N6-methyladenosine (m6A) is the most thoroughly studied type of internal RNA modification, as this epigenetic modification is the most abundant in eukaryotic RNAs to date. This modification occurs in various types of RNAs and plays significant roles in dominant RNA-related processes, such as translation, splicing, export and degradation. These processes are catalyzed by three types of prominent enzymes: writers, erasers and readers. Increasing evidence has shown that m6A modification is vital for the regulation of gene expression, carcinogenesis, tumor progression and other abnormal changes, and recent studies have shown that m6A is important in the development of hepatocellular carcinoma (HCC). Herein, we summarize the nature and regulatory mechanisms of m6A modification, including its role in the pathogenesis of HCC and related chronic liver diseases. We also highlight the clinical significance and future strategies involving RNA m6A modifications in HCC.
N6-methyladenosine (m6A), which is the most abundant epigenetic modification of eukaryotic RNAs, remarkably regulates gene expression at the posttranscriptional level. This modification can emerge in various types of RNAs, including messenger RNA (mRNA) and noncoding RNA (ncRNA), and can regulate splicing, export, translation, degradation and other RNA metabolic processes [1]. These processes are modulated and catalyzed by several specific regulators, such as writers, erasers, and readers, which make m6A modifications dynamic and reversible. Mounting evidence has indicated that the m6A modification plays dominant roles in regulating gene expression, carcinogenesis, tumor progression and other processes.
Hepatocellular carcinoma (HCC) is the most common primary liver tumor worldwide and is characterized by a rapid yet undetectable onset, high incidence, high invasiveness, high recurrence and high mortality [2]. HCC is a typical inflammation-associated tumor, the development of which is tightly related to chronic inflammation in other liver diseases; however, the long-term incidence of HCC is heterogeneous because of the variable prevalence of etiologies and risk factors, such as hepatitis virus infection, alcohol consumption, aflatoxin, tobacco use and metabolic disorders [3], all of which have been relatively well defined. Traditionally, the occurrence of HCC is believed to be related to genetic variations, but increasing evidence shows that epigenetic modifications, especially abnormal m6A modifications and abnormal expression of m6A enzymes related to HCC, play an important role in liver carcinogenesis [4-5].
A better comprehension of the molecular mechanism of HCC is crucial for developing novel prognostic markers and identifying original therapeutic targets, and m6A modifications have attracted increased attention. Here, we will summarize the nature and hepatocellular regulatory mechanisms of m6A modification and its functions and implications in HCC and associated chronic liver diseases. Finally, we will highlight the clinical significance and future strategies that target RNA m6A modifications in HCC.
2The nature of the m6A modification of RNARNA m6A indicates the methylation modification of an RNA adenosine molecule at the N6 position. This modification was first identified in the 1970s [6] and has since been detected in mRNAs from mammals and viruses. M6A is widely distributed among mRNAs and ncRNAs and plays specific roles in various RNAs, including mRNA processing, maturation of microRNAs (miRNAs), posttranscriptional functions mediated by long noncoding RNAs (lncRNAs) and regulation of circular RNAs(circRNAs) [7-9]. This modification results in a dynamic equilibrium status that is maintained by highly conserved proteins, such as methylases (writers), demethylases (erasers) and m6A-binding proteins (readers). In mammals, each mRNA contains 3-5 m6A modification sites, but their distribution is not random. m6A modifications are predominantly distributed in the consensus RRACH motif (R=G, A; H=A, C, U) and enriched near termination codons, in 3′ untranslated regions (3′-UTRs) and within internal long exons [10]. Furthermore, RNA m6A modifications determine the whole life cycle of mRNA by regulating RNA splicing, nuclear export, translation, decay and other metabolic processes [11].
2.1M6A writersThe m6A writer complex is a methyltransferase complex that mediates RNA methylation with S-adenosylmethionine as a methyl donor. RNA methyltransferase exists as a compound composed of two major proteins: methyltransferase-like 3 (METTL3) and methyltransferase-like 14 (METTL14), which form a heterodimer catalytic core that interacts with a regulatory subunit, Wilms' tumor 1-associating protein (WTAP) [12]. METTL3 contains an activated methyltransferase domain, while the function of METTL14 is to specifically promote the recognition of RNA substrates by METTL3. METTL3 and METTL14 are located on nuclear speckles, a subcellular organelle related to mRNA processing, and the localization of these two enzymes is dependent on WTAP. Moreover, VIRMA (also called KIAA1429), RBM15, METTL16, ZC3H13 and CBLL1 are regarded as components of the methyltransferase complex, which also regulates the function of METTL3 and METTL14 [12-15].
2.2M6A erasersDemethylation of m6A is actively performed by the eraser fat mass and obesity-associated protein (FTO) or alkB homolog 5 (ALKBH5), which are the only two m6A demethylases identified that make m6A dynamic and reversible. ALKBH5 belongs to the ALKB dioxygenase family and depends on the cofactors Fe2+ and α-ketoglutarate to execute its catalytic functions [16]. FTO catalyzes the demethylation of m6A through a two-step reaction that produces two intermediates, and abnormal FTO regulation has been linked to obesity, brain malformations and growth delays [17-18]. ALKBH5 can directly catalyze the removal of methyl groups from m6A-methylated adenosine without an intermediate; this suggests its function as an m6A-specific demethylase. It is expressed in various tissues, and of these, the testis showed the highest expression. In one study, male Alkbh5 knockout mice presented with a sperm production disorder and sterility [16]. In addition, ALKBH3 is a novel m6A demethylase that preferentially acts on tRNA over mRNA or rRNA [19]. More m6A demethylases are predicted to exist, and their unknown functions remain to be discovered.
2.3M6A readersAfter the RNA is subjected to m6A modification, interaction with an m6A reader is required to execute the corresponding biological function. Some m6A binding proteins include YTH domain protein family members, heterogeneous nuclear ribonucleoprotein (HNRNP) and insulin-like growth factor binding protein (IGFBP). The YTH domain protein family consists of YTHDF1/2/3 (YTH domain family protein) and YTHDC1/2 (YTH domain containing) [20]. YTHDF1 affects the translation efficiency of m6A-modified genes mainly by recruiting translation initiation factors, such as eIF3, and by promoting ribosome loading to promote protein synthesis [21]. EIF3 can combine with the 5′UTR of mRNA at the m6A modification site to promote mRNA translation, which is a new mechanism by which eIF3 can activate translation initiation in a cap-independent manner [22]. YTHDF2 binds to the 3′UTR m6A site of the targeted mRNA, thereby mediating mRNA degradation and regulating the stability of mRNA [20]. YTHDF3 promotes translation or degradation of m6A-enriched mRNAs in balance with the activities of YTHDF1 or YTHDF2 [23]. YTHDFs share numerous common targets and may affect biological processes in an integrated manner. YTHDC2 can enhance the translation efficiency of the substrate by binding to the conserved m6A motif [24]. However, YTHDC1, which is located in the nucleus, can regulate RNA splicing by recruiting the mRNA splicing factors SRSF3 and SRSF10 in the nuclear speckle and controlling export [25].
The HNRNP family of readers includes HNRNPC, HNRNPG and HNRNPA2B1. These proteins bind to mRNAs according to an “m6A-switch” mechanism, which means that m6A alters the RNA hairpin structure and exposes the HNRNP binding motifs by weakening Watson-Crick base pairing; this enhances the affinity of HNRNP to m6A sites [26]. Furthermore, HNRNPC and HNRNPG may influence the localization and alternative splicing of mRNA by functioning as m6A readers in the nucleus [27]. HNRNPA2B1, also regarded as a nuclear reader, regulates primary miRNA processing and alternative splicing by cooperating with the miRNA microprocessor complex protein DGCR8 [28].
IGF2BPs are a group of conserved m6A readers whose RNA-binding sites include two RNA recognition motif (RRM) domains and four K-homology (KH) domains [29]. IGF2BPs can strengthen mRNA stability, protect m6A-modified RNA from degradation and promote translation by recruiting HuR and MATR3, which are RNA stabilizers and cofactors of IGF2BPs [30].
As mentioned above, RNA m6A plays vital roles in every metabolic process of RNA. Specifically, m6A writers and erasers regulate the m6A status and levels of targeted RNAs, and m6A readers decode targets to control their processes and downstream activities.
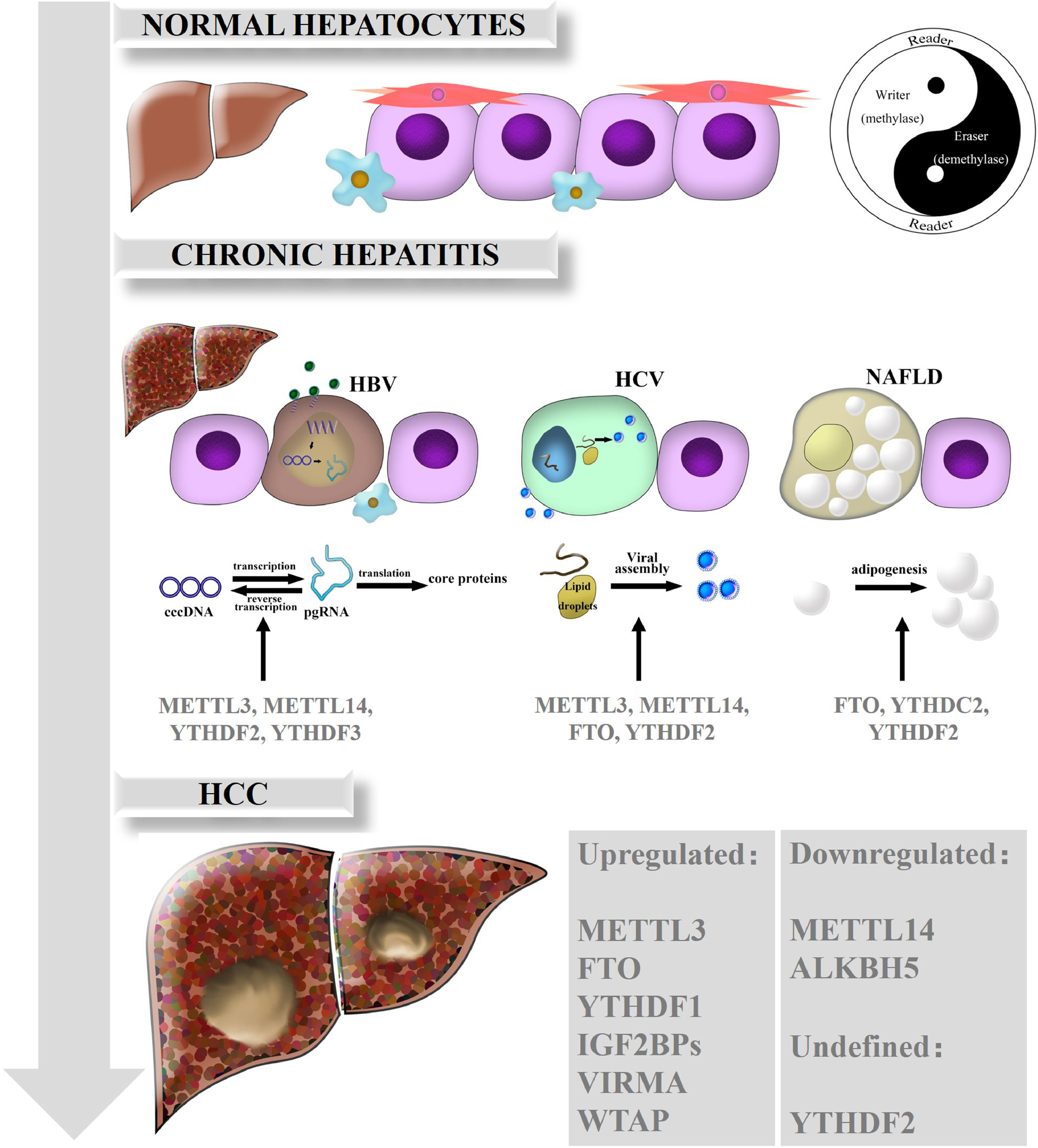
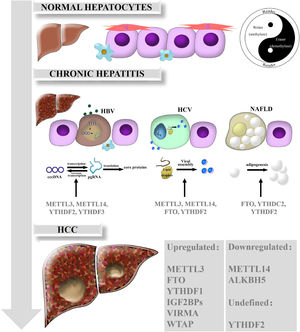
3The mechanisms of m6A in liver diseasesRNA m6A has been illustrated to be dominantly involved in various biological processes and in the occurrence and progression of human diseases. Emerging evidence has gradually clarified the perplexing roles of m6A and the dysregulation of m6A regulators in HCC and chronic liver diseases including nonalcoholic fatty liver disease (NAFLD). Here, we summarize the functions of m6A modification in HCC and related chronic liver diseases, such as chronic hepatitis B (CHB), chronic hepatitis C (CHC), and NAFLD (Fig. 1).
Biological roles of RNA m6A in liver diseases. In normal hepatocytes, m6A modification is in dynamic equilibrium due to the interactions and crosstalk of m6A regulators, just as the theory of Yin and Yang are described in traditional Chinese medicine. While the balance breaks down, m6A is also involved in the different functions of chronic hepatitis such as CHB, CHC and NAFLD. Ultimately, various kinds of m6A regulators are expressed aberrantly and act on downstream target genes, consequently promoting HCC development and progression.
HBV infection is a global public health issue, as over 240 million people have been infected worldwide; HBV is also a leading cause of chronic hepatitis, cirrhosis and HCC [31]. Covalently closed circular DNA (cccDNA), the most immediate evidence of HBV replication and infection, is difficult to detect except through an invasive biopsy procedure. In addition, cccDNA is transcribed into five mRNAs, among which pregenomic RNA (pgRNA) serves as a template for inverse transcription and translates into the pol and core proteins [32]. The epsilon stem loop structure, which is located at both the 5′ and 3′ termini of the pgRNA and the 3′ terminus of all HBV mRNAs, is the area on HBV mRNAs that undergoes m6A modification [33]. Further research found that depletion of METTL3 or METTL14 can inhibit inverse transcription of pgRNA and induce the upregulation of HBc and HBs proteins, which means that m6A on HBV transcripts suppresses the expression of HBV proteins. Knockdown of YTHDF2 or YTHDF3 had similar positive effects. In general, m6A modification is critical to the modulation of HBV infection and associated chronic hepatitis [34].
3.2RNA m6A and CHCHCV is the dominant risk factor for HCC in America, Europe and Japan. Although the incidence rate of HCV is lower than that of HBV, HCV infection develops more easily into chronic hepatitis, and even hepatic cirrhosis and HCC, thus threatening public health [35]. YTHDF2 plays an antiviral role in HCV virus lysis and replication. Further studies revealed that YTHDF2 could bind to viral assembly site lipid droplets, and YTHDF2 depletion profoundly increases the number of infectious viral particles [36]. Depletion of m6A writers (METTL3, METTL14) or erasers such as FTO can increase or decrease infectious HCV particle production, respectively [37]. Intriguingly, m6A modifications in HCV RNA can influence the effect of the innate immune system via the retinoic acid-inducible gene I receptor during the early steps of infection. This system feeds back an instant immune response to viral infections and distinguishes between the host and pathogens to avoid activating autoimmune responses [38]. However, the role of m6A modification in HCV remains unclear and requires further exploration.
3.3RNA m6A and NAFLDNAFLD is characterized by hepatic steatosis in the absence of obvious alcohol intake or other liver disease [39]. Moreover, NAFLD is a potential risk for HCC tumorigenesis in developed countries and is closely related to metabolic syndromes [40]. Recent studies have reported that m6A functions in the regulation of adipogenesis. For example, FTO could facilitate lipogenesis by suppressing the Wnt/β-catenin signaling pathway [41] and by enhancing RUNX1 translocation partner 1 (RUNX1T1)-mediated adipocyte proliferation [42], which could be reversed by YTHDF2-mediated m6A modification [43]. Furthermore, Hu et al. found that glucocorticoid receptor (GR) could mediate transactivation of FTO and thus regulate m6A modification on the mRNAs of lipogenic genes, controlling liver lipid deposition [44]. Clinically, an obvious increase in both the protein and mRNA levels of FTO was confirmed in NAFLD patients, which suggests that FTO is a potential therapeutic target and that inhibition of FTO may alleviate hepatic steatosis [34]. Zhou et al. found that the expression of YTHDC2 in the liver was significantly downregulated in obese mice and NAFLD patients. Furthermore, YTHDC2 could bind to the mRNAs of fatty acid synthase (Fasn) and other lipogenic genes to reduce targeted mRNA stability and inhibit gene expression, which indicates that YTHDC2 is a potential therapeutic target in NAFLD [45].
3.4RNA m6A and HCCHCC is the terminal stage of most chronic liver diseases, including chronic hepatitis and cirrhosis. Existing studies have indicated that m6A is essential for HCC proliferation and progression.
Our preliminary work suggested that the levels of METTL14, FTO and m6A in HCC are reduced in tumor tissues (especially in metastatic carcinoma) compared with those in adjacent tissues and normal liver tissues, which implicates METTL14 as an anti-oncogene in HCC. Apart from these findings, others also reported that the mRNA expression of METTL3, WTAP, ALKBH5 and VIRMA is not remarkably altered in HCC. Low METTL14 expression is related to invasion and metastasis of HCC as well as poor patient prognosis. METTL14 depletion enhances migration and invasion, whereas METTL14 overexpression has the opposite effects. Further exploration revealed that METTL14 acts on the DGCR8 protein to expedite miR-126 processing in an m6A-dependent manner, while miR-126 is regarded as a suppressor of tumor metastasis and thus regulates HCC progression [46].
In contrast, Chen et al. found that METTL3 upregulation in HCC accelerated HCC proliferation and progression in vitro and in vivo, while METTL3 knockdown had the opposite effects. A later study clarified that METTL3 alters the protein expression of SOCS2 via an m6A-YTHDF2 mechanism [47]. SOCS2 is a tumor-inhibiting factor that negatively regulates the JAK/STAT pathway in a variety of cancers [48]. More specifically, METTL3 upregulation increases the level of m6A on SOCS2 mRNA, as YTHDF2 binds to m6A modification sites, mediates SOCS2 mRNA degradation and contributes to HCC progression. This result is inconsistent with the results reported in earlier studies. The reasons for the controversy may involve various aspects, such as the heterogeneity of HCC samples, processing of mRNA transcripts and methodology of m6A detection [49], and thus, further studies are essential to verify these confusions. Li et al. reported that m6A was also increased in epithelial-mesenchymal transition (EMT) and that METTL3 depletion attenuated EMT in HCC both in vivo and in vitro. Additional work revealed that METTL3 works with YTHDF1 to accelerate the expression of Snail, which is a vital transcription factor for EMT. These findings may explain the roles of METTL3 in HCC metastasis [50]. Intriguingly, Lin et al. found that METTL3 was decreased in sorafenib-resistant hepatocellular carcinoma. Depletion of METTL3 decreased FOXO3 mRNA methylation and abolished its stability, which was mediated by YTHDF1, thus enhancing resistance to sorafenib [51].
WTAP was also reported to be upregulated in HCC and to promote tumor development. In addition, WTAP-mediated m6A suppresses ETS proto-oncogene 1 (ETS1) function in the HuR-ETS1-p21/p27 axis, while ectopic ETS1 expression was shown to alleviate HCC growth and rescue the phenotype. WTAP might function in HCC oncogenesis via m6A modification and provide a feasible therapeutic direction for HCC [52]. Cheng et al. found that VIRMA was overexpressed in HCC and upregulated m6A modification of inhibitor of DNA binding 2 (ID2) mRNA to downregulate its expression in turn and finally facilitating the migration and invasiveness of HCC [53].
FTO levels are decreased in HCC and are correlated with poor prognosis. Further exploration revealed that FTO demethylates PKM2 mRNA and promotes PKM2 translation. FTO knockout can induce cell cycle arrest in G0/G1 phase, thereby inhibiting proliferation and tumor growth in vivo [54]. However, ALKBH5 acts as a tumor suppressor and inhibits the expression of LY6/PLAUR domain containing 1 (LYPD1), which is an oncogenic factor identified and stabilized by the m6A reader IGF2BP1, thus suppressing HCC development [55].
Numerous m6A readers have been implicated in the development of HCC. Zhao et al. found that YTHDF1 was significantly upregulated in HCC and indicated a poor prognosis [56]. YTHDF1 abolishes the stability of FOXO3 under METTL3 depletion, which consequently promotes the development of sorafenib-resistant HCC [51]. Liu et al. reported that YTHDF1 accelerated the translation rate of FZD5, which was a dominant component in the Wnt/β-catenin pathway, via an m6A mechanism and strikingly facilitated HCC progression [57].
Although multiple studies have been performed, the roles of YTHDF2 are still elusive in HCC. Yang et al. found that YTHDF2 was obviously increased and negatively correlated with the expression of miR-145 in HCC. Inhibition of miR-145 strongly decreases m6A levels, as YTHDF2 can influence m6A levels by mediating mRNA degradation [58]. A luciferase reporter gene assay showed that miR-145 acted on the binding sites of YTHDF2 mRNA to abolish protein expression, which suppressed the proliferation of HCC cells [59]. According to these studies, YTHDF2 may serve as an oncogenic regulator in HCC. Another study found that YTHDF2 increased cancer stem cell properties and promoted metastasis by mediating the m6A methylation of POU5F1(also called OCT4) mRNA, which resulted in a poor prognosis [60]. However, Zhong et al. reported that hypoxia in HCC cells induced a reduction in YTHDF2 expression and that YTHDF2 served as a tumor suppressor by binding to specific m6A sequences of EGFR mRNA to promote degradation, therefore repressing cell proliferation and growth [61].
In another study, Hou et al. found that upregulated m6A levels in HCC may result in a reduction in YTHDF2 expression and consequential mRNA degradation. Further studies demonstrated that YTHDF2 functioned in the degradation of m6A-containing mRNA of interleukin 11 and serpin family E member 2, both of which are responsible for cancer-promoting inflammation [62].
Huang et al. found that IGF2BPs promoted the stability of targeted RNAs, such as MYC, which is an oncogene. Knockdown of IGF2BPs reduced the stability of MYC mRNA and therefore decreased MYC expression in an m6A-dependent manner, which indicates that IGF2BPs play vital roles in promoting HCC growth [63]. Despite recent progress in understanding the function of m6A modification in HCC, the specific mechanism of m6A, especially that of its related enzymes, remains largely unknown.
4Clinical significance of RNA m6A in HCCAs we know, the executors of m6A modification are m6A enzymes, and aberrant expression of m6A regulators and their functions in HCC has been reported in recent studies; the dysregulations in some of these regulators are closely related to tumor development and invasiveness, which indicate a poor prognosis. Thus, m6A profiling has the potential to be a clinical tool used to assess prognosis and provide new therapeutic targets for HCC.
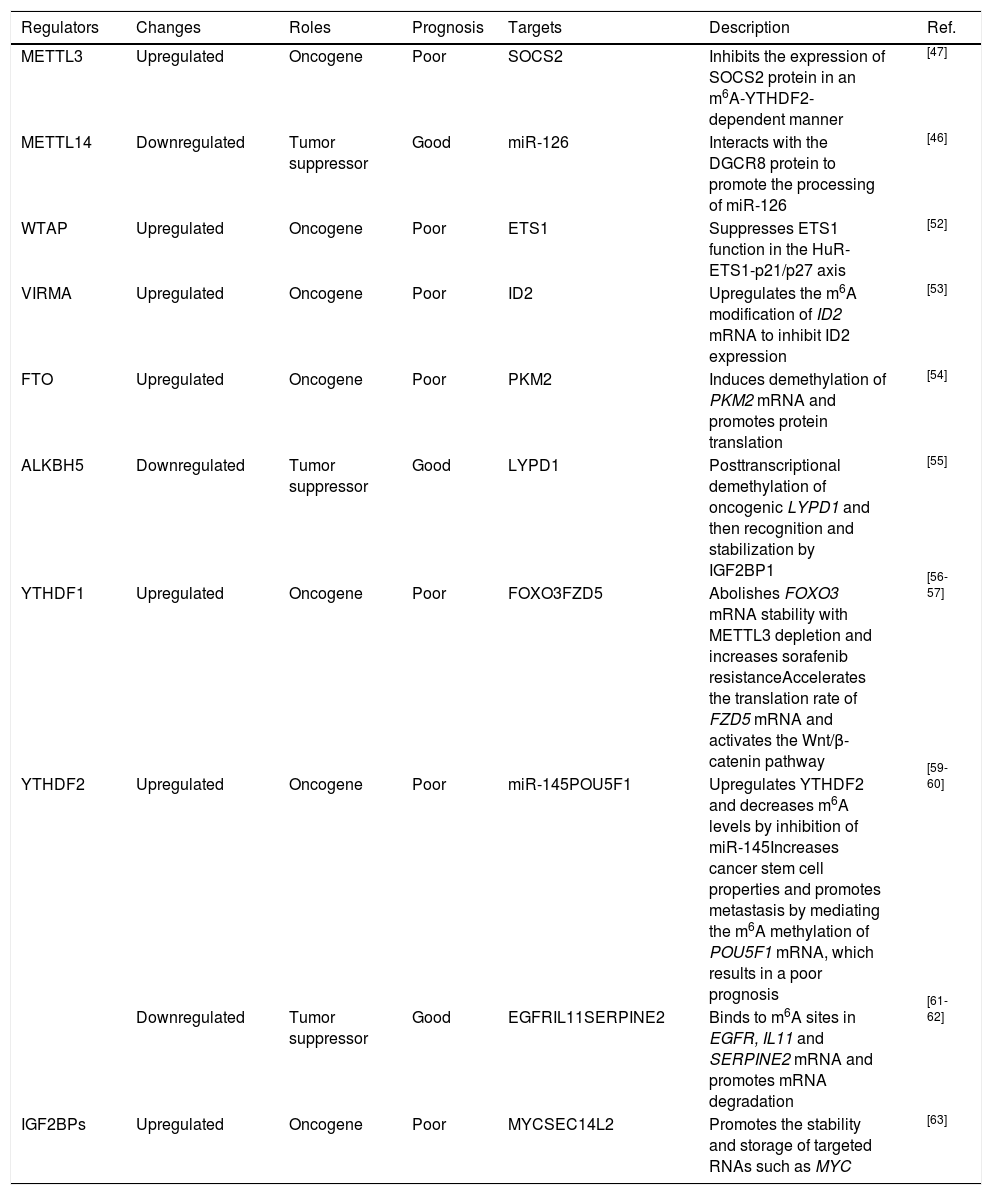
4.1Assessing the prognosis of HCCAccumulating evidence has revealed that dysregulation of m6A regulators can be a prognostic marker. As mentioned above, METTL3, WTAP and VIRMA, the units of the m6A “writer”, are overexpressed in HCC and are tightly correlated with poor survival [47; 52- 53]. Ma et al. reported that downregulation of METTL14 was obvious in HCC and that this downregulation promoted HCC metastasis by decreasing miR-126 expression [46]. Additionally, FTO was shown to be remarkably upregulated and to predict a poor survival [54]. Chen et al. found that ALKBH5 was decreased in HCC and that downregulation of ALKBH5 predicted a poor prognosis [55]. Zhao et al. reported that YTHDF1 was associated with poor survival and poor prognosis [56]. Qu et al. comprehensively analyzed m6A regulators using The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGA) databases and proposed that YTHDF1, YTHDF2, METTL3 and VIRMA were independent prognostic markers of overall survival (OS) [64]. In conclusion, most recent studies have presented evidence that METTL3 and YTHDF1 have oncogenic roles in HCC, but the roles of other m6A regulators, such as YTHDF2, are still controversial and have been found to exert tumor-promoting or tumor-suppressive effects during HCC progression. We have summarized the expression, potential prognostic value and targeted molecules of m6A regulators in HCC (Table 1).
Roles of m6A regulators in HCC.
| Regulators | Changes | Roles | Prognosis | Targets | Description | Ref. |
|---|---|---|---|---|---|---|
| METTL3 | Upregulated | Oncogene | Poor | SOCS2 | Inhibits the expression of SOCS2 protein in an m6A-YTHDF2-dependent manner | [47] |
| METTL14 | Downregulated | Tumor suppressor | Good | miR-126 | Interacts with the DGCR8 protein to promote the processing of miR-126 | [46] |
| WTAP | Upregulated | Oncogene | Poor | ETS1 | Suppresses ETS1 function in the HuR-ETS1-p21/p27 axis | [52] |
| VIRMA | Upregulated | Oncogene | Poor | ID2 | Upregulates the m6A modification of ID2 mRNA to inhibit ID2 expression | [53] |
| FTO | Upregulated | Oncogene | Poor | PKM2 | Induces demethylation of PKM2 mRNA and promotes protein translation | [54] |
| ALKBH5 | Downregulated | Tumor suppressor | Good | LYPD1 | Posttranscriptional demethylation of oncogenic LYPD1 and then recognition and stabilization by IGF2BP1 | [55] |
| YTHDF1 | Upregulated | Oncogene | Poor | FOXO3FZD5 | Abolishes FOXO3 mRNA stability with METTL3 depletion and increases sorafenib resistanceAccelerates the translation rate of FZD5 mRNA and activates the Wnt/β-catenin pathway | [56-57] |
| YTHDF2 | Upregulated | Oncogene | Poor | miR-145POU5F1 | Upregulates YTHDF2 and decreases m6A levels by inhibition of miR-145Increases cancer stem cell properties and promotes metastasis by mediating the m6A methylation of POU5F1 mRNA, which results in a poor prognosis | [59-60] |
| Downregulated | Tumor suppressor | Good | EGFRIL11SERPINE2 | Binds to m6A sites in EGFR, IL11 and SERPINE2 mRNA and promotes mRNA degradation | [61-62] | |
| IGF2BPs | Upregulated | Oncogene | Poor | MYCSEC14L2 | Promotes the stability and storage of targeted RNAs such as MYC | [63] |
Mounting evidence has shown that dysregulation of m6A enzymes is correlated with drug resistance in malignant tumors; for instance, overexpression of METTL3 results in drug resistance in pancreatic cancer [65] and radioresistance in glioma [66]. Knockdown of FTO enhances the sensitivity of melanoma cells to interferon gamma (IFN-γ) and anti-PD1 treatments in mice due to adaptive immunity. The authors proposed that the combinative effect of anti-FTO with anti-PD-1 treatment reduces the tolerance of melanoma to immunotherapy [67]. Furthermore, depletion of YTHDF1 remarkably inhibits the proliferation of B16 melanoma cells. YTHDF1 deficiency induces an elevation in CD8+ T cells, thus promoting antigen cross-presentation and cross-priming mediated by dendritic cells in vivo and sensitizing the immune response to anti-PD-L1 treatment; this indicates that YTHDF1 is a promising immunotherapeutic target [68]. The above studies suggested that targeting m6A modification and m6A enzymes is a possible immunotherapy strategy for HCC. As shown in Table 1, m6A regulators may be therapeutic targets of small-molecule drugs in HCC.
To date, some small-molecule inhibitors targeting m6A-specific enzymes or proteins have been approved for clinical application by the US Food and Drug Administration (FDA). For instance, Huang et al. found that the nonsteroidal anti-inflammatory drug meclofenamic acid (MA) can selectively inhibit the demethylation function of FTO, and its ethyl ester derivative showed even better efficacy than the original molecule in glioma models [69]. Other FTO inhibitors, such as R-2HG and FB23 as well as its derivative FB 23-2, were reported to be validated for the treatment of leukemia [70-71]. Research on small-molecule drugs that regulate the m6A modification of RNA has just begun, and the therapeutic effects of these drugs require additional verification by more clinical studies. RNA methylation modifications will have promising applications in the early diagnosis and accurate treatment of HCC after further studies of m6A modifications and the development of m6A-targeted drugs.
5Future strategies of RNA m6A in HCCAlthough more studies that have explored the profiles of m6A modifications have been performed, many contradictions still need to be resolved. First, it is crucial to further explore the regulators involved in m6A modification in HCC. Both tumor suppressor genes and oncogenic genes require identification. The same regulator may exert completely different effects in different tumors or even in the same tumor (e.g., YTHDF2), and different modulators may exert the same tumor suppressive or oncogenic effects, such as writers and the eraser FTO. Second, the diversity of roles and the intermediate relationships among m6A writer components remain controversial. Further investigations are required to clarify the various functions of constituent elements of the m6A writer complex, especially METTL14, in HCC development. Third, the specific mechanism of abnormal expression and upstream signaling also requires further investigation. Moreover, the effects of m6A modifications on ncRNAs are still vague. The mechanism in different stages of HCC development and the heterogeneity of samples should be considered and further explored. As there exists some inhibitors towards m6A regulator such as FTO approved for clinical use, however, inhibitors towards other enzymes are rare and need to explore. In conclusion, studies of m6A in HCC have highlighted a major problem in the contradictory results related to the expression or effects of diverse m6A regulators, and further investigation of the overall RNA m6A profile is necessary for clarification.
6ConclusionsIn conclusion, RNA m6A is vital in every step of RNA metabolism as well as in important biological processes in various types of RNAs; this modification is mediated by m6A regulators at the posttranscriptional level. Increasing numbers of studies on the roles of m6A modification in HCC have been conducted in recent years, and increasing evidence indicates that m6A modification is tightly correlated with the occurrence and development of liver diseases as a result of modified mRNAs and ncRNAs. More importantly, dysregulation of m6A modulators plays a crucial role in promoting or suppressing the development of HCC; hence, they are considered potential targets for prognostic prediction and molecular therapy in HCC. Since the comprehension of the biological function of RNA m6A is in its infancy, many contradictions still exist, and further explorations of m6A would contribute to a better understanding of its functional role in HCC.
Author contributionsFY designed the study and planned the outline. Y-F W drafted the article. MJ and J-P D collected materials and made the figure and table. C-M G, H-Z Y, Z-H D revised critically the manuscript. All the authors approved the manuscript to be submitted.
This work was supported by the National Natural Science Foundation of China, Grant number 81972657 and 81672345, and the National Key Research and Development Program of China, Grant number 2016YFC1302303.