRAGD knockdown could significantly reduce glucose consumption ratio (>50% reduction) in both Huh-7 and HepG2 cells (Fig. 4A). Similarly, lactate production ratio was dramatically decreased in RRAGD knockdown HCC cells relative to that in controls (Fig. 4B). Consistently, in the absence of RRAGD, the ECAR was significantly reduced in Huh-7 and HepG2 cells compared to control groups (Fig. 4C and D). Collectively, RRAGD regulates glucose metabolism and Warburg effect in HCC cells.
RRAGD promotes the Warburg effect in HCC cells. (A) The glucose consumption ratio measured in Huh-7 and HepG2 cells with or without RRAGD knockdown. (B) Lactate production ratio in Huh-7 and HepG2 cells with or without RRAGD knockdown. (C and D) ECAR of Huh-7 (C) and HepG2 (D) cells with or without RRAGD knockdown were measured by the Seahorse Bioscience XF96 analyzer. Glucose (10 mM), ATP synthase inhibitor oligomycin (1 μM), and glycolysis inhibitor 2-DG (50 mM) were added to the cells at the indicated time points. Data were presented as mean ± S.D. of three independent experiments. ** P <0.001.
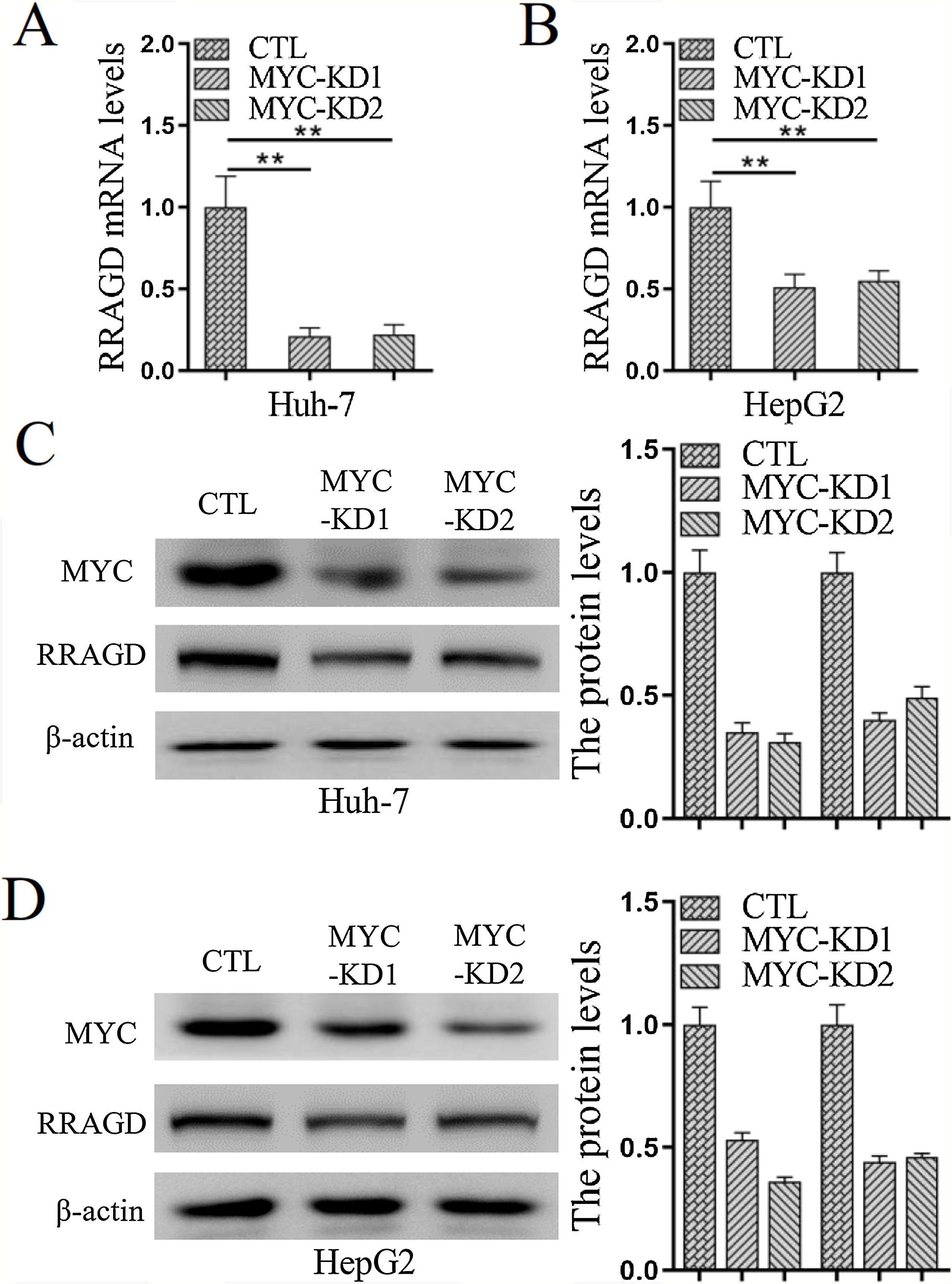
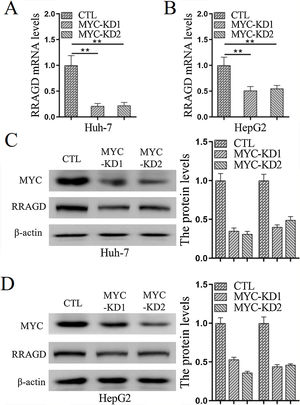
Genomic amplification and overexpression of the MYC oncogene is a common molecular event in HCC, to investigate whether RRAGD contributes to MYC associated tumorigenesis, we created stable MYC knockdown HCC cell lines and then detected the change of RRAGD expression. As predicted, in absence of MYC, the expression of RRAGD were significantly decreased on both mRNA and protein levels in Huh-7 cells compared to that in control group (Fig. 5A and C). Similar pattern of RRAGD reduction after MYC knockdown was observed in HepG2 cells (Fig. 5B and D). These results suggested that RRAGD might be a target of MYC and mediate MYC-induced tumorigenesis.
MYC regulates the expression of RRAGD in HCC cells. (A) The mRNA levels of RRAGD was detected in Huh-7 and HepG2 cells by RT-qPCR. GAPDH was detected as control. (B) Protein levels MYC and RRAGD in Huh-7 and HepG2 cells were detected by western blot and quantified by ImageJ. The β-actin was used as control. Data are mean ± S.D. of three independent experiment and each measured in triplicate, * P <0.05.
Despite there is sufficient levels of oxygen, the rapidly dividing cells, such as tumor cells, convert glucose into lactate, and this cell metabolism known as Warburg effect, which promotes cells to adapt to energetic and anabolic needs for biomass production and to inhibit apoptotic signaling [26–29]. Most of human tumors including HCC display the phenotype of Warburg effect that closely correlates with tumor progression and poor clinical outcomes of patients [30–32]. Interestingly, recent studies demonstrated that Warburg effect might be not a universal feature of proliferating cancer cells, the non-small cell lung cancer cells were remarkably diverse in the rate of glucose utilization and the Lac.Glc ration was as low as 0.3, which indicated that glucose uptake exceeded lactate secretion in these cancer cells [33,34]. All the above findings suggested that the underlying molecular mechanisms between Warburg effect and cancer progression remain largely unclear. Therefore, identifying molecules that regulate Warburg effect in cancer is particularly relevant for developing new therapeutic strategy.
In this study, we demonstrated that the GTP-binding protein RRAGD promotes cancer progression and Warburg effect in HCC cells. Kaplan-Meier analysis of the prognosis of HCC patients from TCGA showed that patients with higher RRAGD expression had poor prognosis, which indicates the potential cancer-promoting role of RRAGD in HCC patients, and RRAGD might be a potential therapeutic target for the treatment of HCC. Accordingly, silence RRAGD in HCC cells significantly inhibit cancer cell proliferation, migration and invasion, reduce glucose uptake and lactate production, and suppress extracellular acidification rate. Interestingly, in HCC cells, we found that RRAGD expression is up-regulated by oncogenic transcriptional factor MYC [35]. RRAGD sustains the survival and proliferation of HCC cells, and thereby it might be a promising target of HCC treatment.
RRAGD involves in the regulation of mTORC1 signaling which is the central component of promoting cell growth in response to insulin, energy, and amino acids levels and is dysregulated in many cancers [36]. In mice, the transcriptional activation of RRAGD enables cellular adaptation to nutrient availability and supports the energy-demanding metabolism, finally result in cell hyperproliferation and cancer growth [20]. In line with the above reports, this study demonstrated that RRAGD expression level was significantly elevated in HCC patients, and RRAGD expression positively correlate with poor prognosis in HCC patients, the patients with high RRAGD expression had shorter overall survival and worsen recurrence-free survival. This finding indicates that RRAGD levels seem to be have critical clinical implications for HCC, and given the positive correlation between high RRAGD level and HCC progression, it is reasonable to infer that mutated hepatocytes may express RRAGD to rewrite the metabolic program and then promote hepatocarcinogenesis.
Warburg effect is attracting increasing attention as it plays important role in liver cancer spread and in response to different therapies [28,37–39]. The expression level of proliferator-activated receptor-gamma coactivator-1alpha, a key transcriptional coactivator in energy metabolism, is decreased in HCC patients, which activate phosphoinositide-dependent kinase-1 through WNT/β-catenin pathway and then promote Warburg effect and cancer progression [12,40,41]. Here, we demonstrated that RRAGD works as a cancer-promoting factor in HCC through activation of cancer cell proliferation and Warburg effect. Knockdown RRAGD could significantly inhibit proliferation, migration and invasion, and reduce glucose uptake and lactate production, two major features of Warburg effect, in HCC cells. Kaplan-Meier analysis also indicated that RRAGD was closely associated with the overall survival of patients with colon cancer, which might be positively associated with the high invasiveness of tumors [42]. Based on ours and others studies, as a GTP/GDP binding G protein, RRAGD acts as molecular switch in variety of processes, such as proliferation, metabolism, and cellular interaction, which can improve cancer cell survival and proliferation, migration, invasion, and cell-in-cell structure formation. These findings suggested that targeting and blocking RRAGD might be a valuable method to suppress HCC progression through inhibiting Warburg effect. Therefore, screening and identifying effective and specific inhibitors of RRAGD have great significance in the future study.
5ConclusionThis study highlights RRAGD as an important cancer-promoting factor for cancer progression and aerobic glycolysis to promote HCC tumorigenesis, and thereby it is a potential therapeutic target for HCC intervention.
Declaration of conflicting interestThe author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
None.