Introduction. Considering the high prevalence of liver tumors and the impact on patient survival, a greater understanding of the biological behavior of those tumors if of great importance. The multidrug resistance gene (MDR1) may present as single nucleotide polymorphism (SNP) which can affect the expression and activity of P-glycoprotein (Pgp), and high expression of Pgp has been associated with a worse prognosis in affected patients.
Objective. To correlate the C3435T polymorphism in the MDR1 gene with the immunohistochemical expression of Pgp.
Material and methods. A total of 67 samples from patients with diagnosis of hepatocellular carcinoma (HCC), collected in the period from 2000 to 2009, were analyzed. The polymorphism in the MDR1 gene was determined by the technique of allele-specific real time PCR using TaqMan assay, and the expression of protein Pgp was evaluated by immunohistochemistry.
Results. Among the samples evaluated, 56 (83.6%) were from male patients and 11 (16.4%) from females. Mean age was 60.6 years (± 8.8), ranging from 37 to 85 years. The etiology of the HCC was related to hepatitis C virus infection (HCV) in 31 (46.3%) of cases, followed by hepatitis C virus infection + alcohol in 24 cases (35.8%), alcohol in 4 cases (6)%, hepatitis B virus (HBV) in 4 cases (6%) and other factors in 4 cases (6%). Liver transplantation was performed in 48 cases (71.6%) and hepatectomia in 19 cases (28.4%). The genotypes CC, CT and TT showed frequencies of 25.4%, 41.8% and 32.8%, respectively, and the allele frequencies were 46.3% for allele C and 53.7% for allele T. The expression of Pgp in over 75% of the cells was significantly more frequent in tumor tissue. On the other hand, a low expression of Pgp, in less than 25% of the cells, was significantly more frequent in non-tumor tissue. The Pgp expression in more than 50% of tumor cells of individuals with genotypes CC, CT and TT was 15.7%, 51.0% and 33.3%, respectively, and was significantly higher when in the presence of allele T (p = 0.002).
Conclusion. The presence of the polymorphic allele T is related to increased expression of Pgp protein in patients with HCC.
Hepatocellular carcinoma (HCC), is the most common primary liver tumor, and represents more than 85% of liver tumors in adults. HCC is currently considered the fifth most frequent malignant tumor in the world, with an estimated incidence of around 711,000 new cases per year and a mortality nearly equal to its incidence, i.e. 680,000 deaths per year. It thus represents the third most frequent cause of cancer-related death in the world.1,2,3
Curative treatment options currently available include radiofrequency ablation, surgical resection and liver transplantation.4 However, these treatments can be used in around 35% of the patients, so that only palliative treatments, such as chemoembolization and, more recently, Sorafenib,5 are available for the remaining patients. Prognostic indicators are very useful for the adequate treatment choice and patients selection.
The genetic modifications related to HCC have not been completely understood yet. It is known that, as in other tumors, the accumulation of various genetic changes have an important role in the carcinogenesis of HCC.6
Genetic polymorphisms are variations in DNA sequences occurring in a population with at least 2% frequecny. Single nucleotide polymorphisms (SNP), which result from the replacement of one nucleotide by a different one, represent one of the most studied polymorphism types. Genetic polymorphisms can result in a final gene product with functional consequences, that can be measurable and result in changes related to a pathological condition. Therefore, in some cases the genetic polymorphism can increase cancer susceptibility.7
The analyses of how cancer cells evade chemotherapy reveal a variety of genetic changes related to drug resistance. This phenomenon is known as multidrug resistance (MDR).8 The MDR phenotype is defined as the cells ability to resist to drug cytotoxicity, and is considered as the primary failure mechanism in chemotherapy.9 The appearance of the MDR phenotype is related to increased capacity of HCC cells to efflux drugs and the inhibition of apoptosisinducing pathways. Recently, a clinical study have shown that the insensitivity of hepatoma cells to chemotherapy might be associated with high expression levels of MDR1 and P-glycoprotein in tumour lesions.10
Three polymorphisms of the MDR1 gene have been especially studied, C1236T, G2677TA, and C3435T. The polymorphism at position 3435, in exon 26, is correlated with P-glycoprotein (Pgp) expression levels and activity of protein. This polymorphism has two alleles, C and T. Allele C is considered as the wild type.11
Pgp is located in the apical canalicular surface of hepatocytes12 and has a role in the protection of the organism against xenobiotics and toxic, environmental and tumorigenic agents.13,14 This physiological Pgp role can be affected by the MDR1 C3435T polymorphism. The wide tissue distribution suggests that Pgp has a fundamental role in normal cellular metabolism.15 It functions as an active efflux pump, decreasing the concentration of its substrates inside the cell and pumping out many substances, including various drugs.16,17 Available data suggest that high Pgp protein has been associated with a worse prognosis in affected patients.18
The possibility of predicting high relapse risk and worse prognosis among patients will allow surgical definition and chemotherapy treatment according to the individual risk. Thus, the search for markers which may be used as new prognostic factors of can importance, since they will help in the selection of the best candidates for curative treatment, leading to higher rates of long-term disease-free survival.
ObjectiveThe objective of this study was to correlate the MDR1 C3435T polymorphism with the immunohistochemical Pgp protein in patients with HCC.
Material and MethodsPatientsThe group under study included patients diagnosed with hepatocellular carcinoma, submitted to hepatectomia or liver transplantation at the Complexo Hospitalar da Santa Casa de Porto Alegre, Brasil. The patients recruitment took place from 2000 to 2009 in order to obtain samples with less than 10 years in parafin preservation and having undergone fixation with processes suitable for use in molecular biology. Among the 124 cases identified with diagnosis of HCC, 32 samples were not used: eight did not present enough material for analysis and 24 had excessive necrosis. The study therefore included 92 samples, with only 67 which presenting adequate paraffin blocks for molecular analyses. Each sample was provided of only one patient.
All patients signed a free informed consent form. The research project was approved by the Research Ethics Committee of Universidade Federal de Ciências da Saúde de Porto Alegre, protocol number: 47609. In order to maintain sample confidentiality and to facilitate handling and data analysis, all cases were encoded, using the acronym HCC followed by sample number. All data relating to patients were collected by a single researcher.
Preparation of samplesThe paraffin blocks were sectioned at 4-μm thickness with a microtome and blades (EasyPath) sterilized for each sample. The sections were mounted on a glass microscope slide. The remaining material was stored in 1.5 mL vials (Eppendorf) at-20 °C until extraction of DNA.
DNA extractionThe samples were deparaffinized with a 15 seconds rinsing with xylene with agitation, and rehydrated through two rinses in ethanol, also lasting 15 seconds each. After washing the samples twice by centrifugation at 13,000 g for 6 min, 20 mg/mL of proteinase K solubilized in lysing buffer was added to the sample, which was incubated at 60 °C overnight to potentiate the effect of the enzymatic digestion. DNA was then extracted with Bioneer Extraction Kit (Bioneer Corporation, USA), according to instructions of the manufacturer. The integrity and amount of DNA were analyzed with NanoDrop (NanoDrop Technologies, Wilmington, USA), and samples were stored at-20 °C.
GenotypingThe molecular identification of the C3435T polymorphism in the MDR1 gene (GenBank access number: M29445) was performed with the allele-specific real-time PCR technique using TaqMan, with a Step One Plus thermal cycler (Applied Biosystems, Foster City, CA, USA). A pair of primers and two probes, which were designed and validated by Applied Biosystems (SNP: 186105073), were used. The primers were used for the identification of the DNA sequence that contains the polymorphism and the two probes were categorized as “normal probe” –used to amplify the wild allele and the “polymorphic probe”– to amplify the allele with the 3435T polymorphism. These reagents allow the identification of samples from normal homozygous individuals (both alleles without the 3435T polymorphism), polymorphic homozygous (both alleles with the 3435T polymorphism) and heterozygous (one of the alleles with the 3435T polymorphism). Allele discrimination can be determined when the fluorescent probe hybridizes in the complementary target region that should be amplified.
ImmunohistochemistryThe immunohistochemical analysis was performed using the primary antibody Monoclonal Mouse anti-Multi-Drug Resistance 1 - clone JSB-1 - Cat. N° 18-0160Z- (Zymed-CA, USA). DAB (Diaminoben-zidine tetra-hydrochloride – reference: D-5637-Sig-ma) was used as substrate. The antibody was diluted 1:100 and the procedure was performed as described.19 Slides were analyzed according to a score ranging from 1-4 (Table 1). To analyze Pgp expression in relation to molecular findings, the scores were grouped by dividing the results in expression less than or equal to 50% and higher than 50% of the cells.
Classification of Pgp expression. Score Predominant staining pattern
| Score | Predominant staining pattern |
|---|---|
| 1 | Weak staining observed in 1% to 25% of cells. |
| 2 | Weak to moderate staining observed in 26% to 50% of cells. |
| 3 | Moderate to strong staining observed in 51% to 75% of the cells. |
| 4 | Strong staining observed in 76% to 100% of cells. |
Quantitative variables were described as average, standard deviation, minimum and maximum. The qualitative variables were described as absolute and relative frequencies. The McNemar Chi-square test was used to compare Pgp protein expression in non-tumor and tumor tissue. The association between the expression of Pgp in tumor and nontumor tissue with the genotype was analyzed with the Pearson's Chi-square test. In the case of statistically significant differences, adjusted residues were analyzed to identify the localization of the differences. The significance level adopted was 5.0% and analyses were performed with the SPSS software version 18.0.
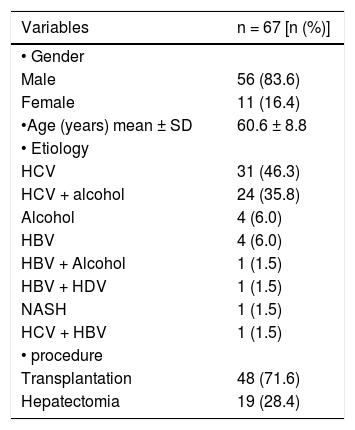
ResultsSample characterizationAmong the 67 cases analyzed, 56 (83.6%) were male and 11 (16.4%) were female. Mean age was 60.6 years (± 8.8), ranging from 37 to 85 years. The etiology of the HCC was related to hepatitis C virus infection in 31 cases (46.3%), followed by hepatitis C virus (HCV) infection with alcohol in 24 cases (35.8%), alcohol in 4 cases (6%), hepatitis B virus in 4 cases (6%) and other causes in 4 cases (6%). Liver transplantation was performed in 48 cases (71.6%) and hepatectomia in 19 cases (28.4%). These data are presented in table 2.
Characteristics of patients with hepatocellular carcinoma.
| Variables | n = 67 [n (%)] |
|---|---|
| • Gender | |
| Male | 56 (83.6) |
| Female | 11 (16.4) |
| •Age (years) mean ± SD | 60.6 ± 8.8 |
| • Etiology | |
| HCV | 31 (46.3) |
| HCV + alcohol | 24 (35.8) |
| Alcohol | 4 (6.0) |
| HBV | 4 (6.0) |
| HBV + Alcohol | 1 (1.5) |
| HBV + HDV | 1 (1.5) |
| NASH | 1 (1.5) |
| HCV + HBV | 1 (1.5) |
| • procedure | |
| Transplantation | 48 (71.6) |
| Hepatectomia | 19 (28.4) |
HBV: hepatitis B virus. HCV: hepatitis C virus. NASH: non-alcoholic steatohepatitis. HDV: hepatitis D virus.
Analysis of the genotype frequencies for the MDR1 C3435T polymorphism in the total sample of HCC patients showed that 17 patients (25.4%) were homozygous for the wild-type allele CC, 28 patients (41.8%) were heterozygous CT and 22 patients (32.8%) were homozygous for the polymorphic allele TT. The analysis of allele frequencies determined a frequency of 53.7% for the polymorphic variant T and 46.3% for allele C.
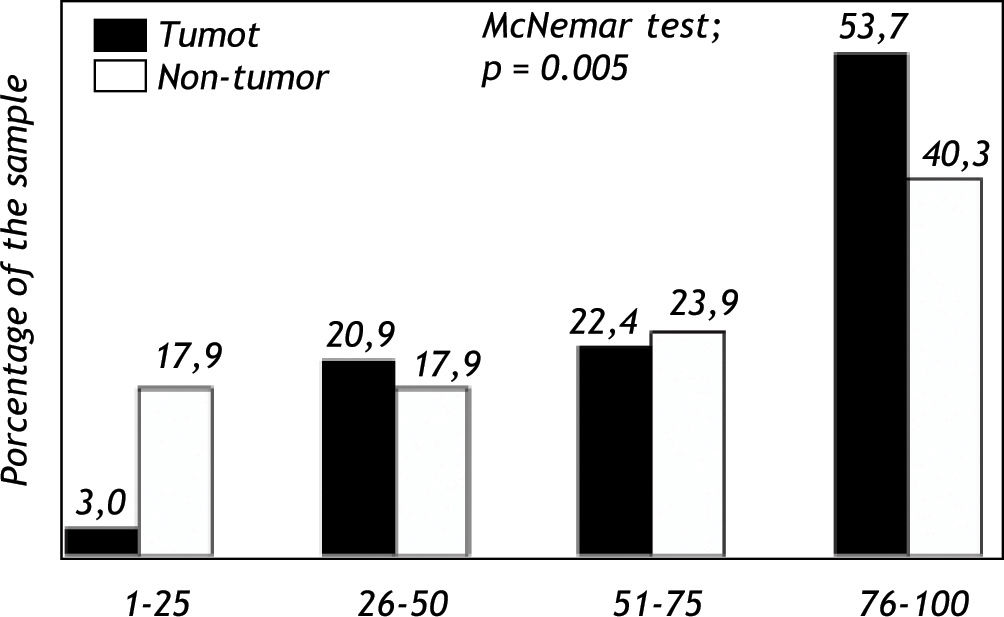
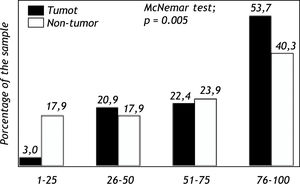
Expression of Pgp protein in tumor and non-tumor tissueImmunohistochemical analyses evidenced expression of Pgp on the apical canalicular surface of hepatocytes in tumor and non-tumor cells. Pgp was found on tumor tissue (TPgp) in 2 patients (3.0%) with score 1, 14 patients (20.9%) with score 2, 15 patients (22.4%) with score 3 and in 36 patients (53.7%) with score 4. Pgp expression in non-tumor tissue (NTPgp) was observed in 12 patients (17.9%) with score 1, 12 patients (17.9%) with score 2, 16 patients (23.9%) with score 3 and in 27 patients (40.3%) with score 4. The expression of Pgp in high levels, ranging from 76-100%, showed a significant difference when tumor and non-tumor tissue were compared (p < 0.05). On the other hand, low Pgp expression levels, between 1 and 25% were significantly observed in non-tumor tissue (p < 0.05) (Figure 1). Pgp expression with scores 2 and 3 showed no significant difference when tumor and non-tumor tissue samples were compared.
Association between genotypes and Pgp protein expression protein in tumor and non-tumor tissueIn 16 samples (23.9%), low Pgp expression on tumor tissue was observed (≤ 50% cells). Nine of these cases (56.2%) had the wild-type homozygous genotype CC, two (12.5%) presented the heterozygous genotype CT, and five (31.3%) were homozygous for the polymorphic T allele (TT genotype). Staining of over 50% cells tumor tissue was observed in 51 samples (76.1%). Among these cases, only eight (15.7%) presented the CC genotype, whereas 26 patients (51.0%) had the heterozygous CT genotype and 17 patients (33.3%) presented the polymorphic homozygous TT genotype.
Low Pgp expression on non-tumor tissue was observed in 24 cases (35.8%), with expression in ≤ 50% cells. Of these 24 cases, eight (33.3%) showed the wild CC homozygous genotype, 10 (41.7%) presented the heterozygous CT genotype and six patients (25.0%) had the polymorphic homozygous TT genotype. Positive staining in over 50% of the cells in non-tumor tissue was observed in 43 samples (64.2%). Among them, nine cases (20.9%) were identified as homozygous CC, 18 patients (41.9%) had the heterozygous genotype CT, and 16 patients (37.2%) presented the polymorphic homozygous TT genotype. These results can be seen in table 3.
Association between genotypes and Pgp protein expression.
The allele C was found in samples from 11 patients (24.4%) with Pgp expression in ≤ 50% tumor tissue, and in 34 patients (75.6%) with Pgp expression in over 50% of the cells. In non-tumor tissue, allele C was observed in 18 samples (40.0%) with ≤ 50% of Pgp expression and in 27 patients (60.0%) with more than 50% of Pgp expression in this tissue.
The C allele was absent in samples from five patients (22.7%) with ≤ 50% of Pgp expression in tumor tissue and in 17 samples (77.3%) with more than 50% Pgp expression in this type of tissue. In non-tumor tissue, the allele C was absent in six samples (27.3%) with ≤ 50% Pgp expression and in 16 samples (72.7%) with more than 50% Pgp expression (Table 1).
Association between allele C presence and Pgp protein expression.
| variables | Allele C absence (n =45) n (%) | Allele C absence (n = 22) | P* |
|---|---|---|---|
| TPgp expression | |||
| ≤ 50% | 11 (24.4) | 5 (22.7) | 1.000 |
| > 50% | 34 (75.6) | 17 (77.3) | |
| NTPgp expression | |||
| ≤50% | 18 (40.0) | 6 (27.3) | 0.454 |
| >50% | 27 (60.0) | 16 (72.7) |
The allele T was found in seven samples (14.0%) with ≤ 50% Pgp expression in tumor tissue and in 43 samples (86.0%) with more than 50% Pgp expression in this tissue. The presence of T allele was significantly increased when Pgp was expressed in more than 50% of the cells (p = 0.002). In non-tumor tissue, allele T was found in 16 samples (32.0%) with ≤ 50% Pgp expression and in 34 samples (68.0%) with more than 50% Pgp expression in this tissue.
Allele T was absent in nine samples (52.9%) with ≤ 50% Pgp expression in tumor tissue and in eight samples (47.1%) with more than 50% Pgp expression in this tissue. In non-tumor tissue, T allele was absent in eight samples (47.1%) with ≤ 50% Pgp expression and in nine samples (52.9%) with more than 50% Pgp expression in this tissue (Table 5).
Association between allele T presence T and Pgp protein expression.
| Variables | Allele presence (T) (n = 50) n (%) | Allele absence (T) (n=17) n (%) | P* |
|---|---|---|---|
| Expression of TPgp | |||
| ≤50% | 7 (14.0) | 9 (52.9) | 0.002 |
| > 50% | 43 (86.0) | 8 (47.1) | |
| Expression of NTPgp | |||
| ≤ 50% | 16 (32.0) | 8 (47.1) | 0.380 |
| > 50% | 34 (68.0) | 9 (52.9) |
The present study identified 124 cases with HCC diagnosis, and DNA was successfully extracted from 67 of the 92 samples collected. A study comparing the efficacy of genotyping DNA samples derived from waxed tissue and blood of patients with HCC diagnosis showed that in 40% of the cases DNA could be extracted with adequate quality for real time PCR amplification.20 In the present study, molecular analyses were concluded in 67 of the 92 paraffin blocks obtained, representing a 73% success rate of in the real-time PCR amplification by specific allele.
Most of the 67 cases included in this study were from men, and the mean age was 60.6 years old. The HCC etiology was mainly related to hepatitis C virus infection. In a similar study conducted in our country region, where the prognosis of cirrhotic patients undergoing liver resection was evaluated, a higher male prevalence was also observed, with mean age of 62.1 years old; the most common chronic hepatopathy etiology was also the HCV.21 In a prospective multinational study to assess the etiology of HCC in Latin America conducted with 240 patients, 174 (72.5%) of the 240 included patients were male, and the average age was 64 years old. The HCC etiology was related to HCV infection in 36.6% of the cases.22 Thus, our results corroborate those of similar studies in our country region.
Genotype frequency analysis for the MDR1 C3435T polymorphism in our HCC patients sample showed that 17 patients (25.4%) were homozygous for the wild C allele, 28 patients (41.8%) were heterozygous CT and 22 patients (32.8%) had the homozygous polymorphic TT genotype. Allele frequency determination showed a frequency of 46.3% for the wild variant C allele and 53.7% for the polymorphic variant T allele. In a study of Huang et al., the frequency of genotypes in a chinese healthy controls of a general population were 37.27% (CC), 46.36% (CT), 16.37% (TT).23 We don't have this date in Brazil.
In a study that assessed the MDR1 C3435T polymorphism in diagnosed breast cancer patients, the homozygous CC wild-type genotype was presented in 12.3% of the cases, the heterozygous CT genotype was identified in 57.9% and the homozygous polymorphic TT genotype in 29.8% of the cases. The C allele showed a frequency of 41.2%, and the allele T frequency was 58.8%. In that study, the allele T frequency in breast cancer patients was significantly higher than in the control group.24 The genotype frequencies were similar to the present study, except for a lower frequency observed for the homozygous CC wild-type genotype. Similarly, our study showed a higher allele C frequency. In a more recent study concerning breast cancer and the MDR1 gene polymorphism, the CC wild-type genotype frequency was 18%, the heterozygous CT genotype was seen in 56% of the cases and 26% of the cases had the homozygous polymorphic TT genotype.25
Another study that evaluated the association between the MDR1 C3435T polymorphism and kidney cancer identified frequencies of 18.4% for the wildtype homozygous CC genotype, 49.2% for the heterozygous CT genotype, and 32.4% for the homozygous polymorphic TT genotype, concluding that the presence of allele T is associated with a higher risk of kidney cancer.26
In a study with ulcerative colitis diagnosed patients, the same SNP for the MDR1 gene resulted in a frequency of 30% for the homozygous CC genotype, whereas the heterozygous CT genotype showed a frequency of 53.3% and the homozygous polymorphic TT genotype had a frequency of 16.7%. The allele frequency evaluated in the patients with ulcerative colitis in that study was 56.7% for C allele and 43.3% for T allele. The allele T frequency was significantly higher in patients with ulcerative colitis compared to controls, suggesting that this allele may be related to the tumor condition.27
Another report that evaluated Pgp expression in 26 paraffin samples of human liver with normal histology identified a lower frequency (9.2%) of the CC genotype, whereas the CT and CC genotypes were observed in 57.7% and 23.1% of the samples, respectively. These frequencies differ from data obtained in the present study. The small sample size of the aforementioned study as well as the fact that it involved the investigation of histologically normal liver tissue might explain these differences.28
The investigation of the MDR1 C3435T polymorphism in HCC patients with recurrence after liver transplantation showed a 36.7% frequency for the CC wild-type genotype, 6.1% for the CT heterozygous genotype, and 57.2% for the TT homozygous polymorphic genotype. These results indicate that individuals carrying the T allele present a higher HCC relapse risk.29 In the present study, the Pgp protein expression was analyzed in tumor and non-tumor tissue, and the results showed a higher expression of this protein in tumor tissue (Figure 1).
Studies describing the molecular investigation of the MDR1 C3435T polymorphism and Pgp protein expression are scarce.11,25,28 Only one study28 analyzed this type of association in liver sample, as well as in HCC patients liver samples. Published reports11,26,28 show controversial results, probably due to the fact that different tissues are analyzed, and also due to small sample sizes. Basic studies demonstrated the involvement of MDR1 gene with others pathways like a NF-KB, and the apoptosis cascade. The study of Sun, et al., showed that MDR1 and breast cancer resistance protein (BCRP) may be the most important factors for drug resistance in hepatocellular carcinoma. Moreover, the positive correlation between their mRNA and protein expression indicates the easy prediction of HCC MDR and possible inhibitive target of drug resistance at multi-levels.30
In the investigation of the relationship between the polymorphism in the MDR1 gene and Pgp protein expression in normal duodenum of21 individuals, a lower Pgp expression was observed in cases with the homozygous TT genotype than in homozygous CC individuals.11 These results differ from the present research, in which individuals with the CC genotype showed lower Pgp protein expression compared to homozygous TT individuals. These differences may be explained by the fact that normal tissue was studied, and that the tissue type investigated was also different.
The presence of the T variant in position 3435 results in an mRNA expression reduction in human liver samples.31 In a total of 36 normal liver samples, allele C was associated with increased Pgp protein expression. However, the results are inconsistent with those reported by Owen et al, who did not observe a correlation between the MDR1 C3435T polymorphism Pgp protein expression in 26 histologically normal liver samples.28
It should be noted that no studies evaluating the Pgp protein expression as classified in the present study (Pgp expression ≤ 50%, and > 50% of the cells stained by immunohistochemistry), nor its association with the MDR1 C3435T polymorphism allele frequencies were found in the literature.
In the present study, analysis of the MDR1 C3435T polymorphism in HCC patients confirmed the higher prevalence of allele T than allele C, as well as the fact that the T allele presented significantly higher frequency in Pgp protein expression samples > 50% in tumor tissue, suggesting that its presence may be associated with an increase in the Pgp protein expression.
Abbreviations- •
MDR1:multidrug resistance gene.
- •
SNP: single nucleotide polymorphism (SNP).
- •
Pgp: P-glycoprotein.
- •
HCC: hepatocellular carcinoma.
- •
HCV: hepatitis C virus infection.
- •
HBV: hepatitis B virus.
- •
MDR: multidrug resistance.
- •
DAB: diaminobenzidine tetra-hydrochloride.
- •
HDV: hepatitis D virus.
- •
NASH: non-alcoholic steatohepatitis.
- •
TPgp: P-glycoprotein in tumor tissue.
- •
NTPgp: P-glycoprotein in non-tumor tissue.