Objectives: This case-control study was done to determine the association and prevalence of p53 codon 249 mutation using cell-free DNA in the plasma of patients with hepatocellular carcinoma (HCC) in South-Western Nigeria. Method: Eighty-five adults with HCC and seventy-seven age and gender matched controls without evidence of liver disease or malignancy involving any part of the body, were recruited. Plasma DNA was analyzed for p53 codon 249 by restriction fragment length polymorphism. Patient evaluation was done by means questionnaire interview, clinical examination, laboratory and radiological tests. The prevalence of the p53 codon 249 mutation was expressed as a percentage amplifiable DNA samples analyzed from HCC patients while that of controls was expressed in the same way. Fisher’s exact test or the student t-test where appropriate were used to assess statistical significance of prevalence between both groups as well as comparison of some characteristics in the HCC cases between those who had codon 249 mutation and those who did not. Associations between the various parameters assessed were determined by odds ratio and significant difference was specified at p < 0.05. Results: p53 codon 249 mutation was present in 6 (7.6%) of the 79 samples from the HCC patients with amplifiable plasma DNA while none (i.e. 0%) of the 73 samples with amplifiable plasma DNA from the controls had this mutation. This prevalence is significantly higher among HCC patients than controls (0.029). The mutation was also found to be significantly associated with HCC (odds ratio = 2.00; 95% C I: 1.70 – 2.35). Conclusion: The prevalence of the p53 codon 249 mutation from plasma DNA of hepatocellular carcinoma patients is significantly higher than among controls in South-Western Nigeria and the presence of this mutation is significantly associated with HCC in this region.
Hepatocellular carcinoma which is a malignant growth of the hepatocytes commonly affects middle-aged men in South-East Asia and sub-Saharan Africa.1-6 The common aetiologic agents associated with this malignancy are hepatitis B virus (HBV), hepatitis C virus (HCV), alcohol and aflatoxin B1 (AFB1). The out-come of this disease is poor as a result of late presentation. To improve the prognosis of this disease, several techniques have been advocated for screening and surveillance of high risk subjects. This is to facilitate early diagnosis at a stage when the disease is potentially curable.2
The only single test that is diagnostic of HCC is histology on liver biopsy specimens or cytology on fine needle aspiration samples from the liver, which are invasive in nature. These procedures are sometimes risky and better avoided in some patients who are not fit to undergo these procedures especially when they present late. Moreover, it may be inconvenient to perform liver biopsy on the same subject at a periodicity of three months which is the time interval advocated presently for screening and surveillance programmes.7,8
Various genetic aberrations have been associated with HCC.9 Of these, the p53 gene mutation is the most studied. A specific mutation in codon 249 of the p53 gene, induced by AFB1 toxicity in which the third base guanine is substituted by thymine (AGG → AGT) is associated with HCC.3,10,11 This leads to the substitution of arginine by serine in the p53 protein causing folding abnormality of the DNA binding domain on the protein. The worldwide prevalence of this genetic abnormality is 11%,9 and has also been found in 13 to 36% of aflatoxin related tumours from South-east Asia.9,12 In sub-Saharan Africa, 36 to 66% of HCC patients had this mutation.13-15In Nigeria, where the rate of food contamination by aflatoxin B1 and the prevalence of HBV viral markers among HCC patients are high, Ndububa et al found this mutation in 1 (5.5%) of the 18 tumour samples taken from HCC patients in South-western Nigeria.16
Most of the genetic analyses done initially were on DNA extracted from tumour tissues because genetic material is stored in the nucleus of nucleated cells. Recent research however, has shown that it is now possible to isolate DNA from body fluids like plasma/serum and urine which carries the same genetic material as the original tumour. Thus, cell-free DNA in the plasma and in the abovementioned body fluids can be used as a surrogate material to detect genetic alterations present in the original tumour.10,11,17 The source of this cell-free DNA is not certain. It is however, thought to be from tumour cells which have undergone necrosis or apoptosis. The objective of this study was to determine the association and prevalence of the p53 codon 249 mutation in plasma DNA of HCC patients.
Materials and methodsEight-five consecutive and consenting adults with HCC who fulfilled the diagnostic criteria for HCC as stated by the European Association for the Study of the Liver (EASL) in Barcelona in the year 2000 were recruited into the study. Seventy-seven age and gender matched adults being managed for ailments other than liver disease or malignancy were also recruited from the Medical Outpatient Department or the Medical wards of the University College Hospital (UCH), Ibadan, Obafemi Awolowo University Teaching Hospital (OAUTH), Ile-Ife and the Lagos University Teaching Hospital (LUTH) Lagos, all in South-western Nigeria.
After obtaining an informed consent from each subject, a questionnaire was administered to obtain socio-demographic data and other relevant clinical history. Clinical examination and relevant laboratory investigations were done.
Genomic DNA was extracted from the plasma using QiAmp DNA blood mini kit according to the manufacturer’s blood and body fluid spin protocol (Qiagen, Hilden, Germany). Purified DNA was eluted from the QiAmp silica column with 200 μL of water (PCR-grade, Sigma, St Louis, MO, USA).
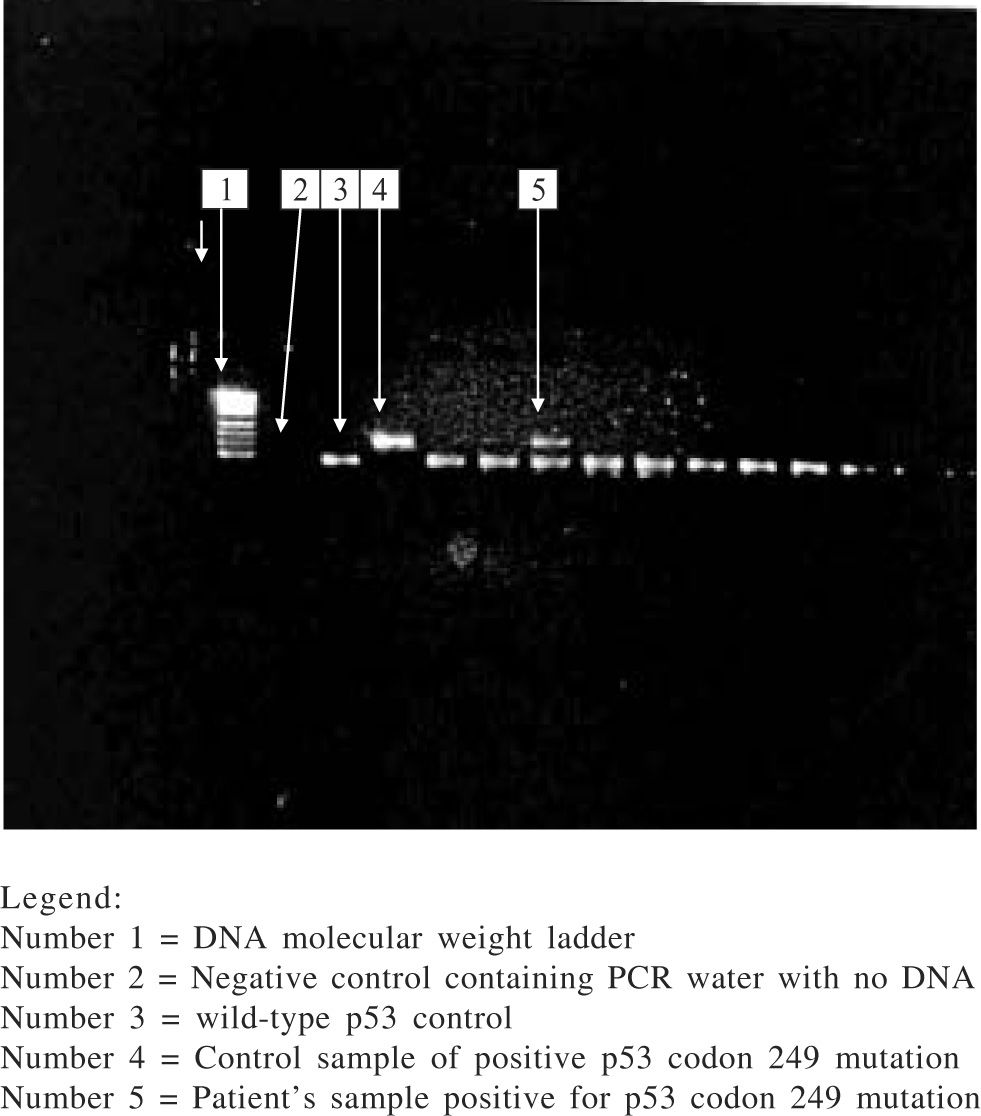
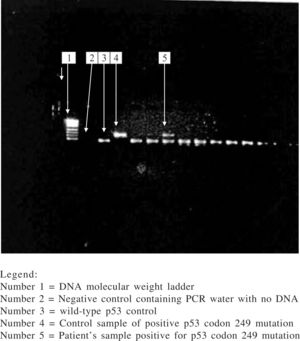
The eluted DNA (8 μL) was amplified by polymerase chain reaction (PCR) for exon 7 of the p53 gene using specific primers: forward primer 5’-CTTGCCACAGGTCTCCCCAA- 3’ and reverse primer 5’ –AGGGGTCACCGGCAAGCAGA- 3’. The protocol for the reaction was 15 minutes denaturation at 95 °C, then 50 cycles of denaturation at 94 °C, primer annealing at 60 °C, and extension at 72 °C at 30 seconds each, followed by a 10-minute final extension step at 72 °C. The amplification product containing 254 base-pairs (bp) was visualized on 3% agarose gel with ethidium bromide. 10 uL of PCR products were then digested with restriction endonuclease Hae III (Boehringer Mannheim GmbH, Mannheim, Germany) which cleaves GG/CC sequence between codon 249 and 250 to generate 92 bp and 66 bp fragments. The presence of codon 249 mutation yields an undigested 158 bp fragment which is identified on 3% agarose gel. Gel bands containing the 249 mutation were excised from samples that had the mutation and following elution of DNA from the gel, nested PCR was done with the primers; (sense) 5' AGGCGCACTGGCCTCATCTT 3' and (antisense) 5' TGTGCAGGGTGGCAAGTGGC 3'. Products of the nested PCR were visualized on 2% agarose gel with ethidium bromide, followed by sequencing of the purified nested PCR product. Sequencing was done by automated, dideoxy sequencing (sequencer AbiPrism 3100, Perkin-Elmer). All PCR and enzyme digestion of PCR products were done twice along with control samples consisting wild-type p53 gene, 249-mutated p53 gene and PCR water as negative control. A third PCR and enzyme digestion was done when discordant results were obtained from the analysis for further clarification.
Statistical analyses were done using SPSS software. The prevalence of p53 codon 249 mutation in HCC Patients was expressed as a percentage of amplifiable DNA samples analysed from HCC patients and that of controls was also expressed in the same way. Association between this genetic mutation and HCC was determined by odds ratio. Fisher’s exact test was used to assess statistical significance of prevalence between both groups. Comparison of some characteristics in the HCC cases between those who had codon 249 mutation and those who did not have this mutation and statistical significance was done using Pearson chi-square, Fisher’s exact test or the student t test where appropriate (p < 0.05).
The University of Ibadan/University College Hospital Institutional Review Committee (UI/UCH IRC) approved the study.
ResultsA total of 162 Nigerian adults were studied. Eightyfive (53.5%) were HCC cases while 77 (47.5) were age and gender matched controls. The mean age of the HCC subjects was 44.82 ± 14.22 years while that of controls was 44.29 ± 14.07 years. Both were statistically similar (p = 0.812). The male: female ratio was similar in both groups (p = 0.693). The peak age incidence was in the 36–45 years age range, with 27.1% of the total HCC subjects belonging to this age range.
Prevalence of p53 codon 249 mutationOne plasma sample in the HCC cases spilled and so the DNA from that sample was not analysed. Of the 161plasma samples analyzed in both groups, exon 7 of the p53 gene could not be amplified in 9 (5.6%) samples; 5 (6.0%) samples in the HCC cases and 4 (5.2%) samples in the control group. More details are shown in table I.
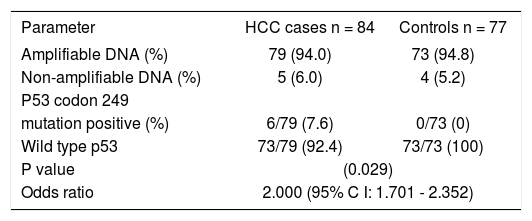
Prevalence of p53 codon 249 mutation.
| Parameter | HCC cases n = 84 | Controls n = 77 |
|---|---|---|
| Amplifiable DNA (%) | 79 (94.0) | 73 (94.8) |
| Non-amplifiable DNA (%) | 5 (6.0) | 4 (5.2) |
| P53 codon 249 | ||
| mutation positive (%) | 6/79 (7.6) | 0/73 (0) |
| Wild type p53 | 73/79 (92.4) | 73/73 (100) |
| P value | (0.029) | |
| Odds ratio | 2.000 (95% C I: 1.701 - 2.352) | |
Among those with amplifiable plasma DNA (i.e.79 HCC cases and 73 controls), 6 HCC subjects had codon 249 mutation (i.e. G → T transversion) in the p53 gene while none of the subjects in the control group had this mutation. The prevalence of p53 codon 249 mutation among HCC subjects was 7.6% while it was 0% among controls and the difference in prevalence between the two groups was statistically significant (p = 0.029). The presence of this mutation was significantly associated with HCC. Those with the p53 codon 249 mutation were twice at risk for having HCC than those who did not have this mutation (odds ratio: 2.000; 95% confidence interval: 1.701–2.352). Direct DNA sequencing of the purified nested PCR product of the codon 249 mutated samples did not show any other mutation in this gene. The electrophoretic pattern of enzyme digestion of PCR product seen on agarose gel is shown in figure 1.
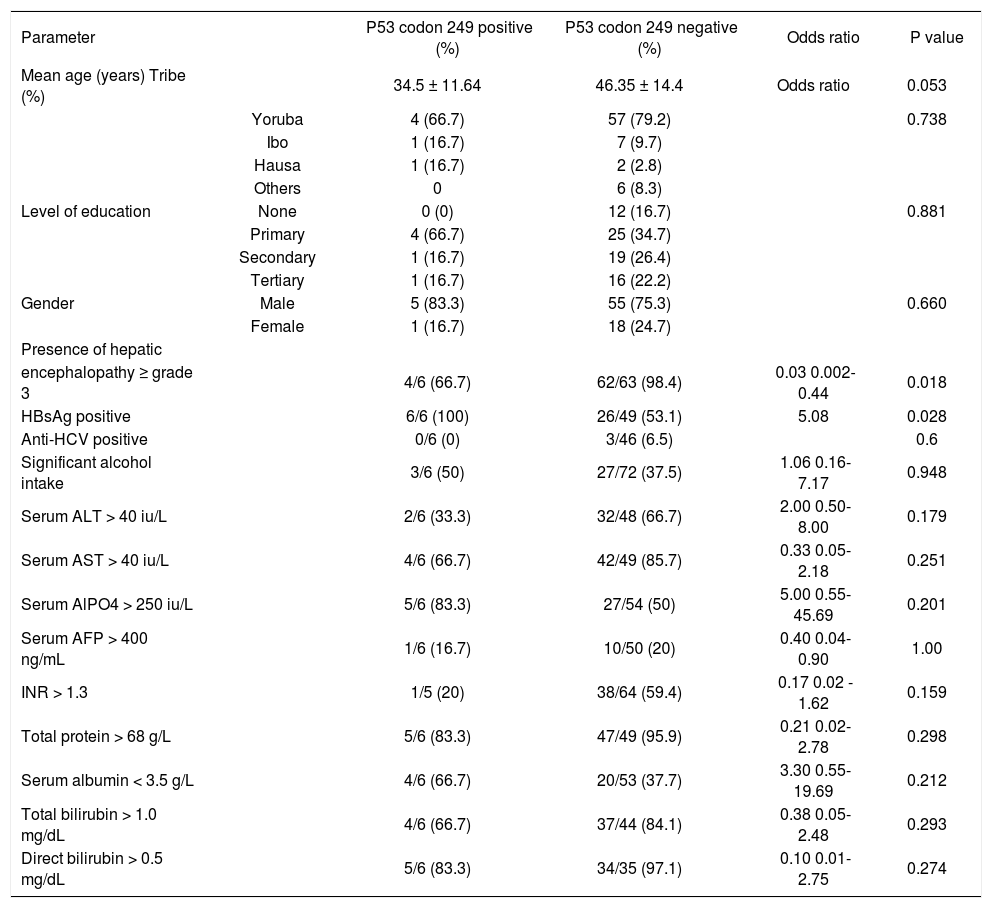
The characteristics of hepatocellular carcinoma patients based on p53 codon 249 statusIn an attempt to further characterize the HCC patients who had p53 codon 249 mutation, comparison was made between this group of subjects and the rest of the HCC patients who were negative for this mutation based on some clinical and biochemical parameters. The patients with this genetic mutation had a mean age of 34.5 ± 11.64 years and seemed to be younger than the HCC patients who were negative for this mutation (p = 0. 053). Five of the 6 HCC patients who had the p53 codon 249 mutation were males while 1 was female. The gender distribution between this group and the rest of the HCC patients was similar (p = 0.660). Both groups of patients were similar in tribal distribution and the level of education attained (p = 0.738 and 0.881 respectively). All the six subjects who had p53 codon 249 mutation were positive for hepatitis B surface antigen (HBsAg) and this proportion of HCC subjects was significantly higher than the rest of the HCC subjects who were HBsAg-positive but did not have this mutation (p = 0.035). It was not possible to evaluate the risk associated with the development of this mutation in HBsAg-positive patients for logistic reasons. The codon 249 mutation-positive patients who drank significant amount of alcohol and those who were anti-HCV positive were similar to the rest of the HCC patients (p = 0.948 and 0.6 respectively). None of these two factors was associated with the development of this genetic mutation. Though, a high proportion of the codon 249 mutation-positive HCC patients had elevated liver enzymes and reversal of the albumin: globulin ratio, they were similar to the rest of the HCC patients who were negative for this mutation. More HCC patients with codon 249 mutation had hepatic encephalopathy ≥ grade 3 on presentation than the rest of the HCC patients (p = 0.018) (Table II).
Characteristics of hepatocellular carcinoma patients based on p53 codon 249 status.
| Parameter | P53 codon 249 positive (%) | P53 codon 249 negative (%) | Odds ratio | P value | |
|---|---|---|---|---|---|
| Mean age (years) Tribe (%) | 34.5 ± 11.64 | 46.35 ± 14.4 | Odds ratio | 0.053 | |
| Yoruba | 4 (66.7) | 57 (79.2) | 0.738 | ||
| Ibo | 1 (16.7) | 7 (9.7) | |||
| Hausa | 1 (16.7) | 2 (2.8) | |||
| Others | 0 | 6 (8.3) | |||
| Level of education | None | 0 (0) | 12 (16.7) | 0.881 | |
| Primary | 4 (66.7) | 25 (34.7) | |||
| Secondary | 1 (16.7) | 19 (26.4) | |||
| Tertiary | 1 (16.7) | 16 (22.2) | |||
| Gender | Male | 5 (83.3) | 55 (75.3) | 0.660 | |
| Female | 1 (16.7) | 18 (24.7) | |||
| Presence of hepatic | |||||
| encephalopathy ≥ grade 3 | 4/6 (66.7) | 62/63 (98.4) | 0.03 0.002-0.44 | 0.018 | |
| HBsAg positive | 6/6 (100) | 26/49 (53.1) | 5.08 | 0.028 | |
| Anti-HCV positive | 0/6 (0) | 3/46 (6.5) | 0.6 | ||
| Significant alcohol intake | 3/6 (50) | 27/72 (37.5) | 1.06 0.16-7.17 | 0.948 | |
| Serum ALT > 40 iu/L | 2/6 (33.3) | 32/48 (66.7) | 2.00 0.50-8.00 | 0.179 | |
| Serum AST > 40 iu/L | 4/6 (66.7) | 42/49 (85.7) | 0.33 0.05-2.18 | 0.251 | |
| Serum AlPO4 > 250 iu/L | 5/6 (83.3) | 27/54 (50) | 5.00 0.55-45.69 | 0.201 | |
| Serum AFP > 400 ng/mL | 1/6 (16.7) | 10/50 (20) | 0.40 0.04-0.90 | 1.00 | |
| INR > 1.3 | 1/5 (20) | 38/64 (59.4) | 0.17 0.02 - 1.62 | 0.159 | |
| Total protein > 68 g/L | 5/6 (83.3) | 47/49 (95.9) | 0.21 0.02-2.78 | 0.298 | |
| Serum albumin < 3.5 g/L | 4/6 (66.7) | 20/53 (37.7) | 3.30 0.55-19.69 | 0.212 | |
| Total bilirubin > 1.0 mg/dL | 4/6 (66.7) | 37/44 (84.1) | 0.38 0.05-2.48 | 0.293 | |
| Direct bilirubin > 0.5 mg/dL | 5/6 (83.3) | 34/35 (97.1) | 0.10 0.01-2.75 | 0.274 |
The prevalence of the p53 codon 249 mutation is significantly higher among hepatocellular carcinoma patients and its presence is associated with a two fold risk for HCC among this group of patients in South-western Nigeria. The observed prevalence in this study is however, lower than those reported earlier in some countries in South-east Asia and sub-Sahara Africa9,12-15 though, consistent with a previous observation among HCC subjects from the same region.16
The reason for this relatively lower prevalence rate compared with previous studies is not likely to be due to the age and gender characteristics of the subjects recruited into this study which is similar to those observed in these studies.9,12-15 The sample size in the index study is not small compared with the number of subjects recruited into those studies and is therefore unlikely to be responsible for the prevalence rate observed in this study. The prevalence of this genetic mutation in the index study may not be related to the fact that genomic analysis was done on plasma DNA. Other studies where the prevalence of this genetic mutation has been determined using plasma DNA have reported higher values than that observed in this study.3,10,11,15 Moreso, previous studies on p53 codon 249 mutation have shown a concordance rate of 88–90% between paired tumour and plasma samples.10,11A more sensitive technique than that utilized in this study such as short oligonucleotide mass analysis (SOMA) may have detected a higher rate than what is currently observed in this study. Such observation was made by Lleonart et al.18 This however, is unlikely to explain the dissimilarity in prevalence rates between the index study and those earlier studies3,11,15 as the observed prevalence rates were obtained by similar technique as that used in this study.
The high prevalence of this genetic mutation among HCC patients which has been implicated as a molecular pathogenetic mechanism for HCC in countries in the above-mentioned regions has also been associated with consumption of food heavily contaminated by Aflatoxin B1 as well as HBV infection endemicity.19,20 The source of aflatoxin is food items which are contaminated by aflatoxin. The relatively lower prevalence value obtained in this study is similar to the prevalence rate of toxic levels of serum aflatoxin of 8.2 and 9% of rural and urban dwellers respectively in this same region (South-western Nigeria).21 It is however, incompatible with the degree of food contamination by aflatoxin reported earlier in South-western Nigeria where most of the food items such as yam, cassava and other processed food derived from these tuber crops as well as palm-oil and smoked-fish which are the staple food consumed have been reported to be heavily contaminated by aflatoxin.22 The food items which are most likely to be contaminated are grains and other items which are processed by drying and stored under poor conditions which favour the growth of Aspergillus flavus, the fungus which produces aflatoxin.
Groundnut has also been reported to be the most heavily contaminated food item by aflatoxin.22,23 Some authors have associated the amount of groundnut ingested daily with significant levels of serum aflatoxin and the presence of this genetic mutation.24 A higher prevalence rate of this genetic mutation may have been observed if this study was done in Northern Nigeria where grains and groundnut are the staple food. One limitation in this study is insufficient data to quantify the daily consumption of groundnuts by the subjects. Based on the tribal distribution of the subjects, those from Northern Nigeria in this study who had this genetic abnormality were similar to those of the same region who had wild type p53 gene. Though the inference from this study may not be reliable as a result of the few subjects who were positive for this mutation, the role of tribal origin and dietary staple in the acquisition of this genetic mutation need to be further evaluated.
Current evidence suggests that following exposure to aflatoxin, the susceptibility to having this genetic mutation and subsequently, HCC is determined by the detoxifying effect of the enzymes responsible for its metabolism; epoxide hydrolase (EPHX) and gluthathione Stransferase Ml (GSTM1) and the efficacy of the DNA repairing process.25 Mutations in the genes coding for these enzymes have been found to be overrepresented in individuals with high serum levels of AFB1-albumin adducts and HCC subjects.26,27 The EPHX and GSTM1 genotypes of the HCC subjects in this environment are not known. The presence or absence of mutations in the genes coding for these enzymes and its contribution to the low prevalence of p53 codon 249 in this region also require further evaluation.
All the HCC patients with p53 codon 249 mutation were HBsAg positive and this was significantly higher than the HCC patients who were negative for this genetic mutation. Significant association between HBsAg positivity and this genetic mutation could not be evaluated statistically for logistic reasons. Previous studies have demonstrated a combined effect of these factors on HCC, where a multiplicative effect of both factors was observed.24 The infection by HCV does not seem to play a role in the development of the p53 codon 249 mutation as none of those who had this mutation was positive for anti-HCV. No study has so far reported an association between this genetic mutation and HCV infection.15,28 There was also no association between significant alcohol ingestion and the p53 codon 249 mutation. This observation is consistent with that in earlier studies where low prevalence of this genetic mutation have been reported in places where significant alcohol ingestion is highly prevalent among HCC patients.15,28
This is the first report of p53 mutation using cell-free DNA in Nigeria. The observed prevalence rate of this genetic mutation among HCC subjects in this study is however, quite similar to that from analysis of 18 tumour samples obtained by liver biopsy, where the same technique of RFLP was employed.16 This study was done in South-western Nigeria, the same geographical location where UCH is also located. The similarity in findings from both studies (i.e. prevalence rates of 5.516 and 7.6% in this study) are not unexpected, as both sample populations were derived from similar ethnic backgrounds and share a common staple diet rich in tuber.
From the observed prevalence rate of the p53 codon 249 mutation in this study, it seems unlikely to be the major molecular hepatocarcinogenetic pathway for HCC in South-western Nigeria.
Despite the low observed prevalence rate of this genetic mutation, it could still be useful as early molecular screening diagnostic tool for HCC because of its significant association with this disease. Moreso, this irreversible genetic mutation has been observed in HCC patients 1 year prior to histological diagnosis.10,24 The use of serological studies such as the antibody to the p53 protein may not be very useful as similar proportions of HCC patients and controls at the UCH, Ibadan have been found to have detectable titre levels of this antibody. It is not surprising because of a possible high rate of false positive antibody reaction. This may be due to the high exposure rate to frequent infections such as malaria which is holoendemic and other parasitic infections with subsequent high circulating antibody titre. A similar explanation has been given for the high rate of false-positive anti-HCV status among sickle cell disease patients by Mutimer et al.30 Otegbayo et al reported that though reduced transferrin and increased alpha 2 macroglobulin in HBV carriers might suggest active liver disease, these two in addition to haptoglobulin lack predictive value for the development of HCC in HBV carriers.31
Since circulating cell-free DNA could be found in the plasma of a large number of HCC patients and controls as observed in this study, genomic analysis using cell-free DNA could be adopted as a convenient method of screening and early diagnosis for HCC. In view of this, more molecular studies on HCC are therefore suggested to identify the major hepatocarcinogenetic pathway which could be a useful biomarker for screening, surveillance and early diagnosis. This would not only improve the out-come of the disease which is poor currently but could also serve as a molecular target for therapy.
Little information was obtained from further characterization of the HCC patients with this genetic abnormality. This may be as a result of the small number of patients who had this mutation. Such information would have been desirable to establish the role of this genetic mutation in the clinical presentation of this group of patients and the prognosis of the disease. A higher proportion of HCC patients with the p53 codon 249 mutation had elevated liver enzymes and a higher grade of hepatic encephalopathy than the rest of the HCC patients whereas Kirk et al observed no difference in the range of liver enzymes in patients with this genetic mutation in Gambia.24 It is therefore suggested that a study involving a larger study population be done to further evaluate the role of this genetic abnormality in HCC in South-western Nigeria.
AcknowledgementWe duly acknowledge the UICC, Switzerland for the award of a travel grant to France for the laboratory analysis of p53 codon 249 mutation. Mrs. Michelle Wrisez, Mrs. Ghyslaine Martel-Planche and other laboratory staff of the Molecular Carcinogenesis Unit, International Agency for Research on Cancer (IARC), are acknowledged for their assistance at various stages of this study. Professor D.O Olaleye and Dr GN Odaibo are appreciated for providing storage facility for the plasma samples. The medical and nursing staff of the Liver Unit in UCH are also appreciated.