To characterize the virological features of hepatitis E virus (HEV) in serum from patients exhibiting chronic liver damage.
MethodsA data-base of 513 unrelated individuals from West-Mexico with liver-disease determined by clinical and biochemical tests and transient elastography between 2011 and 2016 were retrospectively analyzed. According to infectious etiologies, patients were classified as hepatitis B virus (HBV)-, hepatitis C virus (HCV)-infected patients, and patients exhibiting chronic liver damage with non-identified infectious etiological agent (NIIEA). Available serum samples from NIIEA-patients were tested by RT-nPCR for the presence of HEV-RNA and partially sequenced for genotyping.
ResultsOut of the 513 cases, 5.85% were patients infected with HBV, 67.64% with HCV, and 26.51% were NIIEA-patients. Among 76 available samples from NIIEA-cases, 30.26% tested positive for HEV-RNA. Twelve (15.79%) partial HEV sequences allowed phylogenetic analysis, revealing the classification of HEV as HEV-Gt3. Advanced fibrosis (F3–F4 stage) was found in a 26.1% of patients with HEV-active infection.
ConclusionAlthough HCV is the main infectious agent related to chronic liver disease in Mexico, liver damage without an infectious etiology is common. Our findings reveal that an elevated rate of chronic liver disease might be represented by autochthonous infection of HEV-Gt3, whose detection makes Mexico unique in Latin-America with the circulation of HEV strains belonging to three genotypes (Gt1, Gt2, and Gt3). Thus, HEV infection should be a matter of health concern, and mandates for HEV screening to properly handle this commonly undiagnosed disease.
Yearly, about 20.1 million newly hepatitis E virus (HEV) infections occur, constituting an important health concern, particularly in developing countries where demographic, sanitary, and economic conditions predispose enteric virus transmission and infection [1]. Despite being a major cause of large outbreaks of acute hepatitis around the world, and occasionally fulminant liver failure, HEV has also been found causing a rapid development of liver fibrosis and cirrhosis mainly in immunocompromised patients [2]. Routes for HEV transmission include oral–fecal, zoonosis, undercook or raw meat ingestion, blood products, and vertical exposure (mother-fetus) [3].
HEV is a member of the Hepeviridae family within the Orthohepevirus genus [4]. The viral genome consists of a single-stranded, positive-sense RNA molecule, containing three open reading frames (ORFs) [5]. Based on nucleotide sequences, HEV has been classified with at least eight genotypes (HEV-Gt) of which, five infect humans. Limited sequences of HEV-Gt5, 6, and 8 are currently available and were detected only in animals [6], while the HEV-Gt1 and 2 are extensively documented, associated with waterborne outbreaks in developing countries and, have been found mostly in humans. Meanwhile, HEV-Gt3, 4, and 7 are zoonotic, detected mostly in animals (pigs, boars, deer, and camels) and related to chronic infection in humans. HEV-Gt3 is the most extensively studied genotype in the context of chronic infection, being associated with silent chronic liver disease and cirrhosis in co-infections and in immunosuppressed individuals such as solid organ transplant recipients, human immunodeficiency (HIV) patients, and those with hematological diseases [2,7,8]. However, some studies have suggested the capacity of the virus to evolve toward chronic infection in immunocompetent patients [9–11] and even, with a possible development of chronic liver damage [12].
HEV-infections have been found in Latin American countries, mostly associated with sporadic outbreaks of acute hepatitis by HEV-Gt1, 2, and 3 [13,14]. Nevertheless, chronic infections in human have also been reported in Brazil and Argentina due to HEV-Gt3, the most frequent genotype in the region. Likewise, in several countries, including Mexico, HEV-Gt3 has been isolated from pigs and other wild species indicating a potential risk for zoonotic transmission [7,14].
Mexico is considered as a hyperendemic region for HEV-infection, based on an outbreak in the 1980s, where HEV-Gt2 was identified [2,7]; however, thirty-one years after, no new reports on HEV-Gt2 are available in this country. Recently, we reported HEV-Gt1 causing acute hepatitis in children in West-Mexico [13], and HEV-Gt3 has been identified from meat intended for human consumption [15–17]. Interestingly, the highest seroprevalence of HEV in farmed pigs has been found in Jalisco state, located in West-Mexico [18], where a high seroprevalence of HEV in samples from cirrhotic patients with no other etiological agent has also been reported [19]. In Mexico, cirrhosis is considered the fourth major cause of deaths [20], caused mainly by persistent alcohol abuse, non-alcoholic fatty liver disease (NAFLD) and viral hepatitis as hepatitis B and C (HBV and HCV); however, HEV-infections are not reported by the local government health agency. Nation-wide, up to 13% of viral hepatitis are considered as without specific etiology; hence, HEV might represent a non-sufficiently attended hepatic infection in the country [21]. Based on these antecedents, a high prevalence and risk of human HEV-infection are expected, as recently described in HAV (hepatitis A virus)/HEV co-infected pediatric patients with acute hepatitis from West-Mexico [22].
Since 1994, a program to monitor viral hepatitis in West-Mexico was established in a Referral Center in Jalisco at the Civil Hospital of Guadalajara “Fray Antonio Alcalde”, a tertiary healthcare center located in West-Mexico. This program has characterized the most common HCV and HBV genotypes circulating in this region. In the context of this program, herein we carried out the detection and genotyping of HEV in serum samples from patients exhibiting liver injury with no etiological infectious agent previously identified.
2Methods2.1Study populationA retrospective study was carried out by analyzing a database of 513 files from unrelated patients with liver disease admitted between 2011 and 2016 for confirmatory clinical an molecular diagnosis at the Department of Molecular Biology in Medicine, Civil Hospital of Guadalajara “Fray Antonio Alcalde” functioning as a Referral Center for viral hepatitis. The cases were classified according to the infectious etiology in HBV, HCV, and a non-identified infectious etiological agent (NIIEA). HEV-infection was diagnosed using reverse-transcription nested-PCR (RT-nPCR) from available serum samples recovered from 76 NIIEA-patients, and those with viral genome detected were classified as positive to HEV-infection. All samples from NIIEA-patients were negative for antibodies to HAV, HBV, and HCV. The absence of anti-HAV IgM and anti-HAV IgG, the surface antigen of HBV (HBsAg), and anti-HCV antibodies were earlier ruled out as previously reported [23].
Additionally, total anti-hepatitis B core antigen (anti-HBc, total IgM, and IgG) and human immunodeficiency virus (HIV) were ruled out as reported [24,25]. Also, data-files from patients undergoing treatment with hepatotoxic and anti-viral drugs or other diagnosed etiologies of liver damage such as primary biliary cirrhosis, autoimmune hepatitis, primary sclerosis cholangitis, hemochromatosis, Wilson's disease, and α-1 anti-trypsin deficiency, were excluded. Also, decompensated cirrhosis defined as previously [24] was excluded. Clinical and demographic features were recovered from structured questionnaires as previously reported [24,26].
Liver disease was considered in NIIEA-patients with at least one biochemical characteristic or clinical finding including hepatomegaly, jaundice or elevated hepatic function test such as alanine aminotransferase values (>40IU/L), aspartate aminotransferase (>45IU/L), and gamma-glutamyl transpeptidase (>35IU/L) [27]. The hepatic function tests and serum proteins (total proteins, albumin, and globulin) were previously determined by an AU5800 Clinical Chemistry System (Beckman Coulter's INC. USA).
Moreover, concomitant liver disease as alcohol consumption was considered as a risk factor for liver damage. Patients were classified as non-drinker or drinker according to the pattern of alcohol drinking in Mexico and the fifth edition DMS for Alcohol Use Disorder [28].
2.2Measurement of liver stiffness and determining fibrosis stageThe data recovered retrospectively from transient elastography (TE) measurement (Fibroscan) (FibroScan® Echosens, Paris, France) were used to determine liver damage stage. The values of the readings expressed in kilopascals (kPa) were used as an indicator of liver fibrosis that was stratified as absence (F0; <5.8kPa), mild liver fibrosis (F1–F2; 5.9–11.5kPa), and advanced liver fibrosis (F3–F4; >11.6kPa), as previously reported [24].
2.3RNA extraction and RT-nested-PCRRNA extraction was performed from 300μL of patient's serum with QIAmp Viral RNA mini kit (QIAGEN, USA) according to manufacturer's specifications. The purified RNA was fractioned and stored at −80°C until use, 5μL were used immediately for reverse transcription.
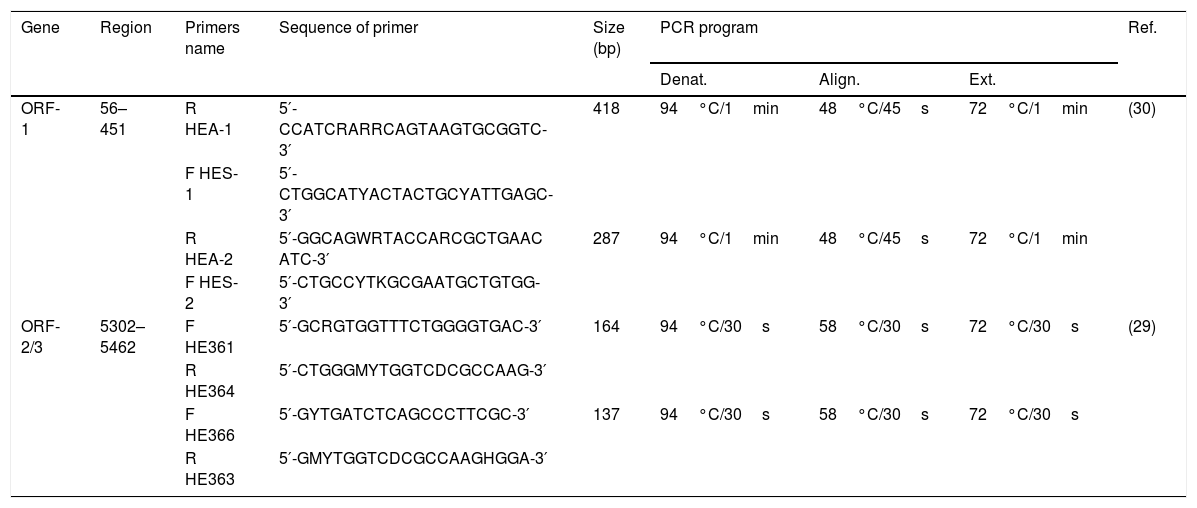
HEV genome detection was performed by targeting the ORF-2/3 (overlapping region) [29], and for genotyping, another method was followed targeting the ORF-1 region (5′ end methyltransferase gene) [30].
For the RT-nPCR, 5μL of purified RNA was used to carry out a two-step 12μL reaction by using 1.75μM of primer for each reaction, 870μM of dNTP MIX (Promega, WI), 3μL of M-MLV 5× Reaction Buffer (Promega, WI) (Tris–HCl pH 8.3 250mM, KCl 375mM, MgCl2 15mM, and DTT 50mM), and 8.5U of M-MLV RT (Promega, WI) as indicated by the manufacturer.
The first round of nested-PCR was carried out in 20μL mix volume containing 4μL of complementary DNA, 1× PCR buffer (10mM Tris–HCl pH 8.8, 50mM KCl), 0.625mM MgCl2, 0.25mM dNTP mix, and 2.5U Taq Polymerase. In the second round, 2μL of the first round were used for amplification under the same cycling conditions. PCR was carried out as previously described [29,30] with modifications in the protocol (Table 1).
Primers and conditions used for the amplification of ORF-1 and ORF-2/3 regions.
| Gene | Region | Primers name | Sequence of primer | Size (bp) | PCR program | Ref. | ||
|---|---|---|---|---|---|---|---|---|
| Denat. | Align. | Ext. | ||||||
| ORF-1 | 56–451 | R HEA-1 | 5′-CCATCRARRCAGTAAGTGCGGTC-3′ | 418 | 94°C/1min | 48°C/45s | 72°C/1min | (30) |
| F HES-1 | 5′-CTGGCATYACTACTGCYATTGAGC-3′ | |||||||
| R HEA-2 | 5′-GGCAGWRTACCARCGCTGAAC ATC-3′ | 287 | 94°C/1min | 48°C/45s | 72°C/1min | |||
| F HES-2 | 5′-CTGCCYTKGCGAATGCTGTGG-3′ | |||||||
| ORF-2/3 | 5302–5462 | F HE361 | 5′-GCRGTGGTTTCTGGGGTGAC-3′ | 164 | 94°C/30s | 58°C/30s | 72°C/30s | (29) |
| R HE364 | 5′-CTGGGMYTGGTCDCGCCAAG-3′ | |||||||
| F HE366 | 5′-GYTGATCTCAGCCCTTCGC-3′ | 137 | 94°C/30s | 58°C/30s | 72°C/30s | |||
| R HE363 | 5′-GMYTGGTCDCGCCAAGHGGA-3′ | |||||||
The PCR products were purified with Wizard® SV Gel and PCR Clean-Up System (Promega, WI) and sequenced by an external provider.
Nucleotide sequences were aligned using Clustal W, and identity matrices were performed with Molecular Evolutionary Genetic Analysis (Mega) software version 7.0. We constructed a phylogenetic tree using Neighbor-Joining algorithm and employing Tamura-Nei as the substitution model of DNA evolution with the 287-bp amplicons for HEV genotyping comparing with GenBank reference sequences according to Nicot et al. [31]. Statistical reliability of the phylogenetic tree was evaluated by bootstrap analysis of 10 000 replicates.
2.5Statistical analysisNormal distribution was analyzed by a Kolmogorov–Smirnov test. Qualitative variables were analyzed using frequencies and percentages. Quantitative variables were expressed as mean±standard deviation. Comparative analyses for variables with normal distributions were determined by t-test and variables with non-normal distribution by Mann–Whitney U-test.
2.6Ethics statementThis study was reviewed and approved by local ethical committees from the Health Science Center of the University of Guadalajara. Patients provided informed consent. This study complied with the ethical guidelines of the 2013 Declaration of Helsinki.
3Results3.1Liver disease without an etiological agent is common in West-MexicoFrom a total of 513 patients files, 30 (5.85%) corresponded to HBV-infections, 347 (67.64%) to HCV-infections, and the remaining 136 (26.51%) were negative for HBV, HCV, HIV, and HAV and categorized as NIIEA-patients, revealing that liver disease without etiology is common in West-Mexico.
3.2HEV was detected in serum from NIIEA-patients with liver diseaseOut of 136 cases corresponding to NIIEA-patients (HAV, HBV, HCV, and HIV negative) with liver disease, 76 serum samples were available. Two RT-nPCR methods directed to ORF-2/3 and ORF-1 were used. The ORF-2/3 specific method, provided the 137-bp expected product in fourteen samples, representing 18.42%. The ORF-1-specific method gave rise to the expected 287-bp DNA fragment in fifteen samples (19.73%), which were subsequently analyzed for genotyping. Twelve samples were available for sequencing, representing 15.79% of the total samples. HEV-genome was consistently detected with both RT-nPCR methods in six samples (7.89%). 23 samples tested positive for any of these methods (30.26%) and were consecutively designated as 1-23|MX|2018.
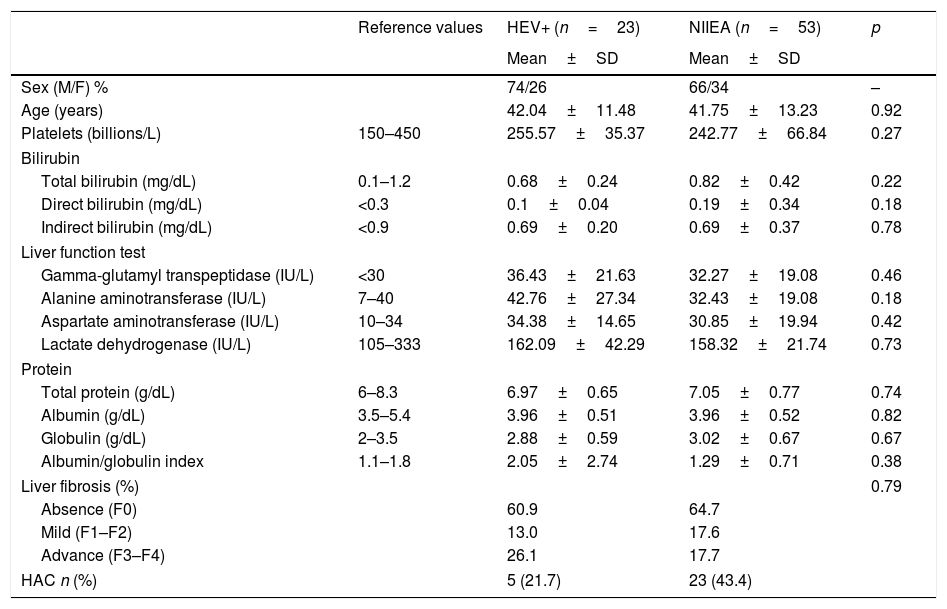
Although liver function was altered in HEV RNA-positive patients and NIIEA-patients, as revealed by biochemistry profile and TE, no significative differences were founded in both groups of patients. However, the group of patients with HEV-active infection showed an increased number of cases with advanced fibrosis (26.1% in F3–F4 stages) relative to the NIIEA-group (17.7%), suggesting a potential role of HEV-infection in disease progression. Moreover, according to previously applied questionnaires, five of the HEV-positive patients (21.74%) admitted a high pattern of alcohol consumption (Table 2).
Summary of the biochemical and demographic comparisons between HEV-RNA positive/NIIEA patients with diverse viral hepatopathies.
| Reference values | HEV+ (n=23) | NIIEA (n=53) | p | |
|---|---|---|---|---|
| Mean±SD | Mean±SD | |||
| Sex (M/F) % | 74/26 | 66/34 | – | |
| Age (years) | 42.04±11.48 | 41.75±13.23 | 0.92 | |
| Platelets (billions/L) | 150–450 | 255.57±35.37 | 242.77±66.84 | 0.27 |
| Bilirubin | ||||
| Total bilirubin (mg/dL) | 0.1–1.2 | 0.68±0.24 | 0.82±0.42 | 0.22 |
| Direct bilirubin (mg/dL) | <0.3 | 0.1±0.04 | 0.19±0.34 | 0.18 |
| Indirect bilirubin (mg/dL) | <0.9 | 0.69±0.20 | 0.69±0.37 | 0.78 |
| Liver function test | ||||
| Gamma-glutamyl transpeptidase (IU/L) | <30 | 36.43±21.63 | 32.27±19.08 | 0.46 |
| Alanine aminotransferase (IU/L) | 7–40 | 42.76±27.34 | 32.43±19.08 | 0.18 |
| Aspartate aminotransferase (IU/L) | 10–34 | 34.38±14.65 | 30.85±19.94 | 0.42 |
| Lactate dehydrogenase (IU/L) | 105–333 | 162.09±42.29 | 158.32±21.74 | 0.73 |
| Protein | ||||
| Total protein (g/dL) | 6–8.3 | 6.97±0.65 | 7.05±0.77 | 0.74 |
| Albumin (g/dL) | 3.5–5.4 | 3.96±0.51 | 3.96±0.52 | 0.82 |
| Globulin (g/dL) | 2–3.5 | 2.88±0.59 | 3.02±0.67 | 0.67 |
| Albumin/globulin index | 1.1–1.8 | 2.05±2.74 | 1.29±0.71 | 0.38 |
| Liver fibrosis (%) | 0.79 | |||
| Absence (F0) | 60.9 | 64.7 | ||
| Mild (F1–F2) | 13.0 | 17.6 | ||
| Advance (F3–F4) | 26.1 | 17.7 | ||
| HAC n (%) | 5 (21.7) | 23 (43.4) | ||
HEV+: hepatitis e virus RNA-positive; NIEAI: non-infectious etiological agent identified; HOMA-IR: homeostatic model assessment insulin-resistant; HAC: high alcohol consumption. Data is represented by mean and standard deviation. Differences between groups were analyzed by t-test and Mann–Whitney U-test with an α=<0.05.
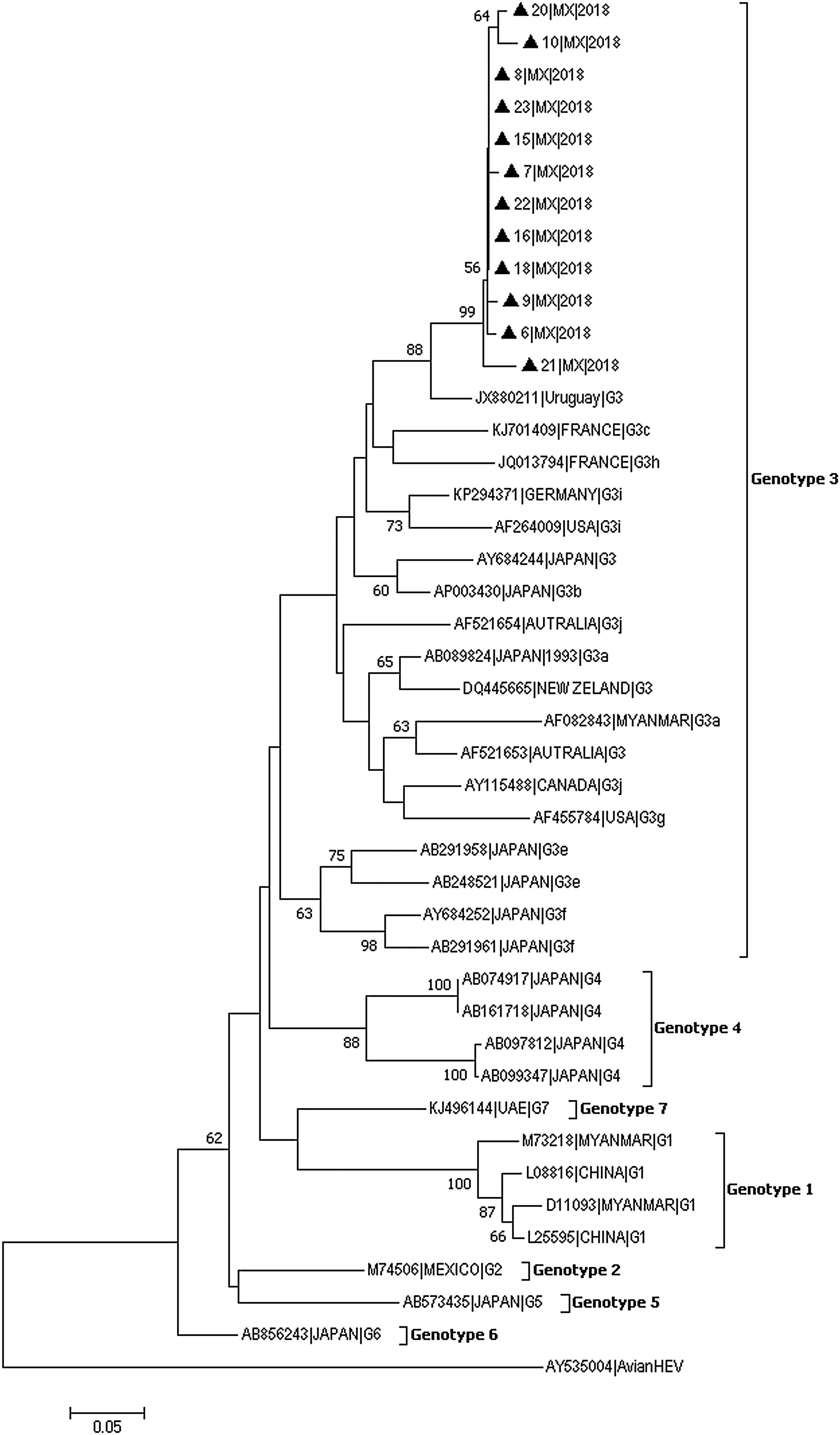
In 12 of the 23 samples in which HEV was detected, the 287-bp amplicon within ORF-1 could be amplified and sequenced (GenBank accession numbers: MH844063-MH844074). Phylogenetic analyses showed that the HEV strains identified in this study clustered together within Gt3 with high bootstrap values (Fig. 1). The nucleotide identity between these isolates and other strains from Gt3 ranged from 96% to 99.9%. Furthermore, a very close phylogenetic relationship was observed between the Mexican strains and isolates from Uruguay (Accession number JX880211) (90–94% nucleotide identity).
Genotyping phylogenetic tree constructed by neighbor-joining method based on sequence analysis of the 287-nt 5′ region of the ORF-1, corresponding to the putative methyltransferase gene. Bootstraps values were determined with 10000 resamplings of the datasets (only values over 50% are shown). Mexican strain (represented with black triangle) was compared with 30 strains whose partial or entire sequence have been reported and belonged to the different HEV genotype. As an outgroup, an avian HEV was included. The reference sequences are identified by GenBank accession number.
Globally, HCV and HBV are the major infectious etiological agents related to liver damage; whereas, HEV is mainly related with acute infection, causing more than 20.1 million new infections every year worldwide [1]. However, the study of HEV impact in the development of chronic liver illness in immunocompetent individuals is still in progress, although the influence in immunosuppressed patients is extensively documented [2].
In Latin America, the presence of HEV has been reported in different countries. HEV-Gt1 has been found circulating in humans from Cuba, Venezuela, Argentina, Uruguay, and recently we reported it in Mexico [13,32–34]. HEV-Gt2 has only been reported in Mexico [35]. Therefore, the current study is the first evidence of HEV-Gt3 in patients with liver disease in the country.
Herein, the partial sequencing of the 5′ end region within viral ORF-1 indicated a very high percentage of nucleotide identity among the strains detected in the same region and, no attempts were done for sequencing the short 137-bp from the ORF-2/3 fragment due to the limited amount of information to be obtained. Moreover, according to plausible differences in the performance of PCR assays, some samples were positive in one or other detected region as previously reported [30,36]. This outcome is consistent with the fact that remarkable differences in the sensitivity and efficiency of detection have been observed among different HEV genome regions [37]. It is important to consider that analytical methods for HEV detection and genotyping are not harmonized [37]. Thus, validation of the diagnostic and genotyping tests should be performed to enable the comparison of epidemiological data. Currently, Central African Republic, Chad, and Tunisia are the only countries who have reported HEV-Gt1, 2, and 3 [6], and the present study becomes Mexico as the only country in America with the presence of these genotypes.
In Mexico, liver illnesses are the fourth major cause of deaths [20] due to alcohol, NAFLD, and viral hepatitis [21] Epidemiological data from the National Health Agency of Mexico reveal that between 2011 and 2016 a total of 4513 viral hepatitis cases were caused by HBV (15.31%), 13,124 by HCV (44.53%), and 11,839 by other not well-defined hepatitis (40.16%) [38]. The high percentage of NIIEA cases is noteworthy, given that in our study a similar distribution was observed, and considering the recent detection of HEV-Gt3, a possible role of this virus in the development of chronic liver disease nationwide should be considered.
Previous epidemiological studies reported the circulation of HEV in Mexico through the detection of antibodies anti-HEV for specific population groups [7]. Based on a high seroepidemiology among the rural population from North-Mexico [7], in swine from farms intended to human consumption [15,17,18], and in a pediatric population from West-Mexico [22], a hyperendemicity of HEV could be expected. In the current study, we reported a total of 23 confirmed adult cases of HEV-infection in patients with liver damage, representing 30.26% of the samples analyzed and confirmed the expected results. Also, the mean age of positive cases was 42 years, which is consistent with previously published works around the world that report a higher seroprevalence in the older population [2].
Herein, no common symptoms related to acute hepatitis were found in HEV-positive cases (jaundice, abdominal pain, vomiting, and others) [7] or in NIIEA; nevertheless, some factors related to liver function were altered in both groups (see Table 2). Thus, although no antibodies determination was carried out in this work, previous contact with the virus cannot be ruled out in NIIEA-patients negative for HEV-RNA detection. Moreover, the high prevalence of HEV-infection in patients with mild or advanced liver disease is noteworthy. Previous studies have also reported a high seroepidemiology, reaching the 30.8%, of HEV in patients with cryptogenic or idiopathic hepatitis liver damage [39–41]. Hence, if HEV is the causative agent of liver damage or the infection is a consequence of the underlying liver disease, requires further studies.
The development of cirrhosis as a consequence of HEV chronic infection has mainly been associated with immunosuppressive conditions [36]. In this study, the retrospective analysis of clinical records did not reveal immunosuppression in the studied patients (data not shown). Interestingly, HEV-RNA was detected in patients with advanced liver damage (see Table 2), this is particularly relevant because HEV-Gt3, 4 and 7 are associated with liver damage by chronic infection [7] and in our study, the HEV-Gt3 was the only one identified in viremic patients, who were also affected with liver damage. Remarkably, a persistent course of HEV viremia has also been described in immunocompetent individuals [9–11], suggesting that immunosuppression could not be the only factor responsible for the development of liver illness, as suggested previously [42]. Also, co-morbidities as alcohol consumption cannot be ruled out, since this behavior is the main cause of chronic liver disease in Mexico [20].
In this study, 21.7% of patients with HEV-infection also consumed alcohol; whereby, the liver damage in this group it is not explained by alcohol consumption for all individuals. If alcohol consumption predisposed to HEV infection in the studied patients or conversely, is unknown, considering that it has been suggested that alcohol consumption might prompt for higher infection risk and clinically overt disease [43,44], and HEV infection in patients with underlying chronic liver disease by alcohol consumption carries a development of acute-on-chronic liver failure [45] and liver decompensation. Also, according with previous information, patients with liver cirrhosis that adquired a superinfection by HEV are susceptible to the development of liver failure in up to 100% of the individuals, increasing the death risk [46]. Moreover, co-morbidities as NAFLD, a condition highly prevalent in Mexico due to the high levels of obesity in the country [21], cannot be discarded. The impact of obesity in liver damage in the context of HEV infection should be tested since we previously reported high percentages of obese patients having antibodies anti-HEV [19]. Thus, future studies should include measurements as body mass index, waist circumference, waist-to-hip ratio, and ultrasonography for efficiently ruled out NAFLD [47] in the context of HEV infection. Also, celiac disease-related liver disease, a condition less frequent than NAFLD in Mexico [48] was not discarded in this study.
In conclusion, data presented herein showed that liver disease without an infectious etiology is common in Mexico and suggests that among adult patients, particularly in those exhibiting chronic liver disease, HEV could be playing a role in the pathogenesis and clinical outcomes. Therefore, in West-Mexico, the circulation of HEV strains of Gt3 is recommended to be considered as another cause of viral hepatitis affecting public health and requiring surveillance.
AbbreviationsHEV hepatitis E virus non-identified infectious etiological agent hepatitis E virus genotype 1 hepatitis E virus genotype 2 hepatitis E virus genotype 3 non-alcoholic fatty liver disease open reading frame retro-transcriptase nested-PCR transient elastography
- •
Oliver Viera-Segura: main author; samples processing; analysis and interpretation of data; drafting of the manuscript; critical revision of the manuscript.
- •
Mauricio Realpe-Quintero: study concept and design; critical revision of the manuscript, interpretation of data.
- •
Arturo Panduro: coordinator of the Viral Hepatitis Clinic; medical liver disease assessment; acquisition of samples; clinical database files; critical revision of the manuscript.
- •
Sonia Roman: acquisition of samples; viral hepatitis serum bank organization; interpretation of data; critical revision of the manuscript.
- •
Alexis Jose-Abrego: diagnosis of HBV; critical revision of the manuscript.
- •
Karina Gonzalez-Aldaco: diagnosis of HCV, critical revision of the manuscript.
- •
Jorge L. Trujillo-Ochoa: analysis and interpretation of data; critical revision of the manuscript.
- •
Nora A. Fierro: study concept and design; critical revision of the manuscript for important intellectual content.
This study was funded by grants from the Consejo Nacional de Ciencia y Tecnología (CONACYT, No. 246839, to Nora A. Fierro). Red Mexicana de Virología (to Oliver Viera-Segura).
Conflicts of interestThe authors declare that they have no conflict of interest.
The authors thank Ana B. Campos Juárez for technical assistance.