Hepatocellular carcinoma (HCC) is one of the sixth most common malignancies worldwide and is accompanied by high mortality. Homeobox B13 (HOXB13) has been shown to be involved in the development of various cancers. This study aimed to investigate the role of HOXB13 in HCC progression.
Materials and MethodsThe expression of HOXB13 in HCC tumor tissues was analyzed using qRT-PCR and immunohistochemical staining . After overexpression or downregulation of HOXB13 in HCC cell lines, cell proliferation was detected by CCK8 assay and Ki67 staining and cell invasion ability were tested by transwell assay. Western blot assay was applied to analyze the effect of HOXB13 on related signaling pathways. In addition, the role of HOXB13 on HCC in vivo was explored using a HCC mouse model. IF and WB were performed to detect cell proliferation, apoptosis and related protein expression in mice tumor tissues.
ResultsThe results showed that the expression of HOXB13 was significantly increased in HCC tissues compared with adjacent tissues and positively correlated with the tumor stage and survival of HCC patients. Overexpression of HOXB13 promoted the proliferation and invasion of HCC cells and up-regulated the protein expression of AKT, mTOR and MMP2. In contrast, the downregulation of HOXB13 resulted in the opposite results. In vivo experiments, HOXB13 significantly promoted tumor growth in mice bearing HCC by promoting cell proliferation and inhibiting cell apoptosis.
ConclusionsThis study suggested that HOXB13 can facilitate HCC progression by activation of the AKT/mTOR signaling pathway. HOXB13 may be a novel target for HCC therapy.
According to the latest data from International Agency for Research on Cancer (IARC), liver cancer is the sixth most common cancer globally and ranks third in mortality. Hepatocellular carcinoma (HCC) accounts for most primary liver cancers [1]. In recent years, significant progress has been made in the clinical treatment of HCC, but the prognosis of HCC patients remains poor, with a 5-year survival rate of only 18% [2]. The etiology and pathogenesis of HCC have not been fully elucidated, which may result from the synergy of multiple factors. Therefore, exploring the molecular mechanism of HCC is crucial for improving the diagnosis and treatment of HCC.
Homeobox B13 (HOXB13) is a sequence-specific transcription factor that binds preferentially to methylated DNA and is a part of the developmental regulatory system [3]. Studies have shown that HOXB13 is predominantly expressed in the developing genitourinary tract, including the prostate and distal colon and HOXB13 acts as a regulator of terminal cell differentiation in adult tissues that normally express HOXB13 [4–6]. In recent years, many studies have demonstrated that abnormal expression of HOXB13 may lead to the development of various tumors, such as breast cancer [7, 8], ovarian cancer [9], prostate cancer [10–14], gastric cancer [15, 16] and so on. For example, HOXB13 was mutated in families with an increased risk of early-onset hereditary prostate cancer and was considered a prognostic indicator for prostate cancer [10]. Currently, there are few studies on HOXB13 in HCC. Zuo L et al. found that HOXB13 was highly expressed in liver fibrotic tissues, and the expression level of HOXB13 was positively correlated with the degree of liver inflammation and fibrosis [17], suggesting that HOXB13 may play a role in the development of HCC.
In this study, we analyzed the HOXB13 expression in tumor tissues of HCC patients. In addition, we explored the effects of HOXB13 on the biological function of HCC cells and the signaling pathway regulating HCC progression in vitro and in vivo. The study provides an essential basis for understanding the pathogenic mechanism of HCC and exploring new therapeutic targets for HCC.
2Methods2.1Bioinformatics analysisIntrahepatic RNA sequencing profiles of 369 HCC patients and 50 healthy controls were acquired from the Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov/). Differentially expressed gene (DEGs) were analyzed with the DESeq2 package and edgeR package and presented as heatmap by Pheatmap. The false discovery rate (FDR) of genes was obtained by P value correction using package multitest. Later, DEGs were screened out with the threashold of FDR <0.05 and |log2 (fold change)|>1. DEGs were identified using Database for Annotation, Visualization and Integrated Discovery (DAVID) online database (http://david.ncifcrf.gov/). Gene ontology (GO) analysis was used for pathway enrichment using Cytoscape (ClueGo) with P<0.01.
2.2Clinical samples and cell linesA total of 50 pairs of human HCC tissues and the adjacent normal tissues were collected from Aug 2015 to Sep 2017 in the Second Hospital of Nanjing University of Chinese Medicine. The detailed clinical information of these patients is shown in Table 1.
The human hepatic cancer cell line Hep3B was purchased from American Type Culture Collection (ATCC), and the HL-7702 and Huh-7 cell lines were purchased from the Japanese Collection of Research Bioresources Cell Bank (JRBC). All cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco) with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin/streptomycin mixture.
2.3Immunohistochemical (IHC) stainingThe 4 μm-thick paraffin-embedded slides were used to detect the protein expression of HOXB13 by IHC staining. Briefly, the slides were incubated with anti-HOXB13 antibody (Abcam, ab201682, 1:100) overnight at 4 °C, followed by incubation of secondary antibody for 1 hour. The expression of HOXB13 was classified into negative (−), weak (+), moderate (++) and intense (+++) according to the proportion of immunostained positive cells and the staining intensity. The results were scored by two technicians blinded to the clinical data.
2.4RNA extraction and quantitative RT-PCR (qRT-PCR) assayTotal RNA was extracted from each 50 mg tissue sample by using TRIzol reagent (Invitrogen, USA) according to the reagent instructions. Then, the extracted RNA was reversed transcribed into complementary DNA (cDNA) using PrimeScript™ II 1st Strand Synthesis Kit (Takara, 6210B). Quantitative Real-time PCR reactions were performed on a Step-One Plus instrument (Applied Biosystems 7500) using SYBR® Premix Ex Taq™ Kit (Takara, RR420A). The relative gene expression level was normalized to β-actin and calculated utilizing the 2−ΔΔCt method. The primer sequences are as follows: β-actin, forward 5′-CATGTACGTTGCTATCCAGGC-3′, reverse 5′- CTCCTTAATGTCACGCACGAT-3′; HOBX13, forward 5′-AGCTCCCGTGCCTTATGGTTA-3′, reverse 5′- GGCTGGTAGGTTCCCGGAT-3′; AKT, forward 5′-AGCGACGTGGCTATTGTGAAG-3′, reverse 5′- GCCATCATTCTTGAGGAGGAAGT-3′; mTOR, forward 5′- ATGCTTGGAACCGGACCTG-3′, reverse 5′- TCTTGACTCATCTCTCGGAGTT-3′; MMP2, forward 5′-TACAGGATCATTGGCTACACACC-3′, reverse 5′- GGTCACATCGCTCCAGAC-3′; MMP9, forward 5′- TGTACCGCTATGGTTACACTCG-3′, reverse 5′- GGCAGGGACAGTTGCTTCT-3′.
2.5Plasmids and siRNA transfectionThe HOXB13 overexpression plasmids were synthesized by Genscript (Nanjing, China), and the corresponding empty plasmid pLVX-puro was used as negative controls. The small interfering RNAs (siRNAs) targeting HOXB13 (siHOBX13) were synthesized by Ribobio Co., Ltd. (Guangzhou, China). The sequences for siHOXB13 are as follows: siHOXB13–1, 5′-GCATTTGCAGGTACCTCTA-3′; siHOXB13–2, 5′-GCTCCTCCCTTGGTTATTA-3′; Negative Control siRNAs, 5′- GCAGTACGGATCCTTTCTA-3′. The transfection was performed with lipofectamine 2000 (Invitrogen, USA) or lipofectamine RNAiMAX (Invitrogen, USA) according to the manufacturer's protocols.
2.6Cell proliferation assayAbout 4 × 103 cells per well were seeded into 96-well plates to detect the cell proliferation ability using Cell Counting Kit-8 (CCK8) (Beyotime, C0038, China) and the absorbance values were measured at 450 nm with a microplate reader (Bio-Rad, USA).
2.7Cell invasion assayThe invasion assays were performed using Boyden Transwell chambers (8-μm pore size, BD Biosciences, USA). 5 × 104 cells were cultured with 150 μL serum-free medium in the upper chamber and 600 μL medium containing 10% FBS was added into the lower chamber. After 24 h of culture, the cells were fixed with paraformaldehyde and stained with crystal violet to observe the cell invasion.
2.8In vivo experimentSix-week-old C57BL/6 female nude mice were purchased from Beijing Weitong Lihua Biological Co., Ltd. and raised in a specific pathogen-free (SPF) environment. The mice were divided into four groups, including the control group (Lentivector), lentivirus overexpressing HOXB13 group (Lenti-HOXB13), MK2206 treatment group (MK2206), and lentivirus overexpressing HOXB13 combined with MK2206 treatment group (Lenti-HOXB13+ MK2206). 100 μL of cell suspension containing 3 × 105 cells (mixed with Matrigel at a ratio of 1:1) were injected subcutaneously in the right armpit of nude mice. The therapeutic dose of MK-2206 was 10 mg/kg subcutaneously in mice. During the experiment, the body weight and tumor size of the mice were measured every week and the tumor tissues were collected for qPCR, WB and other detections after five weeks of tumor formation.
2.9Western blotting (WB)Cells and tumor tissues were lysed in RIPA buffer containing protease inhibitors (Sigma, St. Louis, MO. USA) and phosphatase inhibitors (Roche, Sigma, St. Louis, MO, USA). The protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) and transferred to PVDF membranes (Bio-Rad, CA, USA). After blocking with blocking solution for 1 hour, the membranes were incubated with the primary antibodies overnight at 4 °C, including HOXB13 (Abcam, ab201682), p-Akt (CST, 4060), p-mTOR (CST, 5536), MMP2 (CST, 40,994), MMP9 (CST, 13,667) and β-actin (Abcam, ab8227). Then the membranes were incubated with horseradish peroxidase-conjugated secondary antibody for final exposure with the ECL chemiluminescence kit (Absin, abs920). The Image J program (http://rsbweb.nih.gov/ij/download.html) was used for densitometric analysis of the bands.
2.10Statistical analysisSPSS 20.0 software (SPSS, Chicago, IL) was used for statistical analysis. P-values were analyzed by two-sided Student's t-test, Mann-Whitney U test and χ2 test. A Kaplan-Meier curve and log-rank test were used to determine differences in survival between the two groups. A P-value < 0.05 was considered statistically significant.
2.11Ethical statementWritten informed consent was obtained from each patient included in the study and the study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the Ethics Committee of the Second Hospital of Nanjing University of Chinese Medicine (Approval ID: 2,008,022). Furthermore, all animal experiments were carried out in accordance with the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978) and the protocol approved by the Research Ethics Committee of Nanjing Drum Tower Hospital.
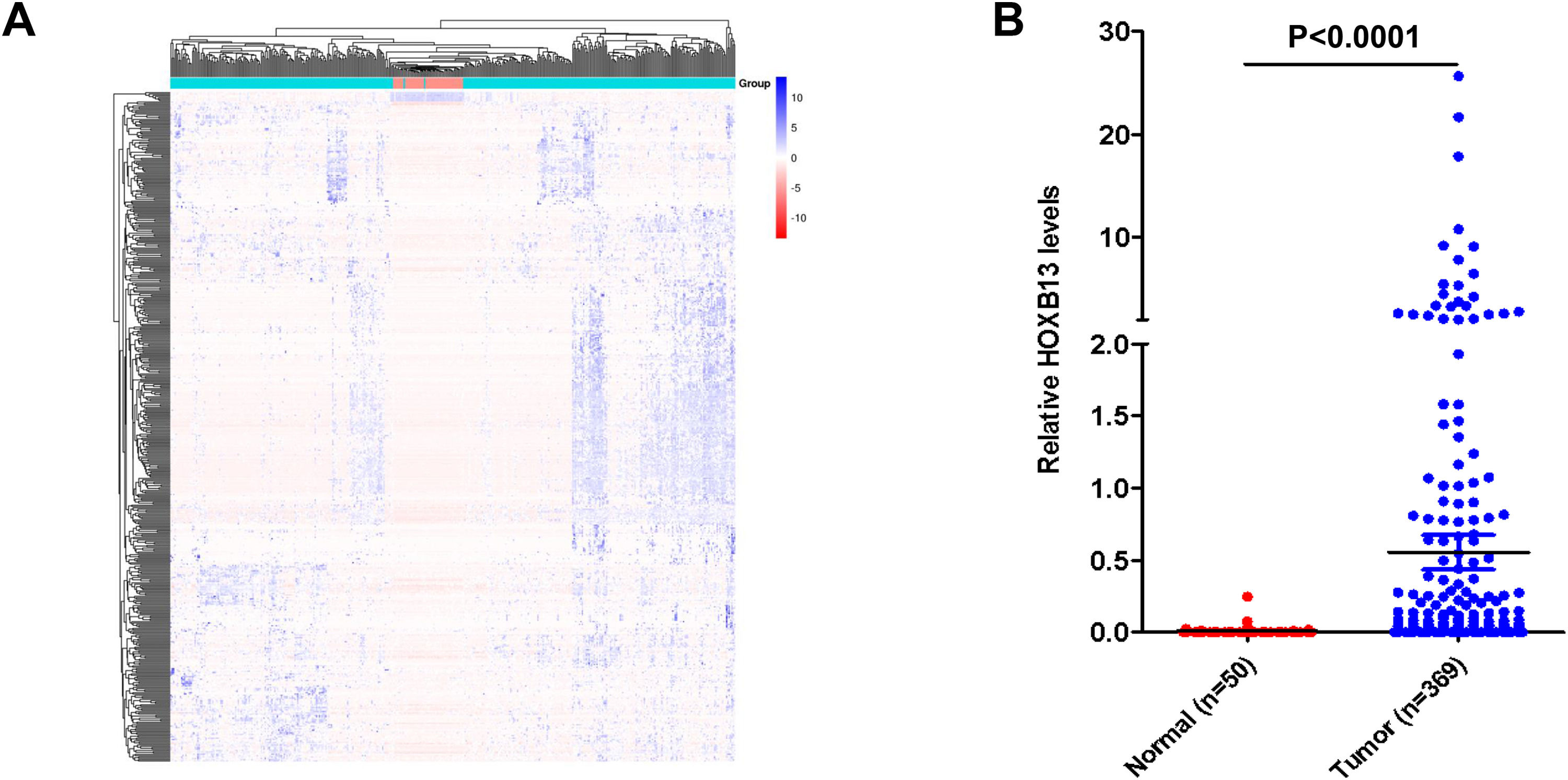
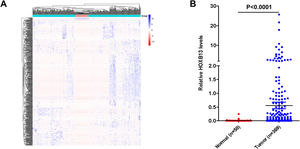
3Results3.1HOXB13 mRNA expression was up-regulated in HCC tumors by bioinformatics analysisWe screened a total of 5431 up-regulated genes and 1355 downregulated genes from 60,482 genes, including mRNA and lncRNA in the TGCA database, and the heatmap of the top 500 genes by differential fold as shown in Fig. 1A. Among the up-regulated genes, the expression level of HOXB13 in HCC tumors (n = 369) was 55.5 times higher than that in normal controls (n = 50) (P = 4.16E−11) (Fig. 1B). It is suggested that HOXB13 may be involved in the development of HCC.
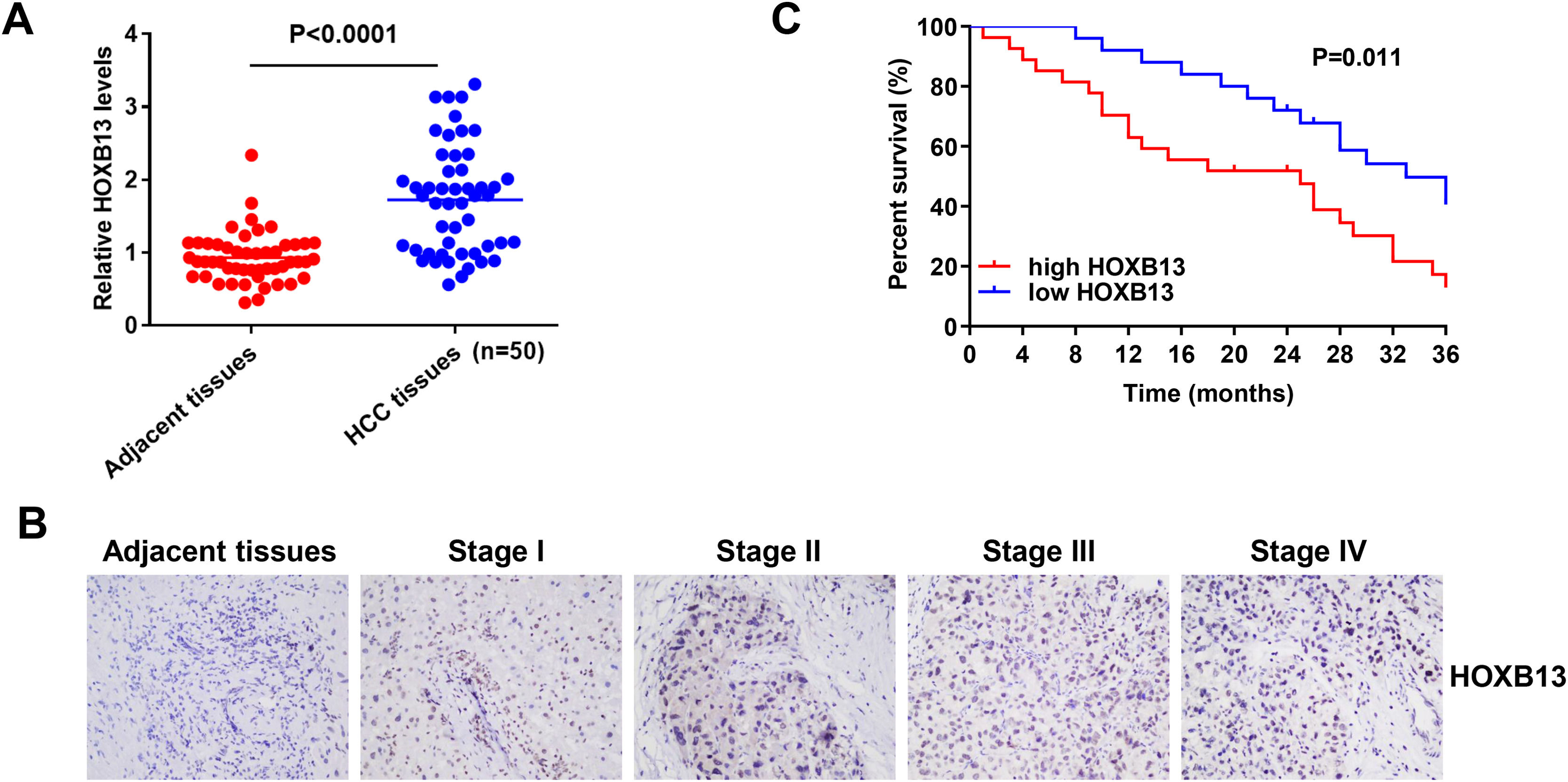
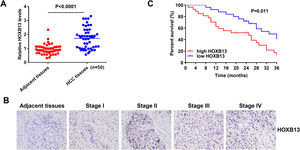
3.2HOXB13 is overexpressed in HCC patient tissues and correlated with HCC stage and survivalTo verify whether HOXB13 is highly expressed in HCC patient tissues, 50 pairs of HCC tumor tissues and their adjacent tissues were collected for qRT-PCR and IHC detection. The results of qRT-PCR demonstrated that the mRNA expression level of HOXB13 was significantly up-regulated in HCC tissues compared with adjacent tissues (Fig. 2A). The IHC results showed that HOXB13 was minimally or weakly positive in HCC stage I/II and strongly positive in stage III/IV (Fig. 2B) and the positive proportion of HOXB13 in HCC tissues was significantly higher than that in adjacent tissues (Table 1). Furthermore, we found that HOXB13 was associated with tumor size (P = 0.0207) and TNM stage (P = 0.0065) but not with sex, age and alpha fetoprotein (AFP) levels (Table 2). In addition, the survival analysis based on the HCC database using Project Betastasis (http://www.betastasis.com) confirmed that high expression of HOBX13 in HCC tissues was associated with poor survival (P = 0.011) (Fig. 2C). These results suggest that high HOXB13 expression may be related to the development and survival of HCC.
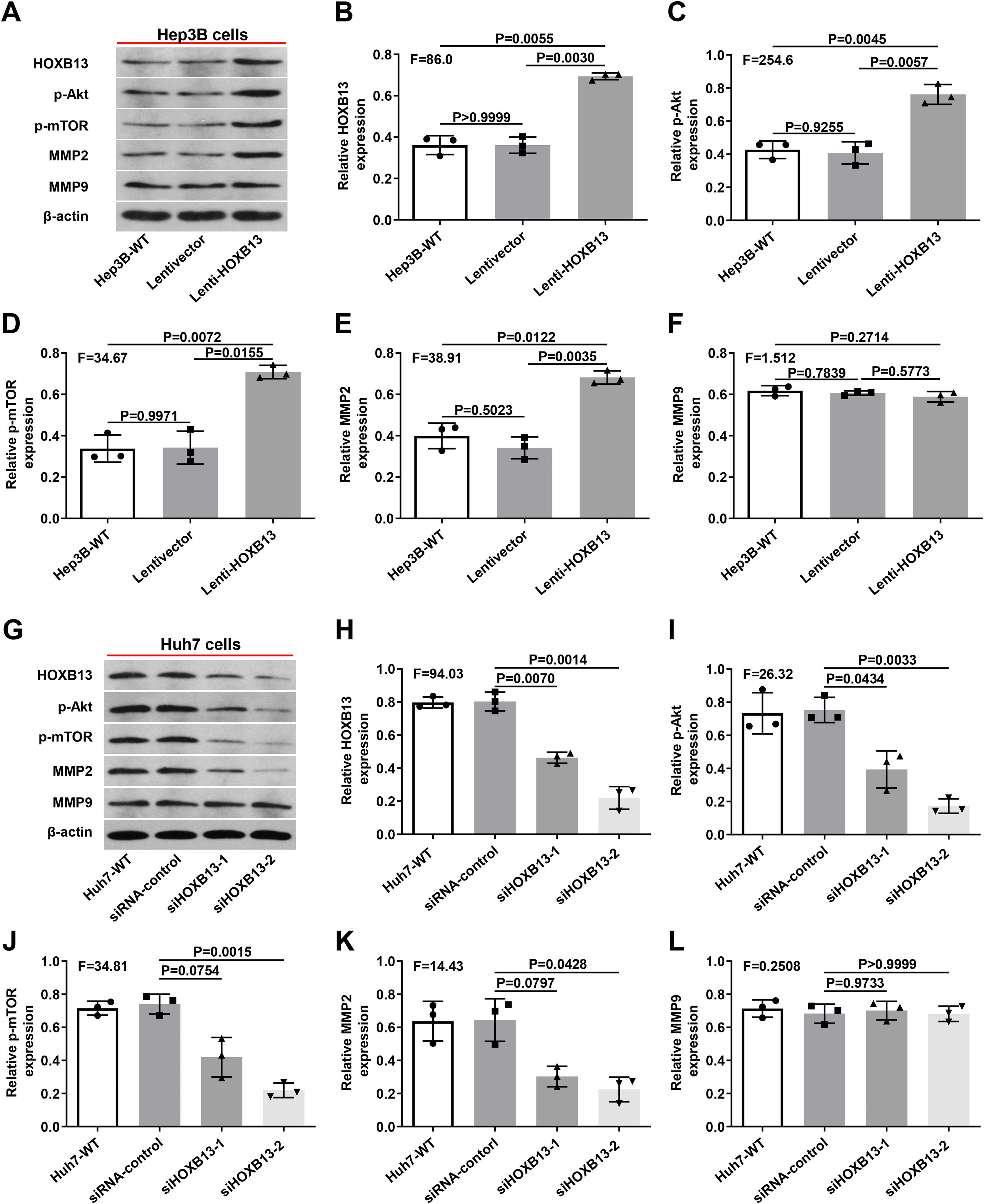
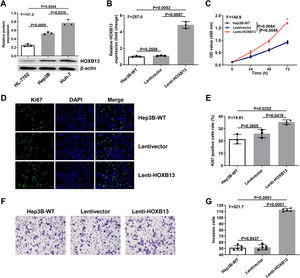
The biological function of HOBX13 was further investigated in vitro. We first analyzed the protein expression of HOBX13 in normal liver epithelial HL-7702 cells and hepatoma cell lines Hep3B and Huh-7. The WB results showed that the expression of HOBX13 was significantly increased in Hep3B and Huh-7 cells compared with HL-7702 cells (Fig. 3A). Then, Hep3B cells stably overexpressing HOXB13 (Lenti-HOXB13) were constructed using the lentivirus system. From the results of qRT-PCR, the expression of HOXB13 in Lenti-HOXB13 cells was up-regulated by nearly five times compared with the WT group and Lentivector group (Fig. 3B) (P<0.005). As shown in Fig. 3C, the proliferation ability of Lenti-HOXB13 cells was significantly higher than that in the other two control groups, especially after 72 h of culture. Further, the cells in each group were stained with Ki67 (Santa, sc-23,900) and DAPI (Santa, sc-359,850) by immunofluorescence, and it was found that the proportion of Ki67 positive cells in Lenti-HOXB13 group was approximately two times higher than that in the control groups, indicating overexpression of HOXB13 can effectively promotes Hep3B cell proliferation (Fig. 3D-E). In the invasion assay, the cell invasion ability in the Lenti-HOXB13 group was also greatly enhanced compared with the WT and Lentivector groups (Fig. 3F-G).
Overexpression of HOBX13 in Hep3B cells promotes cell proliferation and invasion
a. The protein expression of HOXB13 in HL-7702, Hep3B and Huh-7 cells detected by WB. b. HOXB13 mRNA expression was analyzed by qRT-PCR. c. CCK8 assay to detect cell proliferation ability. d-e. Cell growth ability is determined by Ki67 and DAPI staining.
f-g. Invasion assays in different cell groups.
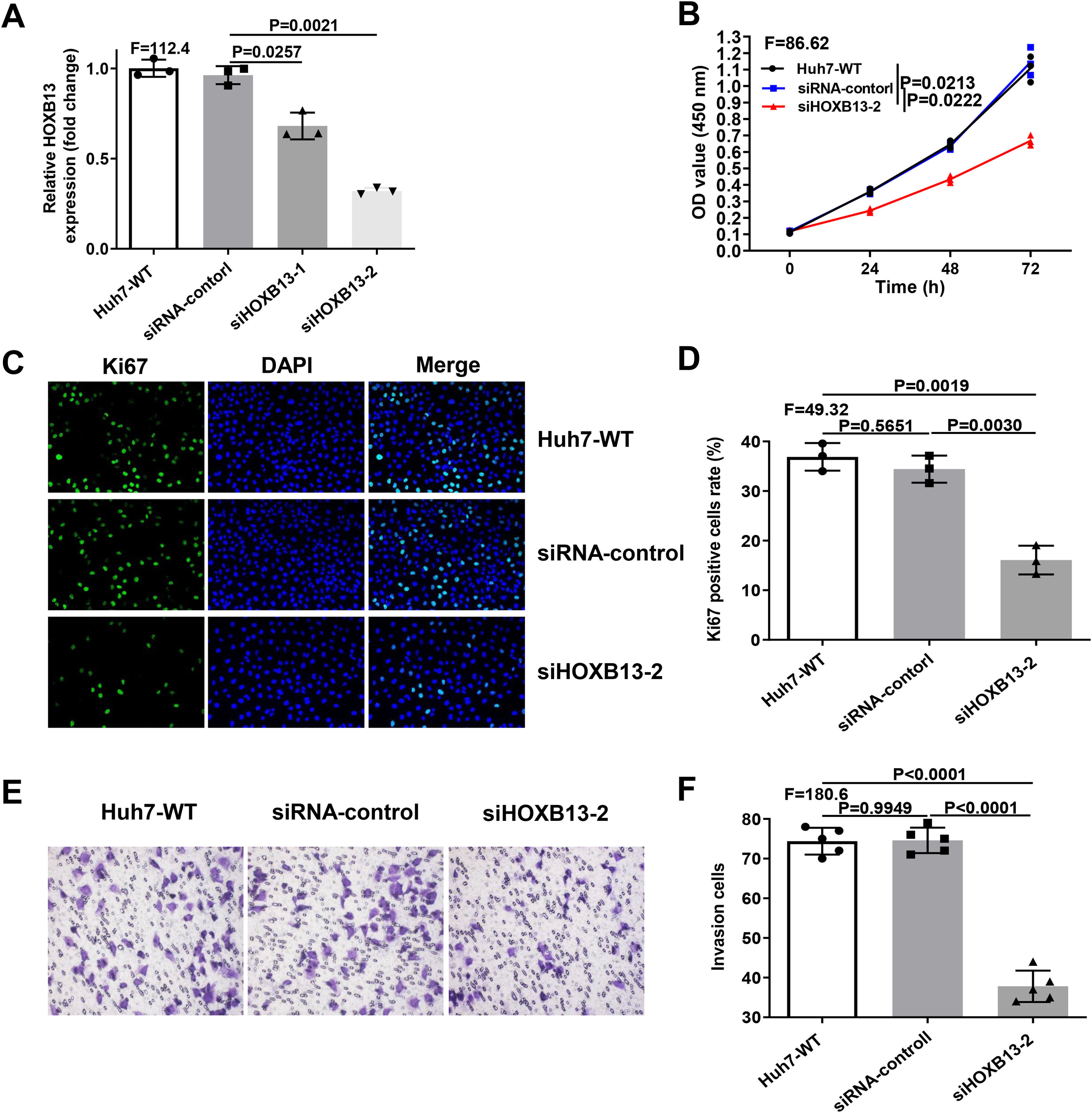
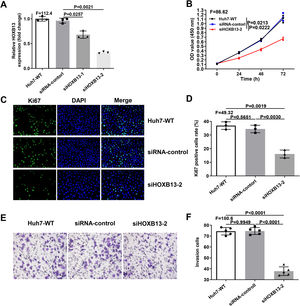
Since overexpression of HOXB13 promotes cell proliferation and invasion, we performed loss-of-function assays in Huh-7 cell line to explore whether downregulation of HOXB13 would affect cell functions. The Huh-7 cells were transfected with the siRNAs of HOXB13 (siHOXB13–1 and siHOXB13–2) and siRNA control, and the expression of HOXB13 was confirmed by qRT-PCR. As shown in Fig. 4A, both siHOXB13–1 and siHOXB13–2 could reduce the HOXB13 expression in Huh7 cells, and siHOXB13–2 with a better downregulation effect was used for subsequent experiments. The results of cell function experiments showed that the proliferation and invasion abilities of Huh7 cells transfected with siHOXB13–2 were significantly inhibited, which were contrary to the results of HOXB13 overexpression in Hep3B cells (Fig. 4B-F). The differences were statistically significant (P<0.005).
Downregulation of HOBX13 in Huh7 cells inhibits cell proliferation and invasion
a. HOXB13 mRNA expression analyzed by qRT-PCR. b. CCK8 assay to detect cell proliferation ability. c-d. Cell growth ability is determined by Ki67 and DAPI staining.
e-f. Invasion assays in different cell groups.
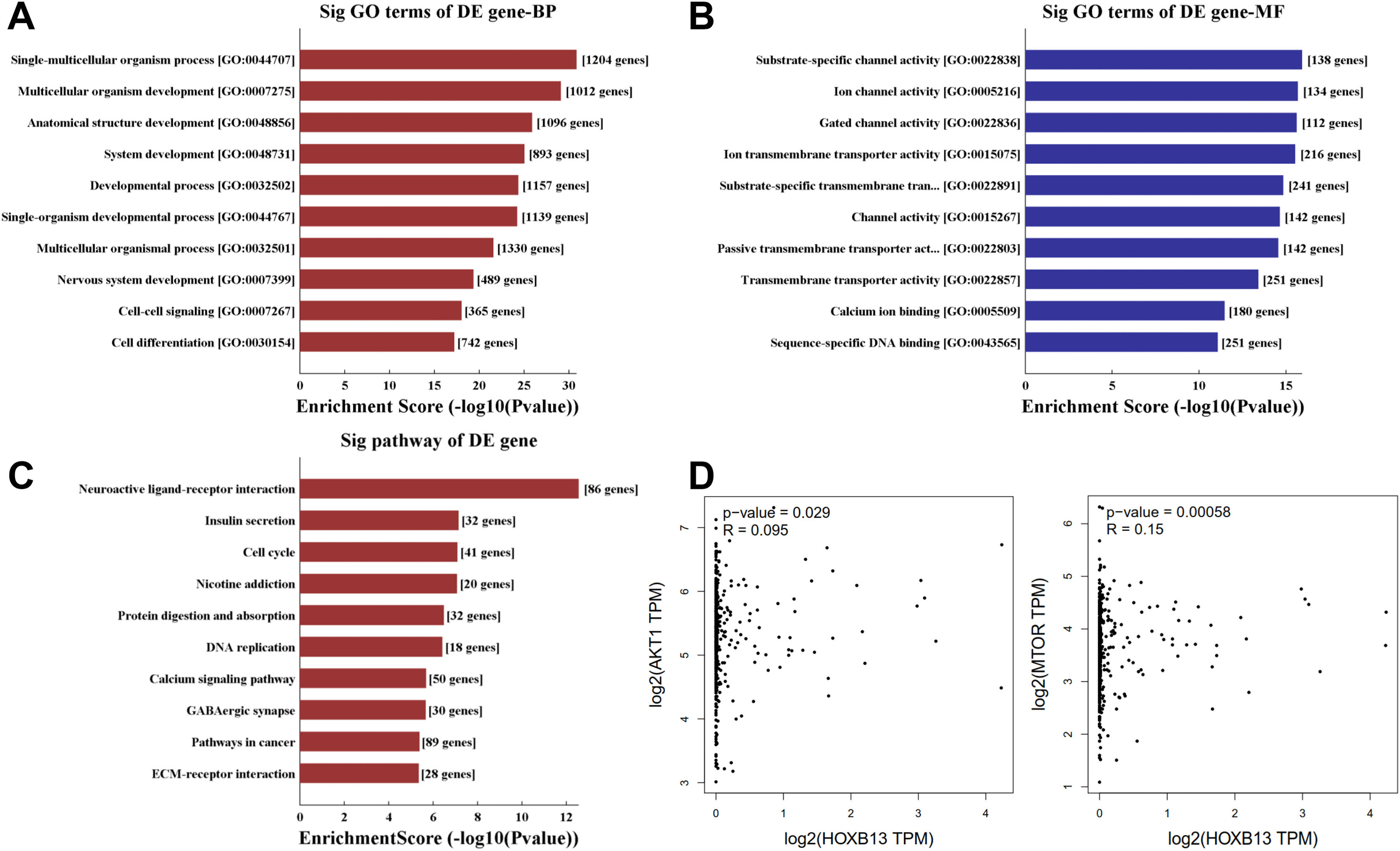
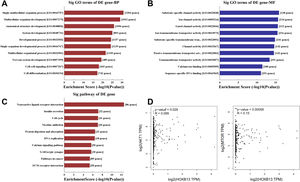
To explore the molecular mechanism of HOBX13 in HCC regulation, GO pathway analysis was performed on the differential genes, including biological processes, molecular functions and signaling pathways (Fig. 5A-C). The results showed that HOXB13, as a sequence-specific transcription factor, may be involved in most biological processes of HCC, such as developmental process, cell differentiation, DNA binding and so on. The AKT/mTOR pathway plays an important role in the occurrence, treatment and prognosis of malignant tumors [18–22]. It was further predicted that HOXB13 was positively correlated with the expression of AKT and mTOR (P<0.05) (Fig. 5D), suggesting that HOXB13 may regulate HCC development through the AKT/mTOR pathway.
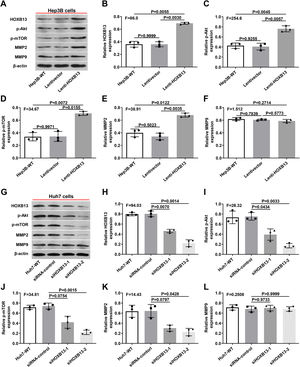
Therefore, we examined the expression of p-AKT, p-mTOR, MMP2 and MMP9 in Lenti-HOXB13 and siHOXB13 cells to confirm whether HOXB13 is involved in regulating the AKT/mTOR/MMP signaling pathway. As shown in Fig. 6A, after overexpression of HOXB13 in Hep3B cells, the protein expression levels of PI3K, AKT and MMP2 were significantly up-regulated compared with the control groups, and the differences were statistically significant (Fig. 6B-E). However, after downregulating HOXB13 in Huh7 cells with siHOXB13–1 and siHOXB13–2, the expression levels of the above molecules were inhibited to different degrees (Fig. 6G), and the difference was statistically significant (Fig. 6H-K). The MMP9 expression in each group did not change significantly (Fig. 6F, 6L).
Effects of HOXB13 overexpression or downregulation on protein expression of p-Akt, p-mTOR, MMP2 and MMP9
a. The expression of HOBX13, p-Akt, p-mTOR, MMP2 and MMP9 in Hep3B cells by WB. b-f. Quantification of the expression levels of HOBX13, p-Akt, p-mTOR, MMP2 and MMP9. g. The expression of HOBX13, p-Akt, p-mTOR, MMP2 and MMP9 in Huh7 cells by WB. h-l. Quantification of the expression levels of p-Akt, p-mTOR, MMP2 and MMP9.
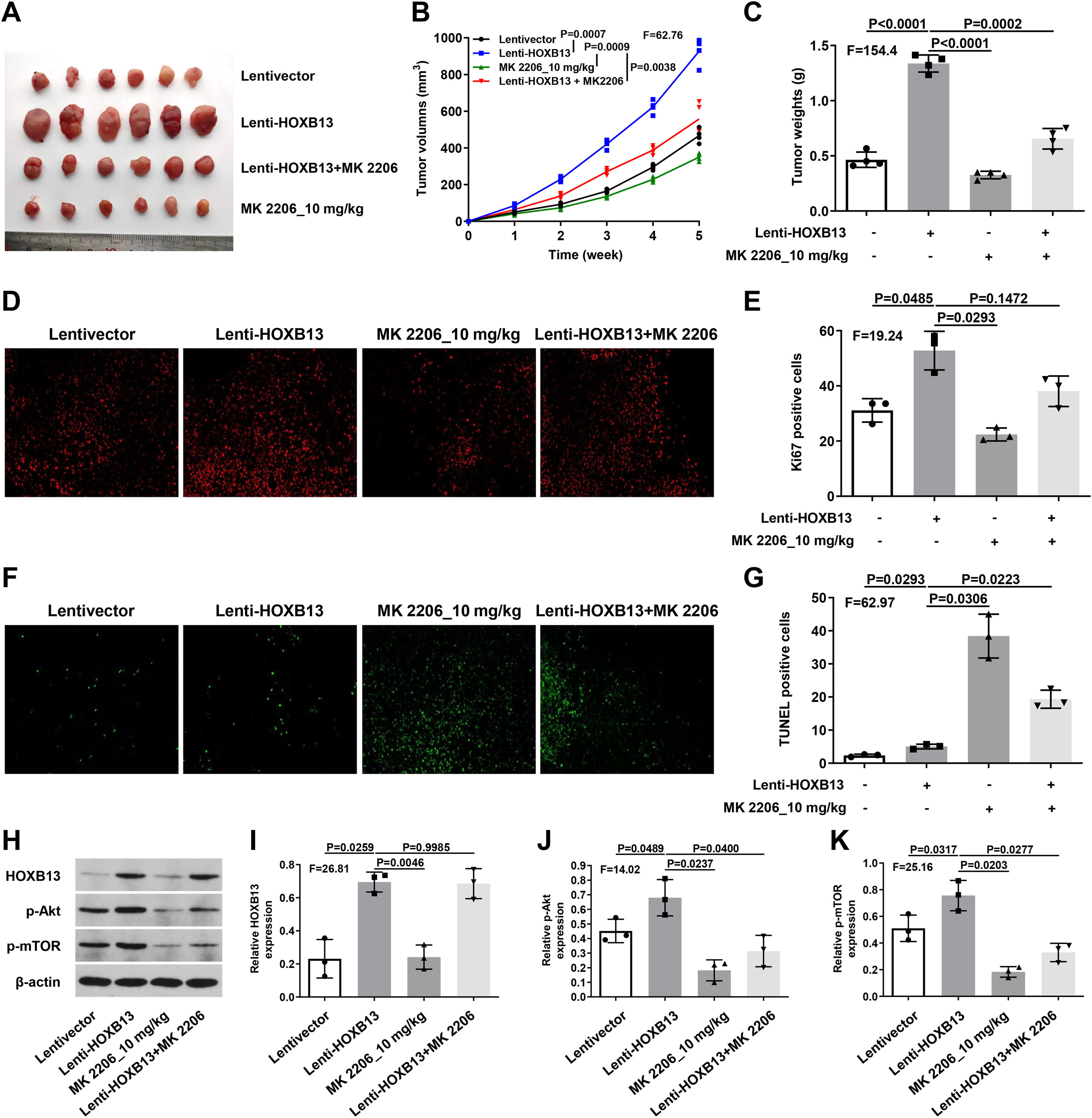
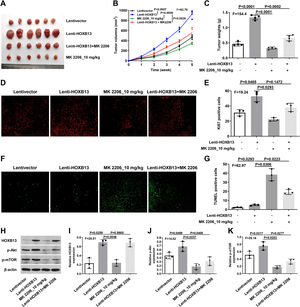
To verify the effect of HOXB13 on HCC in vivo, we constructed the HCC mouse model for the following experiments. There were four groups, including Lentivector, Lenti-HOXB13, MK2206_10 mg/kg and Lenti-HOXB13+MK2206 groups. The MK2206 used in the experiments is an AKT inhibitor that can inhibit tumor growth by blocking the PI3K/AKT pathway. The results showed that the Lenti-HOXB13 group had the largest tumor volume and the tumor growth rate in this group was significantly faster than the other three groups. MK2206 could effectively inhibit the tumor growth of mice in MK2206_10 mg/kg and Lenti-HOXB13+MK2206 groups (Fig. 7A-C).
HOXB13 promotes tumor growth by activating the AKT/mTOR/MMP2 pathway
a. Tumor images of different groups of mice. b. Tumor growth curve of mice in each group. c. Statistical graph of tumor weight in mice. d. Ki67 staining of tumor tissues by IHC. e. Proportion of Ki67+ cells in each group. f. Tunel staining of tumor tissues. g. Proportion of Tunel+ cells in each group. h. The expression of HOBX13, p-Akt, p-mTOR in tumor tissues by WB. i-k. Quantification of the expression levels of HOBX13, p-Akt, p-mTOR.
Further, the IHC results of tumor tissues in each group showed that the proportion of Ki67 positive cells in Lenti-HOXB13 group was the highest and the proportion of Tunel positive cells was the lowest, indicating that overexpression of HOXB13 can effectively promote tumor cell proliferation and inhibit their apoptosis in vivo. And the function of HOXB13 could be partially reversed when given to MK2206 treatment (Fig. 7D-G).
As shown in Fig. 7H, the expression of related protein molecules in tumor tissues was analyzed by WB. The expression levels of p-Akt and p-mTOR were significantly up-regulated in Lenti-HOXB13 group. MK2206 could effectively inhibit the phosphorylation levels of AKT and mTOR, but had no regulatory effect on the expression of HOXB13 (Fig. 7I-K). The in vivo results demonstrated that HOXB13, as an upstream regulator of Akt, could accelerate HCC progression by activating the AKT/mTOR pathway.
4DiscussionHCC remains a global health challenge with increasing incidence and high mortality worldwide. Major risk factors for HCC include long-term alcohol consumption, diabetes, HBV, HCV and so on [23]. The pathophysiology of HCC is a complex multistep process and understanding the molecular mechanisms underlying HCC progress is crucial for early diagnosis and development of rational therapeutic strategies for HCC. Our study found that the expression of HOXB13 was significantly up-regulated in HCC tumors compared with normal controls by bioinformatics analysis and detection of 50 pairs of HCC tumor tissues. Furthermore, we found that HOXB13 expression was associated with tumor stage and survival of HCC patients. These results suggest that HOXB13 may be involved in the occurrence and development of HCC.
HOXB13 is the last identified gene in the homeobox family, which is involved in the regulation of normal developmental systems. For example, HOXB13 has been shown to be a major HOX factor driving the development and differentiation of prostate epithelial cells [24]. HOXB13 can regulate cardiomyocyte maturation and proliferation by cooperating with Meis1 and targeting the calcineurin-HOXB13 pathway was expected to bring promise for the treatment of cardiac injury [25]. In recent years, increasing studies demonstrated that HOXB13 plays different roles in cancer progression. Nakamura H et al. described a novel EWSR1-HOXB13 rearrangement in a fibroblastoma of a young woman [26]. Long noncoding RNA LincIN can promote esophageal squamous cell carcinoma progression by regulating the NF90/microRNA-7/ HOXB13 axis [27]. In breast cancer, the oncoprotein HBXIP enhances HOXB13 acetylation to prevent HOXB13 degradation and co-activates HOXB13 to stimulate IL-6 secretion contributing to tamoxifen resistance [28]. Our findings suggest that HOXB13 can promote HCC progression. Specifically, overexpression of HOXB13 in Hep3B cells could effectively promote cell proliferation and invasion, while downregulation of HOXB13 in Huh7 cells yielded the opposite results. In addition, HOXB13 also contributed to the growth of HCC in mice by promoting cell proliferation and inhibiting cell apoptosis. We further predicted the signaling pathways that HOXB13 may be involved in the regulation of HCC by bioinformatics and found that HOXB13 had a potential relationship with the AKT/mTOR signaling pathway. Then the expression levels of related proteins were detected, and the results showed that HOXB13 could effectively activate the AKT/mTOR/MMP2 axis in vitro and in vivo. In addition, treatment of tumor-bearing mice with MK-2206, an inhibitor of AKT [29], could only partially reverse the effects of HOXB13 on HCC and the expression of HOXB13 in tumors was not affected, indicating that HOXB13 is located upstream of the AKT/mTOR signaling pathway. Therefore, it may be a feasible way to improve drug resistance targeting downstream pathways by developing drugs targeting HOXB13.
Notably, several studies have also reported that HOXB13 can act as a tumor suppressor gene in several cancers [30–32]. Sui et al. demonstrated that HOXB13 expression was significantly downregulated in gastric cancer tissues and was associated with tumor stage and survival, and maybe a potential prognostic factor for gastric cancer [33]. Although our findings suggest that HOXB13 regulates HCC progression by activating the AKT/mTOR/MMP2 pathway, the direct interaction mechanism between HOXB13 and the signaling pathway has not been verified. Therefore, the mechanism of HOXB13 in HCC remains to be further elucidated. In addition, we set up the control groups in vitro experiments, including WT, Lentivector and siRNA-control groups, but no non-HCC cell line was used as a negative control. In vivo experiment, we proved that HOXB13 overexpression significantly promoted tumor growth in mice by promoting cell proliferation and inhibiting cell apoptosis, but lacked the effect of knocking down HOXB13 on tumor growth in mice, making our conclusions not strong enough.
5ConclusionsThe research showed that HOXB13 was overexpressed in HCC tissues and was positively correlated with the tumor stage and survival of HCC patients. HOXB13 could promote the proliferation and invasion of HCC cells and the tumorigenicity of HCC cells in mice by activating the AKT/mTOR/MMP2 pathway. The findings may provide evidence for a novel regulatory mechanism involving HOXB13 in HCC and HOXB13 may serve as a promising therapeutic target for HCC.
FundingThis work was supported by the Nanjing Medical Science and Technique Development Foundation (QRX17141 and YKK19056) and the National Natural Science Foundation of China (82103578).
Author contributionsML, TTT and YG designed the project, conducted experiments and analyzed the data. YT and JP collected clinic samples. JZ and QX organized the figures and tables. HS, LYZ and YXC designed the project and provided administrative support. All authors wrote the manuscript together and approved the final manuscript.
Data sharing statementThe datasets used and analyzed during the current study are available from the corresponding author upon reasonable request.
Declaration of interestNone