Introduction. Hypermethylation of relevant genes may affect the prognosis of patients with cancer. The purpose of this study was to analyze whether methylation of the promoter regions of cell cycle regulators as well as elevated α-Fetoprotein (AFP) levels are useful prognostic factors for patients with hepatocellular carcinoma (HCC).
Material and methods. Nested methylation-specific PCR (nested-MSP) was used to analyze methylation status of the promoter regions of p15, p16, p21, p27, and ras-association domain family 1 (RASSF1A) genes in tumor specimens from 50 patients with HCC.
Results. Promoter methylation was most common in the RASSF1A gene (96%), followed by the p16 gene (56%), the p21 gene (44%), the p15 gene (28%), and the p27 gene (2%). Patients with a serum AFP level < 400 ng/mL and an unmethylated p21 promoter had a better prognosis than patients with a serum AFP level ≥ 400 ng/mL and a methylated p21 promoter (overall survival, p = 0.076; disease-free survival, p = 0.016). In addition, patients with full methylation of the promoter region of RASSF1A had a better prognosis than patients with a partially methylated or unmethylated RASSF1A promoter region if their serum AFP level was ≥ 400 ng/mL (overall survival, p = 0.028; disease-free survival, p = 0.078).
Conclusion. A partially methylated or unmethylated RASSF1A promoter as well as elevated serum AFP level or methylation of p21 in addition to elevated serum AFP level might be associated with poor prognosis in patients with hepatocellular carcinoma.
Hepatocellular carcinoma (HCC) is the sixth most common cancer worldwide and is the third most common cause of cancer-related death.1 Approximately 600,000 people are diagnosed with hepatocellular carcinoma every year. Furthermore, a marked difference has been found in the geographic distribution of HCC, especially in the Far East and Southeast Asia where viral hepatitis is more prevalent.2 Despite improvements in the detection and treatment of HCC, the prognosis of patients with HCC is still very poor.
Promoter hypermethylation has been found in tumor suppressor genes involved in many different signaling pathways in different tumor types.3–6 Epigenetic modification has been identified as a crucial event in carcinogenesis.7 Aberrant methylation of CpG islands in promoters is associated with transcriptional inactivation of genes involved in all aspects of tumor development. Genes involved in DNA damage response, cell cycle control, apoptosis signaling, drug metabolism, detoxification, angiogenesis, DNA repair, intercellular adhesion, and tissue invasion can frequently become methylated and epigenetically silenced in tumors.8 Epigenetic silencing mediated by CpG island methylation is, therefore, a potential therapeutic target as well as a potential prognosticator.8
Cyclin-dependent kinase inhibitors are potent negative regulators of G1/S transition. There are two families of CDK inhibitors. One is the INK4 family, which comprises p15, p16, p18 and p19, and the other is the KIP/CIP family, which comprises p21, p27 and p57.9 Hypermethylation of p15 and p16 is frequently detected in hepatocellular carcinoma.10–12 Negative expression of p21 protein has been shown to be associated with poor prognosis, and it was suggested that p21 protein is an independent survival prognostic factor for HCC.13 The expression of p27 protein alone was shown to predict disease recurrence, indicating that it could be used as an independent prognostic marker for disease-free survival in HCC.14 Mutations within the coding regions of the p21 and p27 genes were not detectable in a large series of human tumors.15,16 Therefore, under-expression of both p21 and p27 proteins in tumor tissues might not be due to mutations in the structural genes. Rather, under-expression might be due to hypermethylation of the genes. RASSF1A (Ras association domain family 1 isoform A) is a tumor suppressor.17 However, hypermethylation of the RASSF1A promoter has long been demonstrated in many liver diseases,18–21 including HCC, cirrhosis, and hepatocellular nodules (HN).22
To the best of our knowledge, no studies have examined whether methylation status of cell cycle regulators and elevated serum α-Fetoprotein (AFP) levels are prognostic factors for patients with hepatocellular carcinoma. The purpose of this study was to analyze whether methylation of the promoter regions of cell cycle regulators (including p15, p16, p21, p27 and RASSF1A) as well as elevated AFP levels are useful prognostic factors for patients with HCC.
Materials and MethodsPatients and specimensHCC tumor specimens were obtained by surgical excision from 50 patients (35 males and 15 females) between 2002 and 2004 at the Taichung Veterans General Hospital, Taichung, Taiwan. Institutional Review Board (IRB) approval and informed consent were obtained. The patients with HCC ranged in age from 36 to 80 years and had a mean age of 58.2 ± 11.9 years. Genomic DNA was extracted with Trizol® Reagent (Invitrogen) according to the manufacturer’s instructions.
Sodium bisulfite modificationGenomic DNA was treated with sodium bisulfate using the MethylEasy™ Xceed Rapid DNA Bisulphite Modification Kit (Human Genetic Signatures Pty Ltd) according to the manufacturer’s instructions.
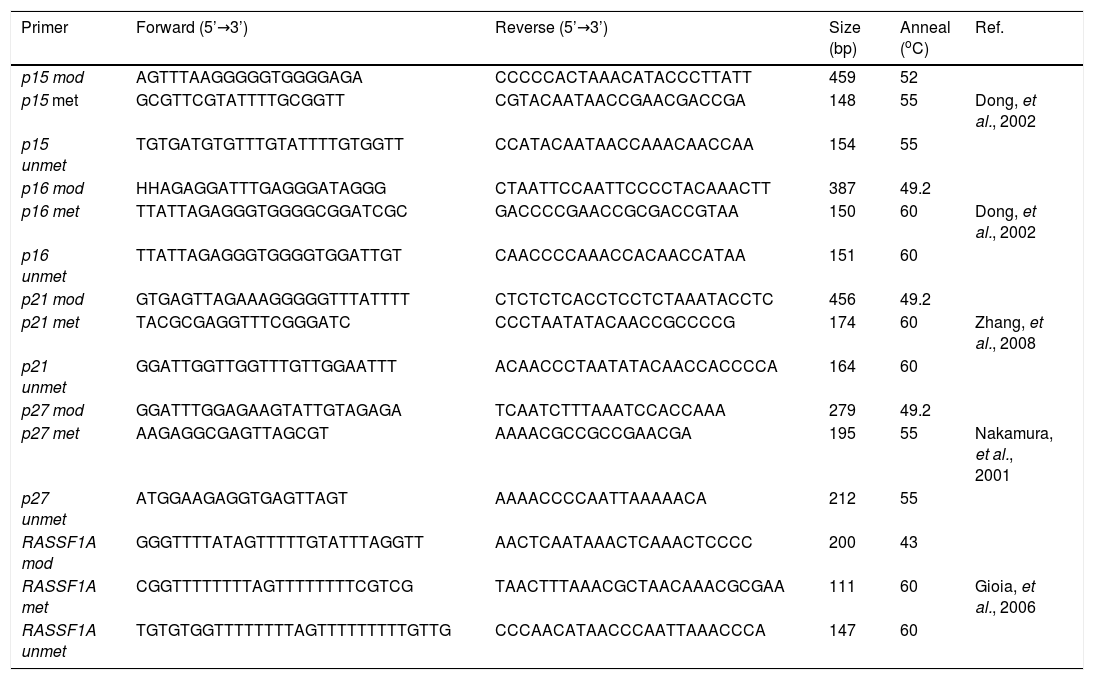
Nested methylation-specific polymerase chain reaction (Nested-MSP)The p15, p16, p21, p27 and RASSF1A promoter regions were subjected to nested methylation-specific polymerase chain reaction (nested-MSP) in a GeneAmp PCR system 2,400 thermal cycler (Applied Biosystems, Foster City, CA, USA). For the first-round, PCR reaction was carried out in a total volume of 50 with 100 ng modified DNA. The PCR mixture contained 200 µΜ dNTP, 5 µΜ primer, 100 ng DNA, 20 mM Tris-HCl (pH 8.4), 50 mM KCl, and 1 unit of Platinum® Taq DNA Polymerase (Invitrogen™). The primers for amplification of the promoter regions were designed to distinguish between bisulfite-sensitive and bisulfite-resistant modifications of unmethylated and methylated cytosines, respectively (Table 1). Thermal cycling conditions were as follows: initial heat denaturing step was 5 minutes at 96 oC, the second step was 25 cycles of 96 oC for 30 sec, then 43-50 oC for 30 sec, then 72 oC for 30 sec, and the final extension at 72 oC for 10 minutes. The PCR products were purified with the PCR Clean Up Kit (GeneMark, Inc., Tai Chung Hsien, Taiwan, R.O.C) according to the manufacturer’s instructions. For the second round, PCR reaction was carried out in a total volume of 25 µL with 50 ng purified PCR products. The PCR mixture contained 100 μΜ dNTPs, 2.5 µΜ primer, 50 ng DNA, 10 mM Tris-HCl (pH 8.4), 25 mM KCl, and 0.5 unit Super-Therm Taq DNA Polymerase (Hoffman-La-Roche). Thermal cycling conditions were as follows: initial heat denaturing step was 5 minutes at 96 oC, the second step was 30 cycles of 96 oC for 30 sec, then 55-60 oC for 30 sec, then 72 oC for 30 sec, and the final extension at 72 oC for 10 minutes. The second PCR products were separated by 2.5% agarose gel electrophoresis with 0.5 X TBE and stained with SYBR® Green I solution for visualization under UV illumination. Epi-Tect® control DNA (human), methylated DNA, and unmethylated DNA (QIAGEN®, Taipei, Taiwan) were used as positive and negative controls. Water was also used as a negative control in the nested-MSP.
Primer sequences used for nested-MSP analysis.
| Primer | Forward (5’→3’) | Reverse (5’→3’) | Size (bp) | Anneal (oC) | Ref. |
|---|---|---|---|---|---|
| p15 mod | AGTTTAAGGGGGTGGGGAGA | CCCCCACTAAACATACCCTTATT | 459 | 52 | |
| p15 met | GCGTTCGTATTTTGCGGTT | CGTACAATAACCGAACGACCGA | 148 | 55 | Dong, et al., 2002 |
| p15 unmet | TGTGATGTGTTTGTATTTTGTGGTT | CCATACAATAACCAAACAACCAA | 154 | 55 | |
| p16 mod | HHAGAGGATTTGAGGGATAGGG | CTAATTCCAATTCCCCTACAAACTT | 387 | 49.2 | |
| p16 met | TTATTAGAGGGTGGGGCGGATCGC | GACCCCGAACCGCGACCGTAA | 150 | 60 | Dong, et al., 2002 |
| p16 unmet | TTATTAGAGGGTGGGGTGGATTGT | CAACCCCAAACCACAACCATAA | 151 | 60 | |
| p21 mod | GTGAGTTAGAAAGGGGGTTTATTTT | CTCTCTCACCTCCTCTAAATACCTC | 456 | 49.2 | |
| p21 met | TACGCGAGGTTTCGGGATC | CCCTAATATACAACCGCCCCG | 174 | 60 | Zhang, et al., 2008 |
| p21 unmet | GGATTGGTTGGTTTGTTGGAATTT | ACAACCCTAATATACAACCACCCCA | 164 | 60 | |
| p27 mod | GGATTTGGAGAAGTATTGTAGAGA | TCAATCTTTAAATCCACCAAA | 279 | 49.2 | |
| p27 met | AAGAGGCGAGTTAGCGT | AAAACGCCGCCGAACGA | 195 | 55 | Nakamura, et al., 2001 |
| p27 unmet | ATGGAAGAGGTGAGTTAGT | AAAACCCCAATTAAAAACA | 212 | 55 | |
| RASSF1A mod | GGGTTTTATAGTTTTTGTATTTAGGTT | AACTCAATAAACTCAAACTCCCC | 200 | 43 | |
| RASSF1A met | CGGTTTTTTTTAGTTTTTTTTCGTCG | TAACTTTAAACGCTAACAAACGCGAA | 111 | 60 | Gioia, et al., 2006 |
| RASSF1A unmet | TGTGTGGTTTTTTTTAGTTTTTTTTTGTTG | CCCAACATAACCCAATTAAACCCA | 147 | 60 |
mod: modification. met: methylation. unmet: unmethylation.
The χ2 test, hazard ratios (HR), and the Fisher’s exact test were used to compare differences in methylation of the promoters of the p15, p16, p21, p27 and RASSF1A genes. The overall survival and disease-free survival of HCC patients were examined by the Kaplan-Meier method and the log-rank test. Two-tailed p values of < 0.05 were considered statistically significant. All statistical analyses were carried out using SPSS 17.0 software.
ResultsFrequency of methylation status of tumor suppressor genes in patients with HCCNested-MSP was performed on HCC specimens from 50 patients with HCC to investigate methylation of the promoter regions of cell cycle regulators, namely p15, p16, p21, p27 and RASSF1A. The nested-MSP results were defined as follows: methylation was defined in products that showed evidence of complete methylation, partial methylation was defined in products that showed evidence of both methylated and un-methylated PCR products, and no methylation was defined in products that showed no evidence of methylation. Representative examples of nested-MSP results are presented in figure 1 and the overall results are summarized in figure 2. The frequency of promoter methylation of five genes in 50 HCC specimens varied from 2% to 96%. Promoter methylation was most common in the RASSF1A gene (96%), followed by thep16 gene (56%), thep21 gene (44%), thep15 gene (28%), and the p27 gene (2%).
Representative nested-MSP for methylation analysis of p15, p16, p21, p27 and RASSF1A genes. PCR products amplified with methylated (M) and unmethylated (U) sequence-specific primers. Methylated DNA (MD) and unmethylated DNA (UD) were used as positive controls. Distilled water (W) without DNA was used as a negative control. Positive and negative controls were used for each PCR run.
Summary of methylation of p15, p16, p21, p27 and RASSF1A in 50 HCC samples. The frequency of proximal promoter methylation of five genes in 50 HCC specimens varied from 2% to 96%. Methylation was detected in 28% for p15, 56% for p16, 44% for p21, 2% for p27 and 96% for RASSF1A. Patient identification numbers are given. Filled boxes, presence of methylation; open boxes, presence of unmethylation; shadow boxes, presence of partial methylation.
The results of our analysis of the association between methylation status and clinicopathological parameters in patients with HCC are presented in table 2. We found that the frequency of p16 methylation was significantly higher in patients with HCV-related HCC than in patients with other types of HCC (p = 0.01) and that the frequency of unmethylated p16 was significantly higher in patients with HBV-related HCC than in patients with other types of HCC (p = 0.02). There was no association between clinicopathological parameters in patients with HCC and the frequency of methylation of promoter regions of p15,p21,p27 and RASSF1A genes (Table 2).
Association between methylation status and clinical characteristics of 50 HCC patients.
| Characteristic | Patients | p15 | ρ16 | ρ21 | ρ27 | RASSF1Α | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M (n = 14) | U (n = 36) | Ρ | M (n =28) | υ (n = 22) | Ρ | Μ (n = 22) | υ (n = 28) | Ρ | Μ (n = 1) | υ (n = 49) | Ρ | Μ (n = 48) | υ (n = 2) | Ρ | ||
| Age (years)* | ||||||||||||||||
| < 58 | 25 | 5 | 20 | 0.34 | 13 | 12 | 0.57 | 12 | 13 | 0.57 | 1 | 24 | 1.00 | 24 | 1 | 1.00 |
| ≥58 | 25 | 9 | 16 | 15 | 10 | 10 | 15 | 0 | 25 | 24 | 1 | |||||
| Sex | ||||||||||||||||
| Male | 35 | 9 | 26 | 0.73 | 18 | 17 | 0.49 | 17 | 18 | 0.49 | 1 | 34 | 1.00 | 33 | 2 | 1.00 |
| Female | 15 | 5 | 10 | 10 | 5 | 5 | 10 | 0 | 15 | 15 | 0 | |||||
| HBV | ||||||||||||||||
| Positive | 22 | 4 | 18 | 0.50 | 8 | 14 | 0.02 | 11 | 11 | 0.59 | 1 | 21 | 0.46 | 21 | 1 | 1.00 |
| Negative | 26 | 8 | 18 | 19 | 7 | 11 | 15 | 0 | 26 | 25 | 1 | |||||
| Unknow | 2 | 2 | 0 | 1 | 1 | 0 | 2 | 0 | 2 | 2 | 0 | |||||
| HCV | ||||||||||||||||
| Positive | 21 | 6 | 15 | 0.75 | 17 | 4 | 0.01 | 8 | 13 | 0.59 | 0 | 21 | 0.57 | 21 | 0 | 0.32 |
| Negative | 28 | 8 | 20 | 11 | 17 | 14 | 14 | 1 | 27 | 26 | 2 | |||||
| Unknow | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | |||||
| AFP (ng/mL) | ||||||||||||||||
| < 400 | 29 | 8 | 21 | 0.90 | 17 | 12 | 0.97 | 15 | 14 | 0.46 | 1 | 28 | 1.00 | 28 | 1 | 1.00 |
| ≥400 | 20 | 5 | 15 | 11 | 9 | 7 | 13 | 0 | 20 | 19 | 1 | |||||
| Unknow | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | |||||
| Cell differentiation | ||||||||||||||||
| Well | 3 | 1 | 2 | 0.96 | 1 | 2 | 0.64 | 1 | 2 | 0.26 | 0 | 3 | 0.71 | 3 | 0 | 0.85 |
| Moderately | 30 | 8 | 22 | 18 | 12 | 16 | 14 | 1 | 29 | 29 | 1 | |||||
| Poorly | 17 | 5 | 12 | 9 | 8 | 5 | 12 | 0 | 17 | 16 | 1 | |||||
| Size (cm) | ||||||||||||||||
| < 5 | 25 | 10 | 15 | 0.16 | 17 | 8 | 0.09 | 11 | 14 | 1.00 | 0 | 25 | 0.50 | 24 | 1 | 1.00 |
| ≥5 | 25 | 4 | 21 | 11 | 14 | 11 | 14 | 1 | 24 | 24 | 1 | |||||
Μ: methiylation (included partially methylation). U: unmethylation. HBV: hepatitis Β virus (according to appearance of serum HBsAg). HCV: hepatitis C virus (according to appearance of serum anti-HCV).
Cox proportional hazards analysis was used to analyze the significance of clinicopathologic characteristics (Tables 3 and 4) and p15, p16, p21, p27 and RASSF1A methylation status (Tables 5 and 6) in predicting overall survival and disease-free survival. We found that age ≥ 58 years was associated with lower rates of overall survival than age < 58 years for patients with HCC (HR, 2.42; 95% CI, 1.04 to 5.63; p = 0.04; Table 3). Although the overall mortality rate among individuals with serum AFP levels ≥ 400ng/mL was twice as high as that among individuals with serum AFP levels < 400 ng/mL, there was no significant difference in overall survival between the two groups (HR, 2.16; 95% CI, 0.98 to 4.73; p = 0.06; Table 3). We also found that there was no significant difference in disease-free survival between patients with serum AFP levels ≥ 400 ng/mL and patients with serum AFP levels < 400 ng/mL (HR, 2.04; 95% CI, 0.98 to 4.23; p = 0.06; Table 4). In addition, no significant associations were found between methylation status of the five genes and clinicopathologic characteristics and overall survival and disease-free survival rates (Table 5 and Table 6).
Univariate analysis of clinicopathologic characteristics for overall survival.
| Characteristic | HCC patients (n = 50) | Overall survival | HR (95% CI) | p value | |
|---|---|---|---|---|---|
| 5-y rate (%) | Median (mo) | ||||
| Age, (years)* | 58.2 ± 11.9 | ||||
| < 58 | 25 | 68.0 | 66.0 | 0.04 | |
| ≥ 58 | 25 | 44.0 | 45.0 | 2.42(1.04-5.63) | |
| Sex | |||||
| Male | 35 | 54.0 | 59.0 | 0.53 | |
| Female | 15 | 60.0 | 66.0 | 0.75(0.31-1.81) | |
| HBV | |||||
| Negative | 26 | 62.0 | 66.0 | 0.34 | |
| Positive | 22 | 45.0 | 44.0 | 1.46(0.67-3.22) | |
| HCV | |||||
| Negative | 28 | 53.6 | 61.0 | 0.90 | |
| Positive | 21 | 57.1 | 59.0 | 1.05(0.48-2.31) | |
| AFP (ng/mL) | |||||
| < 400 | 29 | 65.5 | 65.0 | 0.06 | |
| ≥ 400 | 20 | 40.0 | 38.0 | 2.16(0.98-4.73) | |
| Cell differentiation | |||||
| Well | 3 | 66.7 | 75.0 | ||
| Moderately | 30 | 63.3 | 65.0 | 1.31(0.17-10.07) | 0.79 |
| Poorly | 17 | 41.2 | 44.0 | 2.17(0.28-16.91) | 0.46 |
| Size (cm) | |||||
| < 5 | 25 | 56.0 | 65.0 | 0.61 | |
| ≥ 5 | 25 | 56.0 | 59.0 | 1.23(0.56-2.70) | |
HBV: data are missing for 2 patients. HCV: data are missing for 1 patient. AFP: data are missing for 1 patient.
Univariate analysis of clinicopathologic characteristics for overall survival.
| Characteristic | HCC patients (n = 50) | Disease-free survival | HR (95% CI) | p value | |
|---|---|---|---|---|---|
| 5-y rate (%) | Median (mo) | ||||
| Age, (years)* | 58.2 ± 11.9 | ||||
| < 58 | 25 | 56.0 | 59.0 | 0.21 | |
| ≥ 58 | 25 | 40.0 | 24.0 | 1.61(0.77-3.37) | |
| Sex | |||||
| Male | 35 | 46.0 | 36.0 | 0.32 | |
| Female | 15 | 53.0 | 55.0 | 0.66(0.29-1.50) | |
| HBV | |||||
| Negative | 26 | 50.0 | 55.0 | 0.54 | |
| Positive | 22 | 40.9 | 12.0 | 1.26(0.60-2.63) | |
| HCV | |||||
| Negative | 28 | 50.0 | 43.0 | 0.73 | |
| Positive | 21 | 42.9 | 28.0 | 1.14(0.60-2.37) | |
| AFP (ng/mL) | |||||
| < 400 | 29 | 58.6 | 59.0 | 0.06 | |
| ≥ 400 | 20 | 30.0 | 7.0 | 2.04(0.98-4.23) | |
| Cell differentiation | |||||
| Well | 3 | 66.7 | 77.0 | ||
| Moderately | 30 | 53.3 | 56.0 | 0.95(0.21-4.24) | 0.94 |
| Poorly | 17 | 42.0 | 12.0 | 1.31(0.28-6.06) | 0.73 |
| Size (cm) | |||||
| < 5 | 25 | 44.0 | 41.0 | 0.85 | |
| ≥ 5 | 25 | 52.0 | 58.0 | 0.93(0.45-1.94) | |
HBV: data are missing for 2 patients. HCV: data are missing for 1 patient. AFP: data are missing for 1 patient.
Univariate analysis of methylation status of the five genes for overall survival.
| Characteristic | HCC patients (n = 50) | Overall survival | HR (95% CI) | p value | |
|---|---|---|---|---|---|
| 5-y rate (%) | Median (mo) | ||||
| p15 | |||||
| U | 36 | 52.8 | 59.0 | 0.33 | |
| M | 14 | 64.3 | 66.0 | 0.62(0.23-1.64) | |
| p16 | |||||
| U | 22 | 63.6 | 64.0 | 0.16 | |
| M | 28 | 50.0 | 53.0 | 1.82(0.79-4.23) | |
| p21 | |||||
| U | 28 | 60.7 | 64.0 | 0.24 | |
| M | 22 | 50.0 | 52.0 | 1.60(0.73-3.50) | |
| p27 | |||||
| U | 49 | 55.1 | 64.0 | 0.61 | |
| M | 1 | 100.0 | 59.0 | 0.05(0-5894.83) | |
| RASSF1A* | |||||
| U | 39 | 51.3 | 58.0 | 0.11 | |
| M | 11 | 72.7 | 71.0 | 0.37(0.11-1.24) | |
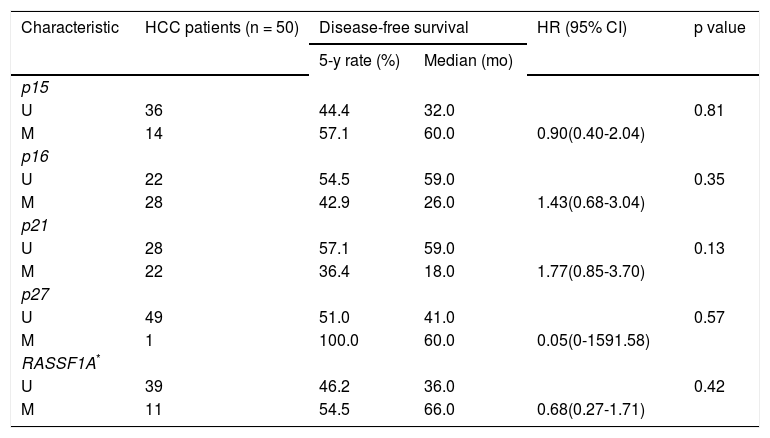
Univariate analysis of methylation status of the five genes for disease-free survival.
| Characteristic | HCC patients (n = 50) | Disease-free survival | HR (95% CI) | p value | |
|---|---|---|---|---|---|
| 5-y rate (%) | Median (mo) | ||||
| p15 | |||||
| U | 36 | 44.4 | 32.0 | 0.81 | |
| M | 14 | 57.1 | 60.0 | 0.90(0.40-2.04) | |
| p16 | |||||
| U | 22 | 54.5 | 59.0 | 0.35 | |
| M | 28 | 42.9 | 26.0 | 1.43(0.68-3.04) | |
| p21 | |||||
| U | 28 | 57.1 | 59.0 | 0.13 | |
| M | 22 | 36.4 | 18.0 | 1.77(0.85-3.70) | |
| p27 | |||||
| U | 49 | 51.0 | 41.0 | 0.57 | |
| M | 1 | 100.0 | 60.0 | 0.05(0-1591.58) | |
| RASSF1A* | |||||
| U | 39 | 46.2 | 36.0 | 0.42 | |
| M | 11 | 54.5 | 66.0 | 0.68(0.27-1.71) | |
Patients were divided into four groups based on serum AFP levels and p21 methylation status: group A p21 (serum AFP level < 400 ng/mL and p21 unmethylation, n = 14), group B p21 (serum AFP level < 400 ng/mL and p21 methylation, n = 15), and group C p21 (serum AFP level ≥ 400 ng/mL and p21 unmethylation, n = 13) and group D p21 (serum AFP level ≥ 400 ng/mL and p21 methylation, n = 7). The median overall survival rate in groups B p21(64.0%), C p21 (48.0%), and D p21 (8%) was shorter than that in groups A p21 (65.0%). No significant difference in overall survival rate was found between groups A p21 and B p21 or between groups A p21 and C p21 (p > 0.05); however, a significant difference in overall survival was found between groups A p21 and D p21 (p = 0.02). The median disease-free survival rate in groups B p21 (36.0%), C p21 (28.0%), and D p21(3%) was also shorter than that in group A p21(60.0%). No significant difference in disease-free survival rate was found between groups A p21 and B p21 or between groups A p21 and C p21 (p > 0.05); however, a significant difference in disease-free survival was found between groups A p21 and D p21 (p = 0.004) (Table 7).
Univariate analysis of p21 methylation status and AFP level for overall survival and disease-free survival.
| Characteristic | HCC patients (n = 50) | Overall survival | HR (95% CI) | p value | Disease-free survival | HR (95% CI) | p value | ||
|---|---|---|---|---|---|---|---|---|---|
| 5-y rate (%) | Median (mo) | 5-y rate (%) | Median (mo) | ||||||
| P21 and AFP (ng/mL) | |||||||||
| AFP < 400 and p21 U | 14 | 71.4 | 65.0 | 71.4 | 60.0 | ||||
| AFP < 400 and p21 M | 15 | 60.0 | 64.0 | 1.99(0.60-6.63) | 0.26 | 46.7 | 36.0 | 1.70(0.60-4.82) | 0.32 |
| AFP ≥ 400 and p21 U | 13 | 46.2 | 48.0 | 2.64(0.80-8.78) | 0.11 | 38.5 | 28.0 | 1.97(0.68-5.71) | 0.21 |
| AFP ≥ 400 and p21 M | 7 | 28.6 | 8.0 | 5.07(1.35-19.05) | 0.02 | 14.3 | 3.0 | 5.49(1.74-17.30) | 0.004 |
AFP: data are missing for 1 patient
Patients were divided into four groups based on serum AFP levels and RASSF1A methylation status: group A RASSF1A (serum AFP level < 400 ng/mL and methylated RASSF1A, n = 7), group B RASSF1A(serum AFP level < 400 ng/mL and partially methylated or unmethylated RASSF1A, n = 22), and group C RASSF1A (serum AFP level ≥ 400 ng/mL and methylated RASSF1A, n = 4) and group D RASSF1A(serum AFP level ≥ 400 ng/mL and partially methylated or unmethylated RASSF1A, n = 16). The median overall survival rates in groups A RASSF1A, B RASSF1A,C RASSF1A and D RASSF1A were 71.0%, 62.0%, 70.0% and 19.0%, respectively. No significant differences in overall survival were found between groups A RASSF1A and B RASSF1A or between groups A RASSF1A and C RASSF1A (p > 0.05); however, a significant difference in overall survival was found between groups A RASSF1A and D RASSF1A (p = 0.05). The median disease-free survival rates in groups A RASSF1A, B RASSF1A, C RASSF1A and D RASSF1A were 71.0%, 59.0%, 59.0% and 5.0%, respectively. No significant differences in disease-free survival were found between groups A RASSF1A and B RASSF1A, between groups A RASSF1A and C RASSF1A or between groups A RASSF1A and D RASSF1A (p > 0.05) (Table 8).
Univariate analysis of RASSF1A methylation status and AFP level for overall survival and disease-free survival.
| Characteristic | HCC patients (n = 50) | Overall surval | HR (95% CI) | p value | Disease-free survival | HR (95% CI) | p value | ||
|---|---|---|---|---|---|---|---|---|---|
| 5-y rate (%) | Median (mo) | 5-y rate (%) | Median (mo) | ||||||
| RASSF1A and AFP (ng/mL) | |||||||||
| AFP < 400 and RASSF1A M | 7 | 71.4 | 71.0 | 57.1 | 71.0 | ||||
| AFP < 400 and RASSF1A U* | 22 | 63.6 | 62.0 | 1.83(0.40-8.40) | 0.44 | 59.1 | 59.0 | 1.00(0.31-3.20) | 1.00 |
| AFP ≥ 400 and RASSF1A M | 4 | 75.0 | 70.0 | 0.89(0.08-9.78) | 0.92 | 50.0 | 59.0 | 0.91(0.17-5.01) | 0.92 |
| AFP ≥ 400 and RASSF1A U* | 16 | 31.3 | 19.0 | 4.58(1.02-20.59) | 0.05 | 25.0 | 5.0 | 2.57(0.82-8.09) | 0.11 |
AFP: data are missing for 1 patient.
The results of the Kaplan-Meier analysis revealed that serum AFP level was associated with overall survival. We found that the five-year overall survival rate among patients with serum AFP levels < 400 ng/mL was 65.5% and that the five-year overall survival rate among patients with serum AFP levels ≥ 400 ng/mL was 40.0% (p = 0.049; Figure 3A). However, the five-year disease-free survival rate was 58.6% among patients with serum AFP levels < 400 ng/mL and 30.0% among patients with serum AFP levels ≥ 400 ng/mL (p = 0.051; Figure 3B). We also found that low serum AFP level combined with unmethylated p21 status was associated with disease-free survival. Patients with serum AFP levels < 400 ng/mL and unmethylated p21 promoters had a better prognosis than patients with serum AFP level ≥ 400 ng/mL and methylated p21 promoters (overall survival, p = 0.076; disease-free survival, p = 0.016; Figure 4). In addition, patients with fully methylated RASSF1A promoter regions had a better prognosis than patients with partially methylated or un-methylated RASSF1A promoters if their serum AFP level was ≥ 400 ng/mL (overall survival, p = 0.028; disease-free survival, p = 0.078; Figure 5).
Kaplan-Meier survival analysis for all patients according to serum AFP level. Survival time was defined as the time from diagnosis to death or last known follow-up. Crosses represent censored values. The log-rank method was used to test for differences between groups. (A) Five-year overall survival was 65.5% and 40.0%, respectively (log-rank test, p = 0.049). (B) Five-year disease-free survival was 58.6% and 30.0%, respectively (log-rank test, p = 0.051).
Kaplan-Meier survival analysis for all patients according to AFP level and methylation status of the p21 proximal promoter region. Survival time was defined as the time from diagnosis to death or last known follow-up. Crosses represent censored values. The log-rank method was used to test for differences between groups. (A) Five-year overall survival rates were 71.4%, 60.0%, 46.2%, and 28.6% respectively (log-rank test, p = 0.076). (B) Five-year disease-free survival rates were 71.4%, 46.7%, 38.5%, and 14.3% respectively (log-rank test, p = 0.016).
Kaplan-Meier survival analysis for all patients according to AFP level and methylation status of the RASSF1A proximal promoter region. Survival time was defined as the time from diagnosis to death or last known follow-up. Crosses represent censored values. The log-rank method was used to test for differences between groups. (A) Five-year overall survival rates were 75.0%, 71.4%, 63.6%, and 31.3% respectively (log-rank test, p = 0.028). (B) The five-year disease-free survival rates were 59.1%, 57.1%, 50.0%, and 25.0% respectively (log-rank test, p = 0.078).
The rate of survival of patients with HCC is low, mainly because of the high rate of recurrence after curative surgical resection. Therefore, it is important to identify groups at a higher risk for recurrence. Although there have been many reports on the prognostic significance of various factors associated with HCC, the results of these studies are controversial. Some groups have reported that methylation of p15 or p16 genes is not an indicator of prognosis for patients with HCC.23,24 However, Wong et al. found that 9 of 12 (75%) patients with methylation of p15 and p16 genes were more likely to develop recurrent disease following resection.25 In addition, Ko, et al. reported that promoter hypermethylation of the p16 gene was associated with poor prognosis in recurrent early-stage hepatocellular carcinoma.26 However, in the present study, p15 and p16 methylation status was not associated with poor prognosis of patients with hepatocellular carcinoma (data not shown).
Kao, et al. reported that methylation of the p21 promoter was an independent predictor of survival for patients with hepatocellular carcinoma after resection.13 In our study, methylation of the p21 promoter and elevated serum AFP levels were found to be associated with poor prognosis in patients with hepatocellular carcinoma (Figure 4). Roman-Gomez showed that p21 methylation status was associated with poor disease-free survival of patients with acute lymphoblastic leukemia (p = 0.0001).27 The result suggested that under-expression of p21 protein might be due to hypermethylation of the promoter of p21 and that hypermethylation plays an important role in the progression of certain tumors.
We found that serum AFP level ≥ 400 ng/mL and promoter methylation of RASSF1A were associated with better overall survival in patients with HCC (p < 0.028). Similar results have been reported in HCC patients in Thailand, although no significant differences in overall survival were observed (p = 0.12).28 However, Chan, et al. found that patients with higher serum RASSF1A methylation concentrations at diagnosis or at 1-year follow-up after tumor resection showed poorer disease-free survival (p < 0.01).29 These conflicting observations may be due to differences in etiologic and underlying antecedent factors of hepatocellular carcinoma. It may be that a prognostic marker for HBV-related HCC is not a prognosticator for HCV-related HCC. Therefore, a study containing a mixed population may fail to take account of this unless appropriate sub analysis is performed.9 In addition, these conflicting observations may be due to the fact that DNA methylation is associated with environmental exposure and dietetic habits.30 Zhang, et al. reported that hypermethylation of RASSF1A and p16 promoter regions is common and that hypermethylation is associated with aflatoxin B1-DNA adduct levels in patients with HCC.31 These conflicting observations may also be due to the different methodologies between studies.
In conclusion, our data indicate that partially methylated or un-methylated RASSF1A promoters as well as elevated serum AFP levels or methylation of the promoter of the p21 gene and elevated serum AFP levels might be associated with poor prognosis among patients with hepatocellular carcinoma. Methylation of RASSF1A and p21 as well as serum AFP level may serve as potential prognosticators in patients with HCC.