Background. Non-alcoholic fatty liver disease (NAFLD) is the most common cause of chronic liver disease. Patients with non-alcoholic steatohepatitis (NASH) have increased plasmatic and hepatic concentrations of bile acids (BA), suggesting that they can be associated with the progression of the disease. Hepatic nuclear receptors are known to modulate genes controlling BA metabolism; thus, in this work we aimed to compare the expression of liver nuclear receptors -farnesoid X (FXR), small heterodimer partner (SHP) and liver X alpha (LXRα) receptors- and BA transporters -sodium+/taurocholate cotransporting polypeptide (NTCP) and bile salt export pump (BSEP)- in liver biopsy samples of patients with simple steatosis (SS) and NASH.
Material and methods. Forty patients with biopsy-proven NALFD were enrolled between 2009 and 2012; liver biopsies were classified as SS (N = 20) or NASH (N = 20) according to the NAFLD activity score. Gene expression of nuclear FXR, LXRa, SHP, NTCP and BSEP was analyzed by real-time reverse transcription polymerase chain reaction and protein level was quantified by western blot.
Results. Gene expression of FXR, SHP, NTCP and BSEP was significantly up-regulated in the NASH group in comparison with SS patients (P < 0.05). In contrast, protein level for FXR, SHP and NTCP was decreased in the NASH patients vs. the SS group (P < 0.05). Gene and protein profile of LXRa did not show differences between groups.
Conclusions. The results suggest that liver nuclear receptors (FXR and SHP) and BA transporters (NTCP and BSEP) are associated with the progression of NAFLD.
Non-alcoholic fatty liver disease (NAFLD) encompasses a spectrum of hepatic pathologies ranging, including simple steatosis (SS) and non-alcoholic steatohepatitis (NASH).1 NASH can progress to fibrosis, cirrhosis, liver failure and hepatocellular carcinoma.2 At present time, NAFLD is considered the most common cause of chronic liver disease and it is a major public health issue.3,4 The prevalence of NAFLD in the general population is around 20%, but certain groups, including patients with type-2 diabetes mellitus, obesity, dyslipidemia, and metabolic syndrome, are at higher risk for developing NAFLD.5–7 In fact, glucose and lipid metabolism disturbances have been clearly implicated as main underlying factors for NAFLD development.8
The Farnesoid X receptor (FXR), a member of the ligand-activated nuclear receptor transcription factors superfamily, has gained importance as a pathogenic factor in NAFLD; it has been shown that this receptor modulates a number of critical genes implicated in the homeostasis of glucose, the metabolism of lipids and bile acids (BA), as well as the immune response.9,10 It has been shown that BA can modify these metabolic pathways through FXR modulation.11–13 Therefore, FXR activity and BA homeostasis are currently considered as key players in the regulation of general metabolism, energy expenditure and inflammatory processes.9 In this connection, it is noteworthy that NASH patients have increased levels of BA, both in plasma and liver tissue,14 suggesting an association between the presence of toxic levels of BA and the development of the disease;15 thus, understanding the contribution of nuclear receptors and BA dysregulation to the pathogenesis and progression of NAFLD is a central issue in hepatology.16–18 BA metabolism genes under regulation of FXR include the nuclear receptor small heterodimer partner17 (SHP, NR0B2), the sodium/taurocholate cotransporting polypeptide (NTCP, SLC10A1), cholesterol 7a-hydroxylase (CYP7A1), and the bile salt export pump (BSEP, ABCB11).18,19 In addition, Liver X receptor alpha (LXRα, NR1H3) has also been implicated in the pathogenesis of NAFLD. LXRα is a nuclear receptor involved in the modulation of cholesterol metabolism and hepatic free acids biosynthesis whose expression has been found altered in NAFLD patients.20,21
In this work, we aimed to compare the expression of liver nuclear receptors (FXR, LXRa and SHP) and BA transporters (NTCP and BSEP) in liver biopsy samples of patients with SS and NASH. We hypothesized that NASH patients could display a differential gene and protein expression profile when compared with SS patients, and that such differences could help to increase our understanding of NAFLD progression.
Material and MethodsStudy designThis is a descriptive study, designed for evaluation of gene and protein expression of components of intrahepatic BA metabolism, including FXR, LXRa, SHP, NTCP and BSEP.
PatientsForty patients with biopsy-proven NAFLD were enrolled in the study at the Liver Research Unit of the Medica Sur Clinic & Foundation over the period from 2009 to 2012. For each patient, demographic, clinical, and biochemical variables were recorded. Exclusion criteria included age younger than 18 or older than 65, history of liver injuries and infective pathologies (hepatitis B, hepatitis C, or human immunodeficiency), organ transplantation, malignancy, autoimmunity, genetic disorders, therapy with immunosuppressive agents or excessive alcohol consumption (> 10 g/day in women and > 20 g/day in men). Informed written consent was obtained from all subjects for use of clinical and tissue materials for research purposes.
Ethics statementThe study protocol complied with the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Ethics Committee (Comité de Ética en Investigación de Médica Sur, S.A.B. de C.V.) of our hospital.
Human sample preparation and processingBiopsy specimens were formalin fixed, sectioned, and stained with hematoxylin and eosin and Masson’s trichrome. The NAFLD activity score, NAS, was used for the histological assessment of NAFLD.22 Patients who had NAS < 4 were considered to have SS; NASH was defined as the presence of steatosis, lobular inflammation, and ballooning degeneration with or without Mallory-Denk bodies, and with or without fibrosis (NAS ≥ 4).
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysisTotal RNA was extracted from liver biopsies by using the RNeasy FFPE Kit (Qiagen). The RNA purity and integrity was corroborated by spectrophotometry (260/280 ratio) and electrophoresis (agarose gel 1 %). RT-qPCR assays were performed in a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA). Targets RNAs were quantified with the QuantiTect SYBR Green RT-qPCR Kit (Qiagen). Gene-specific primer, table 1, were designed and synthesized at the Unidad de Biología Molecular of the Instituto de Fisiología Celular on the Universidad Nacional Autónoma de México. One step RT-qPCR was conducted with reverse transcription at 50 °C for 30 min, denaturation at 95 °C for 15 min and 40 cycles at 94 °C for 15 s, 55 °C for 30 s, and 72 °C for 30 s. The specificity of gene amplification was confirmed by independent end-point PCR analysis. The gene expression data are presented as relative gene expression using ubiquitin C (UBC) and hydroxymethylbilane synthase (HMBS) as endogenous RNAs reference; UBC and HMBS are been found to be the most accurate normalization factors for real time RT-qPCR analysis in liver samples.23 Values were collected for the threshold cycle (Ct) for each gene, and only Ct values less than 40 were considered for further analysis; the results shown are the mean of duplicate experimental determinations.
Sequences of oligonucleotides employed in RT-qPCR assays.
| Gene | Sequence |
|---|---|
| FXR | F: 5’-ACAGAACAAGTGGCAGGTC-3’ R: 5’-CTGAAGAAACCTTTACACCCCTC-3’ |
| LXR-a | F: 5’-GAAGAAACTGAAGCGGCAAGA-3’R: 5’-ACTCGAAGCCGGTCAGAAAA-3’ |
| SHP | F: 5’-GGCTTCAATGCTGTCTGGAGT-3’R: 5’-CTGGCACATCGGGGTTGAAGA-3’ |
| BSEP | F: 5’-GGAACCAGTGTTGTTTGCCT-3’R: 5’-AAAATCATGCAGCTGAGCCT-3’ |
| NTCP | F: 5’-CCCAAGAAGCCTCACCTATC-3’R: 5’-TTGGGTCACAAAACTTGGAA-3’ |
| UBQ | F: 5’-CCTGGTGCTCCGTCTTAGAG-3’R: 5’-TTTCCCAGCAAAGATCAACC-3’ |
| HMBS | F: 5’-GGCAATGCGGCTGCAA-3’R: 5’-GGGTACCCACGCGAATCAC-3’ |
Total protein was extracted from paraffin tissue blocks using the protein isolation Qproteome FFPE Tissue Kit (Qiagen). Protein was quantified by the bicinchoninic acid method using the Micro BCA Protein Assay Kit (Thermo Scientific Pierce), 4-20% SDS-PAGE was performed in Mini-PROTEAN TGX precast gels with 15 g of total protein and electroblotted onto polyvinylidene difluoride membranes (Bio-Rad).
Immunodetection was performed with anti-FXR, LXRa, SHP, BSEP and, NTCP (1:250; GeneTex) and β-actin (1:20,000; Bio-Rad) monoclonal antibodies. Goat anti-rabbit peroxidase conjugated secondary antibody (1:20,000; Bio-Rad) and the Immun-Star WesternC Chemiluminescent Kit (Bio-Rad) were used to visualize protein bands. Images were digitized with a Gel Doc XR+ System and densitometrically analyzed with the Quantity One software (Bio-Rad); protein expression was normalized with respect to the β-actin signal.
Statistical analysisData are given as means ± standard deviations. Statistically significant differences were assessed by one-way analysis of variance. If differences were found, values were compared using the Student’s t-test; P < 0.05 was considered significant. Analyses were performed with SPSS software (20.0.1, v. 2012; IBM SPSS).
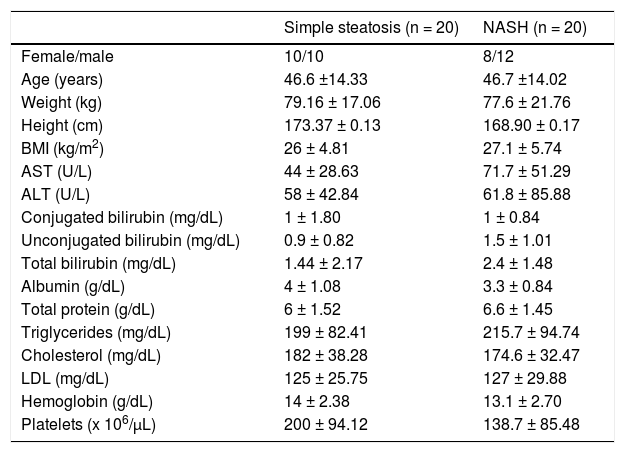
ResultsForty NAFLD patients, 20 classified as SS and 20 as NASH according to the NAS score, were enrolled in the study. The clinical and demographic features of these patients are shown in table 2.
Demographic and clinical data of NAFLD patients.
| Simple steatosis (n = 20) | NASH (n = 20) | |
|---|---|---|
| Female/male | 10/10 | 8/12 |
| Age (years) | 46.6 ±14.33 | 46.7 ±14.02 |
| Weight (kg) | 79.16 ± 17.06 | 77.6 ± 21.76 |
| Height (cm) | 173.37 ± 0.13 | 168.90 ± 0.17 |
| BMI (kg/m2) | 26 ± 4.81 | 27.1 ± 5.74 |
| AST (U/L) | 44 ± 28.63 | 71.7 ± 51.29 |
| ALT (U/L) | 58 ± 42.84 | 61.8 ± 85.88 |
| Conjugated bilirubin (mg/dL) | 1 ± 1.80 | 1 ± 0.84 |
| Unconjugated bilirubin (mg/dL) | 0.9 ± 0.82 | 1.5 ± 1.01 |
| Total bilirubin (mg/dL) | 1.44 ± 2.17 | 2.4 ± 1.48 |
| Albumin (g/dL) | 4 ± 1.08 | 3.3 ± 0.84 |
| Total protein (g/dL) | 6 ± 1.52 | 6.6 ± 1.45 |
| Triglycerides (mg/dL) | 199 ± 82.41 | 215.7 ± 94.74 |
| Cholesterol (mg/dL) | 182 ± 38.28 | 174.6 ± 32.47 |
| LDL (mg/dL) | 125 ± 25.75 | 127 ± 29.88 |
| Hemoglobin (g/dL) | 14 ± 2.38 | 13.1 ± 2.70 |
| Platelets (x 106/µL) | 200 ± 94.12 | 138.7 ± 85.48 |
NAFLD: nonalcoholic fatty liver disease. NASH: nonalcoholic steatohepatitis. BMI: body mass index. ALT: alanine aminotransferase. AST: aspartate aminotransferase. LDL: low density lipoproteins.
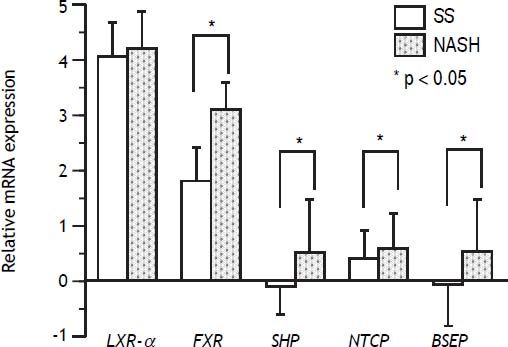
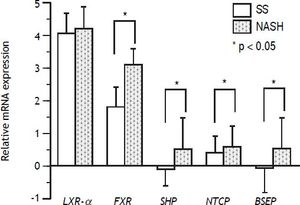
Gene expression studies showed that the FXR mRNA levels in the NASH group were significantly higher than those in the SS group (P < 0.05), in concordance, overexpression was observed for SHP (P < 0.05) and BSEP (P < 0.05). Contrary to expectations, NTCP was overexpressed in NASH patients compared with the SS group (P < 0.05). LXRa mRNA expression levels were high in both groups, but without significant differences between them (Figure 1).
Gene expression of nuclear receptors and bile acid transporters SS and NASH liver biopsies. FXR, SHP, NTCP, and BSEP gene expression was significantly upregulated in NASH vs. simple steatosis (SS) (P < 0.05)*, whereas LXRa expression was not significantly different between NASH and SS patients. FXR, farnesoid X receptor; SHP, short heterodimer partner; NTCP, Sodium+/Taurocholate cotransporting polypeptide; BSEP, bile salt export pump; LXRa, Liver X receptor alpha. Differences were analyzed by Student’s t-test.
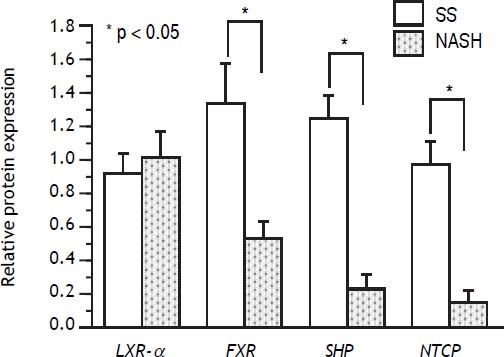
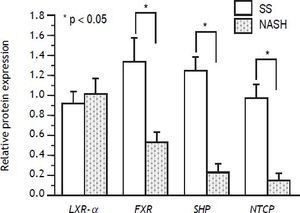
In regard to protein expression, it was found that FXR was diminished in NASH patients compared with SS patients (P < 0.05), and the same behavior was observed for SHP and NTCP (P < 0.05 for both). As for the RT-qPCR assays, LXRa protein expression remained unchanged between both groups (Figure 2). BSEP was under the limit of detection of Western blot assays and could not be further analyzed.
Protein level of nuclear receptors and bile acid transporters in SS and NASH liver biopsies. Protein expression of FXR, SHP and NTCP was significantly diminished in NASH compared with SS patients (P < 0.05)*, while LXRa protein content remained unchanged between the groups. FXR: farnesoid X receptor. SHP: short heterodimer partner. NTCP: Na+ltaurocholate cotransporting polypeptide. LXRα: liver X receptor alpha. Differences were analyzed by Student’s t-test.
The results of this work show that NASH patients possess a different mRNA and protein expression profile of hepatic nuclear receptors (FXR and SHP) and BA transporters (NTCP and BSEP) when compared with SS patients. It was found an up-regulated mRNA expression of FXR, SHP, NTCP and BSEP in liver biopsies from patients with NASH, whereas in contrast, FXR, SHP and NTCP showed a decreased protein content in the same samples. No statistically significant differences were detected for LXRa gene expression and protein level between the SS and NASH groups.
It has been described that FXR regulates, directly or through the nuclear receptor SHP, a wide variety of target genes critically involved in BA metabolism and lipid and glucose homeostasis.9 Regarding BA metabolism, it has been shown that activated FXR induces the expression of SHP in hepatocytes, which in turn blocks the expression of NTCP;24,25 in addition, activation of FXR inhibits the synthesis of BA from cholesterol and decreases its accumulation in liver.26 In this connection, it has been demonstrated that toxic accumulation of liver BA is associated with fibrosis progression.27,28 Finally, animal models of FXR deficiency show the pathologic manifestations of NASH, including macro-steatosis, hepatocyte ballooning and inflammation.29,30 It seems to be clear that FXR and its gene targets play a central role on the pathogenesis of NAFLD, but this conclusion arises mainly from animal models.24,26,28,30 In this respect, the data obtained from patient biopsies are illustrative from the human pathology.
In concordance with the animal model studies, we found a low protein level of FXR in liver samples from NASH patients, which was paralleled by a decreased protein level of SHP and NTCP (Figure 2). Interestingly, however, corresponding mRNA expression of FXR, SHP and NTCP were elevated in the same samples (Figure 1). From these results, two points deserve consideration:
- •
We detected a down-regulation protein expression of FXR, SHP and NTCP; however, we would expect an up-regulation of NTCP with regard to a putative negative feedback regulation by SHP. This inconsistency could be related to the known large inter-individual variability of the NTCP expression (even in healthy controls), and to the high inhibitory effect of BA over NTCP.31 In addition, a dysfunction in the repression pathway of SHP has been recently reported in a group of super-obese NASH patients,32 which may suggest a disturbance on the regulation of NCTP by SHP in NASH patients.
- •
The apparent discordance between mRNA expression and protein level for FXR, SHP and NTCP is intriguing; since mRNA is translated into proteins; it is generally assumed that protein levels must be correlated to the levels of its corresponding mRNAs. However, recent experimental evidence indicates that this assumption may be in many cases erroneous.33 For example, the correlation between RNA levels and its corresponding proteins evaluated by transcriptomic (cDNA and oligo microarrays) and proteomic (immunohistochemistry in tissue microarrays) data obtained for 23 human cell lines indicates that correlation coefficients vary widely; a significant correlation was found only in one third of the cases, with a mean correlation coefficient for the total sample of only ~0.3.33 Along this line, a whole human proteome dataset based on mass spectrometry,34 was used to compare the protein expression of 12 tissues with recently published mRNA values determined by quantitative transcriptomics analysis.35 The results show an average correlation coefficient of 0.41, indicating again, a general poor correlation between protein and mRNA levels. In general, correlation coefficients of ~0.4 have been found in diverse biological systems;36 this implicates that most of the time, as in our case, protein levels may not be proportional to its mRNA. Current data indicates that the relationship between mRNA expression and protein abundance reflects the dynamic balance between diverse transcriptional and translational processes, with a main role for regulation occurring after mRNA synthesis (v.g. post-transcriptional, translational and protein degradation regulation) contributing as much as transcription regulation.36
In concordance with the data obtained in this work, it can be established that mRNA expression changes may not always match with changes in protein levels. The quantification of these two macromolecules is not redundant if not complementary, and both are necessary for a full description of disease mechanisms.
Recently, alterations in BA transport and metabolism, including under-expression of NTCP and BSEP genes and lower protein levels of NTCP, has been described in morbidly obese NASH patients,32 suggesting an important role of FXR in the progression of NAFLD. Compared with that work, lower protein level of NTCP is consistent with our results; however, the results of mRNA expression seem opposites. As indicated, these discrepancies could suggest complex post-transcriptional mechanisms (low translational rate or high protein degradation, for example) regulating the expression of proteins related to BA transport and metabolism. Despite the differences, the data in both works support the connection between BA metabolism dysregulation and NASH development mediated by FXR.
The present study has some limitations, including the low number of patients studied, the lack of quantification of CYP7A1 expression and the fact that protein level of BSEP could not be evaluated. Additional studies surpassing these limitations and extending the analysis of transcriptional factors and nuclear receptors involved in lipid and BA metabolism and its regulation mechanisms in patient samples could increase our understanding of NAFLD progression.
ConclusionsThis work showed overexpression of FXR, SHP, NTCP, and BSEP genes whereas protein level of FXR SHP and NTCP was decreased in liver samples of NASH patients compared with SS patients. Altogether, the results involve to FXR and its associated genes and metabolic pathways with the progression from SS to NASH in NAFLD patients. Future and complementary studies are required to characterize the detailed molecular mechanisms involved in the regulation of BA metabolism and its relationship to NAFLD progression; it is plausible that this information can contribute to found new therapeutic options for this disease.
Abbreviations- •
BA: bile acids.
- •
BSEP: bile salt export pump.
- •
CYP7A1: cholesterol 7a-hydroxylase.
- •
FXR: nuclear farnesoid X receptor.
- •
HMBS: hydroxymethylbilane synthase.
- •
LXRa: nuclear liver X receptor.
- •
mRNA: messenger ribonucleic acid.
- •
NAFLD: Non-alcoholic fatty liver disease.
- •
NAS: NAFLD activity score.
- •
NASH: non-alcoholic steatohepatitis.
- •
NTCP: sodium/taurocholate cotransporting polypeptide.
- •
RT-qPCR: reverse transcription-quantitative polymerase chain reaction.
- •
SDS-PAGE: sodium dodecyl sulfate polyacrylamide gel electrophoresis.
- •
SHP: small heterodimer partner.
- •
SS: simple steatosis.
- •
UBC: ubiquitin C.
Medica Sur Clinic & Foundation funded this study.
Conflict of InterestNone.
AcknowledgmentsMedica Sur Clinic & Foundation, Mexico City, Mexico.