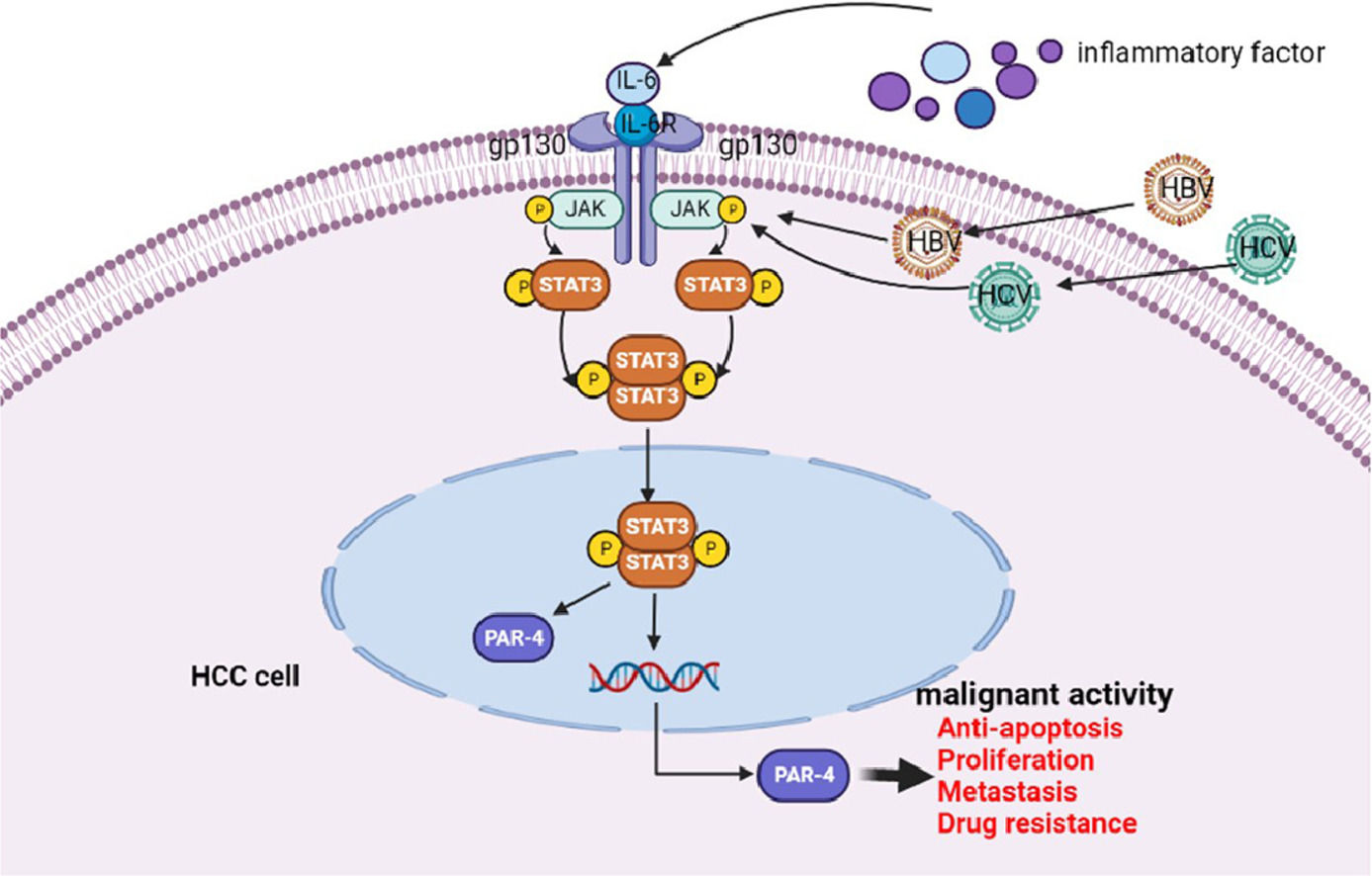
The schematic diagram of IL-6/STAT3 signaling pathway stimulates PAR-4 to promote malignant behavior of HCC cells
Hepatocellular carcinoma (HCC) is the most common pathological type of primary liver cancer (PLC). According to global tumor data for 2021 [1], the number of new HCC cases reached more than 900,000, and the number of deaths was 830,000, ranking third. As a country with a large population, China ranks first in the world in terms of the number of new cancer cases and deaths. The number of new HCC cases is 410,000, ranking fifth, and death cases are 390,000, ranking second [2]. Liver cancer occurs in the middle and advanced stages when it is clearly diagnosed. Approximately 80% of HCC patients are in an advanced stage when diagnosed [3], with a median survival time of approximately nine months and a 5-year overall survival rate of only 10%. There have been great advances in the treatment of HCC, including surgical treatment (such as radical resection) and local treatments, including radiofrequency ablation(RFA) and transarterial chemoembolization (TACE) plus systemic treatments [4], and the treatment of immune checkpoint inhibitors [5,6], etc. these shows a promising prospect for liver cancer therapy. However, it is estimated that there is a 60 to 70% recurrence risk. In addition, chemotherapy, radiotherapy, tyrosine kinase inhibitors and immune checkpoint inhibitors (ICIs) have made great advances and have not been shown to improve clinical outcomes in terms of progression-free survival (PFS) or overall survival (OS) [7]. HCC has a poor prognosis and is prone to relapse, which seriously threatens the lives and health of patients. Therefore, new diagnostic and treatment strategies are urgently required. Therefore, further understanding of the molecular mechanisms underlying the malignant progression of HCC is crucial for improving the therapeutic efficacy and prognosis of HCC.
The occurrence and development of HCC are complex processes involving multifactorial and multistep interactions, which are related to the dysregulation of many signaling pathways. Recent studies on the occurrence and development of HCC have identified IL-6/STAT3, PI3K/Akt/mTOR [8], MAPK [9], NF-κB [10], and other signaling pathways. The IL-6/STAT3 signaling pathway plays an important role in the initiation, development, invasion, and metastasis of HCC [11]. Researchers initially discovered the IL-6/STAT3 signaling pathway in the regulation of downstream signals by interferon (IFN) and IL-6 [12]. Many related studies have focused on the IL-6/STAT3 signaling pathway as a potential wedge in targeted therapy for liver cancer [13,14]. As a classical extracellular stimulator of this pathway, IL-6 is secreted into the microenvironment of liver tissue during hepatogenesis and infection with hepatitis B virus (HBV) or hepatitis C virus (HCV). By targeting the IL-6 receptor(IL-6R) and GP130, the JAK/STAT3 signaling pathway is activated, thereby causing a series of malignant behaviors such as proliferation, invasion, and metastasis of liver cancer cells [15,16].
Prostate apoptosis response protein-4 (PAR-4), also known as PRKC apoptosis WT1 regulator (PAWR), was first identified in apoptotic rat prostate cells [17]. It is a protein containing a leucine zipper domain, which is widely expressed in many cell types and tissues and is ubiquitous in the cytosol and nucleus of many tumor cells. The PAR-4 protein is approximately 40 kDa in size and includes 342 amino acids in humans, 332 amino acids in rats, and 333 amino acids in mice. PAR-4 is evolutionarily conserved across all vertebrates, with rat and mouse PAR-4 proteins showing 93% amino acid homology and rat and human PAR-4 proteins having 75% identical and 84% functionally similar amino acids.
PAR-4 has been found to be down-regulated in most tumor tissues, including breast cancer, pancreatic cancer, prostate cancer, endometrial cancer, etc. Its downregulation is considered one of the key events in tumorigenesis, and low expression of PAR-4 predicts poor prognosis. PAR-4 is widely expressed in human cells, and most of its expression is localized to the cytoplasm and nucleus. In recent years, PAR-4 has become a research hotspot as a potential target for tumor therapy. Several studies have shown that PAR-4 is involved in the development of various tumors and can cause programmed cell death via exogenous and endogenous pathways [18,19]. A large number of studies have reported in prostate cancer [20], breast cancer [21], pancreatic cancer [22], ovarian and endometrial cancer [23] and other tumors that PAR-4 can specifically inhibit the proliferation of tumor cells to play a cancer suppressor. However, there are no relevant reports on PAR-4 expression in HCC. The expression of PAR-4 in HCC cells and tissues and its role of PAR-4 in the occurrence and development of HCC require further study. IL-6 is an important stimulator of the tumor microenvironment and plays a key role in promoting the occurrence and development of HCC [24]. This suggests that IL-6 may downregulate the expression of PAR-4 in HCC, thereby promoting the occurrence of malignant behavior in HCC. To fill the gap in the research on PAR-4 in HCC, this study aimed to explore the mechanism of IL-6 or IL-6/STAT3 signaling pathway regulation of PAR-4 and its effect on the malignant behaviors of HCC at the cellular and molecular levels. The effect of IL-6/STAT3 signaling pathway upregulation of PAR-4 on the malignant behaviors of HCC cells was investigated at the cellular level in vitro and in vivo and may provide a solid theoretical basis for targeted therapy of HCC.
2Materials and methods2.1Collection of experimental tissue specimens and tissue chip technologyThirty patients with liver cancer diagnosed clinically or pathologically at the Second Affiliated Hospital of Hainan Medical College between January 2019 and December 2020 were enrolled. Exclusion criteria included active bleeding within the last six months and a history of blood transfusion within the past month. And acute infection or inflammation in the past three months. HCC staging was performed using the China Liver Cancer Staging (CNLC), as shown in Appendix 1. All subjects provided informed consent, and the collection and use of tissue samples were approved by the Medical Ethics Committee of the Second Affiliated Hospital of Hainan Medical College. Tumor tissues of 30 HCC patients hospitalized at the Second Affiliated Hospital of Hainan Medical College and the corresponding adjacent tissues of the patients were collected. Adjacent tissues were located ≥2 cm from the tumor edge. After removal, all tissue samples were divided into two groups and stored. After removal, group 1 was frozen in liquid nitrogen for more than 5 min, transferred to −80 °C for storage in an ultra-low-temperature refrigerator, and used for tissue protein extraction. In the other group, the tissue chips were prepared by soaking them in 4% paraformaldehyde. Thirty patients with clinically confirmed HCC were included in the study. The age, sex, drinking history, Performance Status (PS) score (see Appendix 2), Child-Pugh grade (see Appendix 3), and alpha-fetoprotein (AFP) levels of the experimental subjects were recorded in detail. AFP concentration, HBV DNA copy number, albumin (ALB), alanine transferase (ALT), tumor stage, histological grading (histological grading, HG), Ki-67 content, tumor number, tumor size, microvascular infiltration, distant metastasis, and other clinical parameters. All 30 patients with HCC were followed up by telephone until December 2021, and the Overall survival (OS) of 30 patients with HCC was recorded. The relationship between PAR-4 and clinicopathological features of HCC patients is described below (diagram).
2.2Tissue microarrays chip techniqueThe tissue microarray chip technique was used to detect protein expression in liver cancer and adjacent tissues. Briefly, paraffin section preparation: The excised liver cancer tissues and corresponding adjacent tissues were fixed with 4% paraformaldehyde for more than 24 h, and the tissues of the desired target parts were repaired, leveled with scissors, and then placed into the dehydration box with labels. Dehydration and wax leaching: Depending on the concentration of ethanol, the dehydration box was placed in a dehydrator for dehydration, which was 75% (4 h)→85% (2 h)→90% (2 h)→ absolute ethanol (30 min × 2)→ xylene (8 min × 2), and then melted at 65 °C (1 h × 3). Embedding: The melted wax was placed in the embedding frame, and the required tissue was removed from the dehydration box before the wax solidified, placed into the embedding frame as required, cooled at −20 °C, removed from the embedding frame after the wax solidified, and trimmed at the same time. Slicing: place the trimmed wax block on the −20 °C freezing table to cool, and then place the cooled wax block on the paraffin slicer to slice the cooled wax block with a thickness of approximately 4 µM. The tissue was flattened in warm water at 40 °C in a spreading machine, mounted on a slide, and the slices were baked in an oven at 60 °C. After the wax was baked, slices were removed and stored at room temperature until further use. Hematoxylin-eosin (HE) staining and localization, tissue chip production, tissue chip HE staining, and immunohistochemistry (IHC). Protein expression was observed with a microscope, and images were collected, saved, and analyzed using Aipathwell software. These methods have been described previously [25,26].
2.3Cell lines and BALB/C nude micePLC/PRF/5 (hereinafter referred to as PLC) HCC cells were purchased from the Shanghai Cell Bank (Shanghai, China), HLE cells were purchased from Kebai Biological Company, and Bel7402 cells were purchased from Bode Biological Company. Qualified cell identification books are available for all cell lines, and cell cultures were performed in accordance with the Guidelines for Cell Experiments. BALB/C nude mice were purchased from Hunan Anshengmei Pharmaceutical Research Institute Co., Ltd.. They were 4–5 weeks old and all were male. The weight of the nude mice was between 18 and 20 g, and they were kept in the animal room of the Hainan Medical College (with an experimental animal use license), which met the SPF standard. The temperature of the living environment of the mice was maintained at 21–23 °C and they lived in an independent ventilated cage (IVC) system. The drinking water of the nude mice was autoclaved, and the bedding and feed purchased were SPF. All animal experiments passed the ethical review of laboratory animals of the Laboratory Animal Committee of Hainan Medical College.
2.4Western blotting analysisTo detect the expression of PAR-4 in liver cancer and adjacent tissues, the influence of IL-6 on the expression of PAR-4, STAT3, p-STAT3, Src, and Ras in HCC cells was analyzed by Western blotting. Briefly, the proteins were probed with the following primary antibodies: mouse anti-PAR-4 (1:500), anti-STAT3 (1:500), anti-p-STAT3 (1:500), anti-Src (1:500), anti-Ras (1:500), anti-tubulin (1:1000), and anti-GAPDH (1:1000)(all from eBioscience and Abcam Inc.). The detailed procedure has been previously described [27,28]. According to the relevant operation process of the Tianeng automatic imaging system, development, photography, and preservation were carried out, and the gray value of the strip was analyzed using ImageJ.
2.5Bioinformatics analysisThe UALCAN database (ualcan.path.uab.edu/home/) can provide TCGA database-related analysis data. On the home page, click TCGA, Enter gene name (PAWR) in Scan by gene(s)/ Enter gene symbol(s), Enter HCC in TCGA Dataset, and click exploration. Expression in HCC and pan-cancer views of various tumors.
The STRING database (https://cn.string-db.org/) can provide the protein interaction relationship in the database: Protein Name, enter the Protein Name and click search. Select Homo sapiens and click Continue; proteins interacting with PAWR can appear. At the interface where the desired protein interactions appeared, click analysis-KEGG pathways were used to query the possible signaling pathways involved between interacting proteins.
2.6RT-PCR detectionThe upstream and downstream primers of PAR-4 and internal reference primers were designed by the “stem-loop method” and synthesized by Shanghai Sangon Biotech (Table 1). Real-time PCR(RT-PCR) was used to detect the mRNA levels of target genes, as previously described [29].
2.7Lentivirus infection and construction of stable cell linesShort hair (sh) clips interfering with RNA lentivirus (Shanghai Jikai Biotech, Limited Company, Shanghai, China), shIL-6R lentivirus, and shPAR-4 lentivirus were used to infect the human hepatoma cell lines, PLC, HLE, and Bel7402. The virus stock solution was added to the cells and mixed according to MOI values of 3, 10, 15, 30, and 100, and an appropriate amount of polybrene was added. Four hours after adding the virus, 50 µL of medium was added to the cells in a 96-well plate. Fluorescence was observed using a microscope 48 h after virus infection. 72 h after virus infection, the group with infection efficiency of approximately 80% and favorable cell growth status was selected as the basis for infection conditions and MOI corresponding to subsequent infection. According to the previous transfection conditions for 72 h, the culture medium was changed to contain purinamycin (the concentration of purinamycin in Bel7402 cells was 2 μg/mL, and the concentration of PLC and HLE was 1 μg/mL), and fresh complete culture medium containing 1 μg/mL purinamycin was changed every 1–2 days according to the cell conditions. After all cells in the blank group were killed, the concentration of purinamycin was halved. After 3 to 5 days of virus culture, the cells in the infected group were removed and replaced with fresh complete culture medium, and first-generation (P1) stable cell lines were obtained.
2.8MTT colorimetric analysisThe MTT Cell Proliferation Assay Kit (colorimetric) was used to analyze the proliferation of PLC, HLE, and Bel7402 cells. PLC, HLE, and Bel7402 cells were digested with trypsin and diluted with fresh medium into a cell suspension diluted to 1.0 × 104/mL. Two hundred microliters of cell suspension was aspirated using a pipetting nozzle, added to a 96-well plate by drop, and placed in the incubator for further cultivation for 24 h. Different concentrations of media containing recombinant human IL-6 protein were prepared according to the following concentrations: 0, 2.5, 5, 10, 20, 40, and 80 ng/mL. Each concentration was tested in five wells. According to the specific requirements of the experiment, the culture medium containing IL-6 was replaced and incubated for 24 h and 48 h. Under darkness, 20 µL of MTT solution with a concentration of 5 mg/mL was added to each well, and the plate was placed back into the incubator for further cultivation for four h. Then, the 96-well plate was removed, the liquid in the wells was sucked up, 180 µL DMSO was added to each well, and then slowly shaken on a shaker for 10 min in the dark. The absorbance was measured at 490 nm using a microplate reader. The absorbance (OD) of each drug-treated group and control group was measured, and the average value of each group was calculated. The experiment was repeated three times. The effect of IL-6 on the proliferation rate of liver cancer cells was calculated using the following formula: proliferation rate (%) =(ODdrug-treated group − ODcontrol group)/ODcontrol group × 100%. These methods have been previously described [30,31].
2.9Immunofluorescence and laser confocal microscope techniquesImmunofluorescence and laser confocal microscopy were used to observe the expression and localization of PAR-4 in the HCC cells. Briefly, 5 × 104/mL HCC cells were cultured in a sterile glassy bottom culture dish and cultured for 48 h; then the cells were fixed with 4% paraformaldehyde for 30 min, followed by the addition of the primary antibody against PAR-4 and the secondary antibody, and the cells were gently shaken at 37 °C for two h. Then, 50 µL of DAPI solution at a concentration of 100 mg/L was added for nuclear staining. Immunofluorescence and laser confocal microscopy were used to observe the images. The specific operation procedure has been described previously [32].
2.10Cell scratch testCell scratching and repair motility were analyzed using a wound-healing assay. One day before scratching, PLC, HLE, and Bel7402 cells were infected with shPAR-4 lentivirus and seeded into 6-well plates to almost total confluence within 24 h. A scratch wound was made by scraping the middle of the cell monolayer using a sterile micropipette tip. After all detached cells were washed away with PBS, the cells were cultured with medium containing 10% FCS, treated with IL-6(20 ng/mL), and images of cell repair in the wound area were captured at 0, 24, 48, and 72 h using an inverted microscope, and their distances were recorded. Cell-repaired motility was evaluated using the following formula: cell repair ratio (%) = (distance 0 h − distance X h)/distance 0 h × 100%. (X: observed time points). The specific operation procedure has been described previously [27].
2.11Plate cloning experimentPLC, HLE, and Bel7402 cells were infected with lentivirus interference and PAR-4 expression was selected and inoculated into 6-well plates after trypsin digestion at approximately 500 cells/well. After cell adhesion, IL-6 (20 ng/mL) was added to the cells, the fresh medium was changed every 2–3 days, and the cells were cultured in an incubator for 14 days. When visible clones were found in the 6-well plate, the culture medium was aspirated and the cells were washed with 1 × PBS. Next, 1 mL of 4% paraformaldehyde was added to the wells to cover the cells, which were then fixed for 30 min. The wells were washed twice with 1 × PBS buffer, and then 1 mL of 0.1% crystal violet was added dropwise and stained for 30 min. The 6-well plates were washed with running water, air-dried, and counted under a microscope.
2.12Transwell migration experimentPLC, HLE, and Bel7402 cells were infected with lentivirus interference and PAR-4 expression was selected for migration and invasion assays, which were performed according to the manufacturer's protocols. Transwell chambers were used to observe the cultured cell inserts to measure cell migration (Transwell chamber, 8-mm pore siz, Costar, High Wycombe, UK). The cells were placed in 12-well culture plates and the upper and lower chambers were separated. HCC cells (5 × 105/mL) were added to the upper chamber, cultured in serum-free DMEM, and treated with IL-6 (20 ng/mL), whereas the lower chamber was filled with a complete medium (containing 20% FCS). After 24, 48, and 72 h of incubation, cells that migrated through the membrane to the lower surface were fixed with 90% methanol and stained with 0.1% crystal violet. The number of cells that migrated through the pores was quantified by counting five independent visual fields under a microscope (Olympus) using a 20 × objective lens. Three independent assays were performed for each sample. The specific operation procedure has been described previously [27].
2.13Trypan blue stainingPLC, HLE, and Bel7402 cells were infected with lentivirus interference PAR-4 expression, and the cell numbers were controlled at 5 × 105/mL when the cells were cultured in a 6-wells plate for 24 h followed treated with sorafenib(5 µg/mL) for 24 h. After trypsinase digestion, the cells were loaded into 15 mL EP tubes and centrifuged at 1800 rpm for 10 min, and the cell precipitate was collected. The cells were then stained with 100 µL trypan blue solution for 2–10 min at room temperature, and the number of living and dead cells was observed under an inverted microscope. Cell viability ratio= number of viable cells/number of total cells.
2.14Flow cytometry detectionPLC, HLE, and Bel7402 cells were infected with lentivirus interference PAR-4 expression, and the cell concentration was controlled at 1 × 106 cells/mL when the cells were cultured in 150 mL culture flask for 24 h followed treated with 5 µg/mL Sorafenib for 24 h. Group Design: untreated group (control), PE group, 7-AAD group, and double staining group. The cell suspension (100 µL) was aspirated using a pipette gun and transferred to 1.5 mL centrifuge tubes. According to the group settings, 5 µL of PE Annexin V and 5 µL of 7-AAD were added, and the cells were gently mixed. The mixture was incubated at room temperature for 15 min (avoid light). In the experimental group, 100 µL of cell resuspended solution was aspirated into a 1.5 mL centrifuge tube with a pipette gun, 5 µL PE Annexin V and 5 µL 7-AAD were added, and the cells were gently mixed and incubated at room temperature for 15 min (in the dark). At the end of incubation, another 400 µL of 1 × Binding Buffer was added to the centrifuge tube, and sample loading analysis was performed by flow cytometry (NovoCyte 2040R). The sample loading analysis was completed within one h, as much as possible.
2.15Subcutaneous tumor-bearing model in nude miceMale nude mice aged 4–5 weeks, weighing 18–20 g, were selected. Nude mice were evenly divided into three groups according to body weight: control, NC (sh-negative control), and shPAR-4, with six nude mice in each group. PLC, HLE, Bel7402 cells, and the corresponding lentiviral stable cell lines PLC-shPAR-4, HLE-shPAR-4, and Bel7402-shPAR-4 were cultured in large quantities, cells with favorable cell states and full cell bottles were selected, and the cells were digested with 0.25% trypsin. Digestion was terminated with a double medium without serum or double antibodies. The cell suspension was transferred to a 15 mL centrifuge tube and centrifuged at 2000 rpm for 5 min. The supernatant was discarded, and the cells were resuspended again in a double medium without antibody. Centrifugation was performed at 2000 rpm for 5 min. The supernatant was discarded and the cell precipitate was collected. Finally, the cell precipitate was made into a cell suspension at a concentration of 5 × 107/mL, and 100 µL of the cell suspension was injected subcutaneously adjacent to the right forelimb of the nude mice using a syringe to establish subcutaneous tumors. After 28 days, the nude mice were sacrificed, the tumors were removed, and the nude mice and tumors were photographed and stored. The length (L) and width (W) of the tumors were measured and the tumor volume was calculated according to the formula (V=π/6 × L × W2). The tumors were weighed using an electronic analytical balance. The tumor tissues were subjected to microarray analysis.
2.16Statistical analysisThe Aipathwell software was used to analyze the ratio of IHC-positive cells, ImageJ software was used to analyze the gray value of the bands, and GraphPad Prism software (version 8.0) was used to draw the charts. Statistical analysis was performed using SPSS 26.0, and the measurement data of experimental data were expressed as the mean standard deviation (SD). A t-test or chi-square test was used for analysis between two groups, ANOVA was used for comparison between multiple groups, and correlation analysis was used for correlation analysis between two variables. The survival curve was plotted and the difference in the survival curves between the two groups was tested using the log-rank test. The experimental data are expressed as the SD of the mean. The differences between two groups were analyzed using the t-test, and differences between multiple groups were compared using ANOVA. Differences were considered statistically significant at P < 0.05.
2.17Ethical statementWritten informed consent was obtained from each patient included in the study and the study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the Medical Ethics Committee of the Second Affiliated Hospital of Hainan Medical College. All animal experiments were approved by the Institutional Animal Care and Use Committee at the Hainan Medical College, Haikou, Hainan Province, PR. China.
3Results3.1Expression and correlation analysis of PAR-4 in biological information databaseTo preliminarily express PAR-4 in HCC tissues, the UACLAN-TCGA database was used to analyze the expression of PAR-4. The expression of PAR-4 was found to be different in different types of tumors and normal tissues. In breast cancer (BRCA), kidney cancer (KICH, KIRC, KIRP), prostate cancer (PRAD), adrenal cancer (PCPG), endometrial cancer (UCEC) and other tumors, the expression level of PAR-4 in cancer tissues was significantly lower than that in normal tissues, as shown in Fig. 1A. Further comparison of 425 samples from TCGA database (371 liver cancer specimens and 50 normal liver tissues) showed that the expression level of PAR-4 was significantly higher in liver cancer tissues (P < 0.001), as shown in Fig. 1B. The protein interaction relationship between PAR-4 and STAT3 was analyzed using the STRING database, and a protein interaction relationship between PAR-4 and STAT3 was identified. Among the proteins with an interaction relationship, PAWR was found to be related to the JAK/STAT signaling pathway by KEGG pathway analysis. Strength was 0.86, P = 0.0297) (Fig. 1C).
Analysis of PAR-4 in biological information database that correlated with cancer. A, UACLAN-TCGA database was used to analyze the expression of PAR-4; the expression level of PAR-4 in cancer tissues was significantly lower than that in normal tissues. B, The expression of PAR-4 was a comparison of 425 samples in TCGA database (371 liver cancer specimens and 50 normal liver tissues), PAR-4 higher expression in liver cancer tissues compared with normal liver tissues(P < 0.001). C, The protein interaction relationship between PAR-4 and STAT3 was analyzed by String database, PAWR was found to be related to JAK/STAT signaling pathway by analysis-KEGG Pathways Analysis. Strength was 0.86, P = 0.0297. The red arrows show the correlation between the PAR-4 and STAT3 signaling pathways.
To further clarify the biological function of PAR-4 in HCC, we detected the expression of PAR-4 in tissue samples from patients with HCC. First, a tissue chip was made from cancer tissue specimens of 30 patients with HCC and their corresponding adjacent tissues. In the tissue chip matrix, T represents the liver tumor tissue and P represents the corresponding adjacent tissues (para-carcinoma tissues), as shown in Fig. 2A. Second, the expression of PAR-4 was detected by immunohistochemistry, and the results showed that the positive expression rate of PAR-4 in cancer tissues (52.75%±0.38%) was significantly higher than that in the corresponding adjacent tissues (8.65%±0.12%), with a statistical difference(P < 0.01), as shown in Fig. 2B. Third, Western blotting results showed that the expression of PAR-4 protein in cancer tissues was significantly higher than that in paired adjacent tissues, and the difference was statistically significant(P < 0.01), as shown in Fig. 2C.
The expression of PAR-4 in HCC tissues. A. Tissue chip was arranged from 30 HCC patients cancer tissue specimens and corresponding adjacent tissues. T represents the cancer tissue and P represents the corresponding adjacent tissues(para-carcinoma tissues). B. The expression of PAR-4 was detected by immunohistochemistry in tissue samples. C. Western blotting is applied to detect the expression of PAR-4 in cancer tissue and paired adjacent tissues; the bar graph on the right shows the statistical histogram of the gray level of the difference in PAR-4 expression between the cancerous tissues and the paired adjacent tissues. ***P < 0.01.
PAR-4 staining was scored in HCC tissues (0: <5%, 1: 5–20%, 2: 20–60%, 3: 60–100%). 0–1 patients were divided into the LOW PAR-4 group (n = 10), and 2–3 patients were divided into the high PAR-4 group (n = 20). The clinical parameters of the two groups (including: There was no significant difference in age, gender, drinking history, HBV-DNA, PS score, AFP concentration, ALT, ALB, Child-Pugh grade, tumor number, tumor stage, microvascular invasion, distant metastasis, etc.), P > 0.05. PAR-4 expression was correlated with tumor size (P = 0.006), histological grade (P = 0.019), and Ki-67 expression (P = 0.045). Patients with HCC with high PAR-4 expression had a larger tumor size, lower histopathological differentiation, and a higher Ki-67 positive index, as shown in Table 2.
The expression of PAR-4 relationship with clinicopathological features of HCC patients.
| Variables | PAR-4 | P | |
|---|---|---|---|
| Low PAR-4 (n = 10) | High PAR-4 (n = 20) | ||
| Gender | 0.704 | ||
| Male | 9 | 18 | |
| Female | 1 | 2 | |
| Age (year) | 57.4 ± 3.99 | 52.5±.34 | 0.357 |
| Drinking | 0.101 | ||
| Yes | 9 | 11 | |
| No | 1 | 9 | |
| PS | 0.633 | ||
| ≤2 | 9 | 15 | |
| >2 | 1 | 5 | |
| AFP (ng/mL) | 0.584 | ||
| <400 | 8 | 12 | |
| ≥400 | 2 | 8 | |
| HBV-DNA (copies/mL) | 0.709 | ||
| <1 × 103 | 6 | 10 | |
| ≥1 × 103 | 4 | 10 | |
| ALB (g/L) | 0.702 | ||
| <35 | 3 | 8 | |
| ≥35 | 7 | 12 | |
| ALT (U/L) | 0.7 | ||
| <40 | 6 | 9 | |
| ≥40 | 4 | 11 | |
| Child-Pugh | 0.563 | ||
| A | 9 | 19 | |
| B + C | 1 | 1 | |
| Tumor stage | 0.55 | ||
| Ⅰ+Ⅱ | 5 | 11 | |
| Ⅲ+Ⅳ | 5 | 9 | |
| Histilogical grade | 0.019a | ||
| Low | 2 | 14 | |
| Medium and well | 8 | 6 | |
| Well | |||
| Tumor numbers | 0.672 | ||
| Single | 9 | 18 | |
| mulpitle | 1 | 2 | |
| Tumor size (cm) | 3.62±1.50 | 6.75±4.21 | 0.006a |
| Ki-67 | 0.045a | ||
| <30% | 7 | 5 | |
| ≥30% | 3 | 15 | |
| Microvascular infiltration | 0.533 | ||
| M0 | 4 | 12 | |
| M1 | 4 | 5 | |
| M2 | 2 | 3 | |
| Distant metastases | 0.682 | ||
| Yes | 2 | 6 | |
| No | 8 | 14 | |
Further analysis of the correlation between the expression levels of PAR-4 and IL-6 in 30 clinical tissue samples showed that the expression of PAR-4 was high in tissues with high IL-6 expression, whereas the positive expression rate of PAR-4 was slightly lower in tissues with low IL-6 expression. SPSS 26.0 correlation analysis showed that the correlation coefficient was r = 0.302, P = 0.019, indicating that the expression of PAR-4 was positively correlated with the expression of IL-6 in patients with HCC, as shown in Fig. 3A and 3B. The survival curve was drawn according to the expression of PAR-4 in the tissues of 30 HCC patients and the survival of patients. The results showed that the median overall survival of HCC patients with high PAR-4 expression was significantly lower than that of patients with low PAR-4 expression (P = 0.013) (Fig. 3C). High PAR-4 expression indicates that the patient may have a poor prognosis.
The relation of PAR-4 with IL-6 in HCC patients, and PAR-4 expresses the prognosis of HCC patients. A, The expression of PAR-4 and IL-6 in 30 samples of clinical tissue was detected by Western blotting. B, SPSS 26.0 correlation analysis of the positive expression rate of PAR-4 with low IL-6 expression, correlation coefficient r = 0.302, P = 0.019. C, Median overall survival of 30 samples of clinical HCC patients with high PAR-4 expression was lower than that of patients with low PAR-4 expression (P = 0.013).
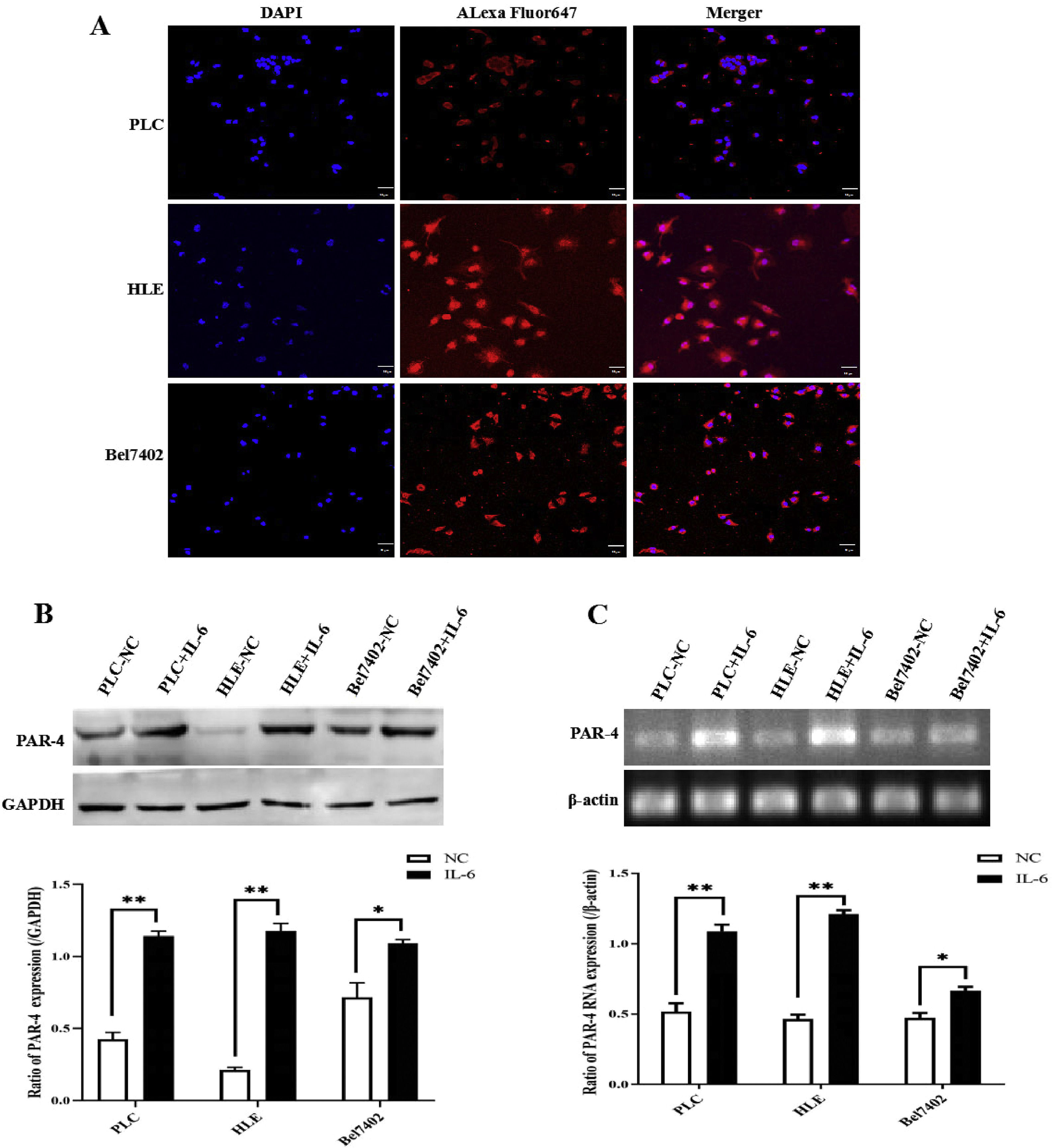
In the present study, laser confocal microscopy was used to observe the expression and localization of PAR-4 in HCC cell lines, and the results indicated that PAR-4 was widely distributed in the cytosol and nucleus of PLC, HLE, and Bel7402 cells (Fig. 4A). When these cells were stimulated with 20 ng/mL recombinant human IL-6 protein for 24 h, they were collected, and the expression of PAR-4 was detected by Western blotting. The results showed that the expression of PAR-4 in HCC cells was significantly increased compared with that in the untreated groups (P < 0.05), as shown in Fig. 4B. At the same time, RT-PCR was used to detect the RNA level, and it was found that the mRNA of PAR-4 in HCC cells was significantly increased (P < 0.05), as shown in Fig. 4C. These results indicated that IL-6 up-regulates the expression of PAR-4 in HCC cells.
Expression and localization of PAR-4 in HCC cells, and the effect of IL-6 on the expression of PAR-4. A, The expression and localization of PAR-4 in HCC cell lines, PLC, HLE and Bel7402 cells were observed by laser confocal microscopy. B and C, PLC, HLE and Bel7402 cells were treated with 20 ng/mL recombinant human IL-6 protein for 24 h, the expression of PAR-4 was detected by Western blotting, the bar graph on the low shows the statistical histogram of the gray level of the difference in PAR-4 expression in HCC cells.**P < 0.05(B). RT-PCR was used to detect the PAR-4 mRNA level, the mRNA of PAR-4 in HCC cells was significantly increased, the bar graph on the low shows the statistical histogram of the gray level of the difference in PAR-4 mRNA in HCC cells. **P < 0.05(C). The images were a representative three independent experiments.
To analyze the effect of the IL-6/STAT3 signaling pathway on the expression of PAR-4 in HCC cell lines, HCC cell lines in PLC, HLE, and Bel7402 cells silenced the expression of IL-6R (Supplementary materials: s-Fig. 1. Stable HCC cell lines with interfered IL-6R expression were successfully constructed). Recombinant human IL-6 protein at a concentration of 20 ng/mL was used to treat the cells. The results showed that the expression of phosphorylated-STAT3(p-STAT3) and PAR-4 in HCC cells did not change significantly after interfering with the expression of IL-6R (P > 0.05). These results suggest that IL-6 may bind to IL-6R in the membrane of HCC cells, causing activation of the downstream molecule p-STAT3 to up-regulate the expression of PAR-4. After interfering with the expression of IL-6R in HCC cells, the IL-6/STAT3 signaling pathway was blocked, thereby inhibiting IL-6 to up-regulate the expression of PAR-4, as shown in Fig. 5A. To further verify that the IL-6/STAT3 signaling pathway can regulate the expression of PAR-4, the STAT3 inhibitor Stattic (concentration of 5.1 µmol/L) was used to treat HCC cells. After treatment with IL-6(20 ng/mL) and Stattic for 48 h, Western blotting was performed to detect the expression levels of STAT3, p-STAT3, and PAR-4 in each group. The expression levels of PAR-4 and p-STAT3 were significantly increased in these cells following IL-6 treatment. The expression levels of PAR-4 and p-STAT3 were decreased after Stattic treatment alone. When HCC cells were co-treated with IL-6 and Stattic at the same time, the expression of PAR-4 and p-STAT3 decreased compared to that in cells treated with IL-6 alone, and the total STAT3 did not change significantly, as shown in Fig. 5B. The results showed that IL-6 activated the IL-6/STAT3 signaling pathway and up-regulated the expression of PAR-4. The STAT3 inhibitor Stattic was able to block IL-6 up-regulated the expression of PAR-4 and inhibited the IL-6/STAT3 signaling pathway, which attenuated the upregulation of PAR-4 induced by IL-6.
The effect of IL-6/STAT3 signaling pathway on the expression of PAR-4 in HCC cells. A, HCC cell lines, PLC, HLE and Bel7402 cells silenced the expression of IL-6R, and then 20 ng/mL of IL-6 was applied to the treatment of these cells; the expression of STAT3, p-STAT3 and PAR-4 were detected by Western blotting, the bar chart right shows statistical differences of these proteins in HCC cells, NS represent no statistical differences (P > 0.05). B, IL-6 (20 ng/L), STAT3 inhibitor Stattic(concentration of 5.1 µmol/L) or IL-6 (20 ng/L)+Stattic was used to treat HCC cells for 48 h, the expression of STAT3, p-STAT3 and PAR-4 in these cells was detected by Western blotting, the bar chart below shows statistical differences of these proteins in HCC cells, NS represents no statistical differences (P > 0.05); *P < 0.05; **P < 0.01. The images were representative of at least three independent experiments.
The main factors contributing to the poor prognosis of HCC are intrahepatic and distant metastases, and an increase in cell motility is the basic element of cancer cell metastasis. However, the effect of PAR-4 on this ability remains unclear. First, we constructed stable HCC cell lines that interfered with PAR-4 expression (Supplementary material: s-Fig. 2. Stable HCC cell lines that interfered with PAR-4 expression were successfully constructed). According to the scratch test results, compared with the negative control (NC) group, the healing and repair ability of liver cancer cells in the IL-6 group was significantly increased, but the healing and repair ability of liver cancer cells in the shPAR-4 group was decreased. Compared to the IL-6 treated group, the healing and repair ability of the shPAR-4+IL-6 group was weakened, indicating that the healing and repair ability of IL-6 in HCC cells may be weakened after interfering with PAR-4 expression. The healing and repair rates were analyzed, and the differences were found to be statistically significant (Fig. 6A).
The role of PAR-4 in the malignant behaviors of HCC cells. A, PLC, HLE, Bel 7402 cells were stable infections of LV-shPAR-4 lentivirus, and then these cells were treated with IL-6(20 ng/L) for 0 h, 24 h, 48 h, and 72 h; the scratch and repair ability of the cells were observed by microscopy, the bar chart below shows statistical differences of the distance. *P < 0.05, ***P < 0.01. B, PLC, HLE, Bel 7402 cells were stable infection of LV-shPAR-4 lentivirus, and then these cells were treated with IL-6(20 ng/L) for five days; the clone formation of the single cancer cells was detected by cloning assay, the bar chart below shows statistical differences of the distance. *P < 0.05. C, PLC, HLE, Bel 7402 cells were stable infections of LV-shPAR-4 lentivirus, and then these cells were treated with IL-6(20 ng/L) for 48 h; the effect of PAR-4 on the migratory ability of HCC cells was analyzed by Transwell chamber migration assay, the bar chart below shows statistical differences of the migratory cell numbers. *P < 0.05, ***P < 0.01. D and E, PLC, HLE, Bel 7402 cells were stable infection of LV-shPAR-4 lentivirus, and then these cells were treated with sorafenib (5 µg/mL) for 24 h, and then trypan blue staining (D) and flow cytometry analysis (E) were performed for observing the survive and apoptosis of HCC cells, the bar chart below shows statistical differences of the viable and apoptotic cells numbers. NS represent no statistical differences(P > 0.05); *P < 0.05. The images were representative of at least three independent experiments.
Distant metastasis of malignant tumors often occurs when a single cell or a small number of cells enter body fluid and are disseminated and planted at other sites. The proliferation of single cancer cells is critical for the formation of metastases. In the present investigation, a single cancer cell cloning assay indicated that compared with the NC group, the clonogenic ability of HCC cells in the shPAR-4 group was decreased, while the clonogenic ability of HCC cells in the IL-6 treated group was significantly increased. However, the clonogenic ability of the shPAR-4+IL-6 treated group was decreased compared to that of the IL-6 treated group, indicating that interference with PAR-4 could inhibit clone formation of HCC cells induced by IL-6. The results are shown in Fig. 6B.
In order to analyze the effect of PAR-4 on the migratory ability of HCC cells, in this study, a Transwell chamber migration assay was applied to observe the migratory ability of HCC cells. The results showed that the migratory numbers of HCC cells in the IL-6 treated group were significantly higher than that in the NC group, and the migratory numbers of the interfered expression of PAR-4 group significantly decreased compared to that in the NC group, suggesting that IL-6 increased the migration of HCC cells. Interference with PAR-4 inhibited the migration of HCC cells. When using IL-6 to stimulate HCC cells with interfered expression of PAR-4, the number of migratory cells was significantly lower than that in the IL-6 treated group, suggesting that interference with the expression of PAR-4 may weaken the role of IL-6 in stimulating the migratory ability of HCC cells (as shown in Fig. 6C).
To determine whether the expression of PAR-4 affects the sensitivity of liver cancer cells to Sorafenib, liver cancer cells (NC and shPAR-4 groups) were treated with 5 µg/mL Sorafenib for 24 h, and trypan blue staining and flow cytometry analysis were performed to observe the survival and apoptosis of HCC cells. The number of surviving cells in the shPAR-4 group was significantly lower than that in the NC group (Fig. 6D). The flow cytometry analysis results also indicated that compared with the NC group (PLC:24.38%±0.79%, HLE:14.17%±1.09%, Bel7402:19.87±0.52%), the apoptosis rate of HCC cells significantly increased, while interference with the expression of PAR-4 (PLC:24.38%±0.79%, Bel7402:19.87±0.52%). 29.21%±1.33%, HLE: 19.52%±0.87%, Bel7402: 31.37±2.72%)(Fig. 6E).
3.8IL-6 stimulates the proliferation of HCC cells and promotes the expression of proliferation- and metastasis-related proteins mediated by PAR-4In this study, MTT assay was used to determine the optimal time and concentration of IL-6 for the proliferation of HCC cells. PLC, HLE, and Bel7402 cells in logarithmic growth phase were treated with different concentrations of recombinant human IL-6 protein. After 24 and 48 h of treatment, the cell proliferation rate at different concentrations of IL-6 was measured using the MTT assay. The proliferation rate of HCC cells was significantly higher when treated with 20 ng/mL IL-6 than when treated with 0, 2.5, 5, or 10 ng/mL, was statistically significant (P < 0.05). When the concentration of IL-6 was greater than 20 ng/mL, growth increased slowly, indicating that the degree of cell proliferation slowed down. There was no significant difference in the proliferation rate between the high-dose of IL-6 and 20 ng/mL concentration (P > 0.05). This study also found that there was no significant difference in the proliferation rate of HCC cells treated with different concentrations of IL-6 at 24 and 48 h (P > 0.05). The results also indicated that proliferation of HCC cell lines PLC (Fig. 7A), HLE(Fig. 7B), and Bel 7402 cells (Fig. 7C) was stimulated by IL-6.
The role of IL-6 in proliferation and the expression of growth- and metastasis-related proteins in HCC cells. PLC (A), HLE (B) and Bel7402 (C) cells were treated with different concentrations(0 ng/mL, 2.5 ng/mL, 5 ng/mL, 10 ng/mL, 20 ng/mL, 40 ng/mL and 80 ng/mL) of recombinant human IL-6 protein for 24 h and 48 h, the proliferation of HCC cells was measured by MTT. **P < 0.05, N = 6. PLC (D), HLE (E), Bel 7402 (F) cells were stable infection of LV-shPAR-4 lentivirus, and then these cells were treated with IL-6(20 ng/L) for 48 h, and the expression of Src and Ras in these cells were analyzed by Western blotting. The right bar chart shows statistical differences in the Grayscale scan value of protein bands. *P < 0.05. The images were representative of at least three independent experiments.
We explored the molecular mechanism by which IL-6 stimulates proliferation, which correlates with PAR-4 in HCC cells. In the present study, Western blotting was used to detect the expression of proliferation- and metastasis-related proteins in the NC, shPAR-4, IL-6, and shPAR-4+IL-6 groups. Compared with the NC group, the expression levels of Src and Ras in HCC cells were significantly decreased, while the cancer cells were interfered with the expression of PAR-4, and the expression of Src and Ras was significantly increased after treatment with IL-6. Similarly, when IL-6 was used to treat HCC cells, which interfered with the expression of PAR-4, the expression of Src and Ras significantly decreased compared to in the IL-6 treated-group. These results proved that interference with the expression of PAR-4 attenuated the upregulation of Src and Ras expression mediated by IL-6 (Fig. 7D–F).
3.9Effect of PAR-4 on subcutaneous tumorigenesis in nude miceTo further explore the effect of PAR-4 on the malignant behaviors of HCC cells, we conducted in vivo experiments on subcutaneous tumorigenesis in nude mice. Nude mice were evenly divided into three groups according to body weight: blank (serum-free medium), NC, and shPAR-4. Twenty-eight days after subcutaneous inoculation with PLC, HLE, or Bel 7402 cells, the nude mice were sacrificed by ampulpotomy, and the tumors were exhaled. Macroscopic observations showed that the tumor volume in the shPAR-4 group was smaller than that in the NC group (Fig. 8A). The tumors were weighed and measured, and it was found that the tumors in sPAR-4 group (volume: 60.83±50.05 mm3, Weight: 0.12±0.028 g) was significantly higher than NC group (volume: 205.98±133.18 mm3, Weight: 0.21±0.058 g), and the difference was statistically significant (P < 0.01), as shown in Fig. 8B. These results suggest that interference with PAR-4 expression may inhibit the tumorigenesis of HCC cells in nude mice.
Effect of PAR-4 on tumorigenesis and tumor growth in nude mice. A and B, PLC, HLE, Bel 7402 cells or these cells were stable infections of LV-shPAR-4 lentivirus; then the nude mice were subcutaneously inoculated with these cells for 28 days (A); the nude mice were sacrificed by ampulpotomy and the tumors were exhaled, then tumors were weighed and measured, the bar chart low shows statistical differences of the tumors volume and weigh (B). **P < 0.01. C and D, tumor tissues were exhaled from nude mice, the expression of PAR-4 and Ki-67 was detected by immunohistochemistry, and the positive rate of staining in the tumors tissue was analyzed by Aipathwell software; the bar chart shows statistical differences of the positive rate of PAR-4 and Ki-67 in control group and interference the expression of PAR-4 groups. **P < 0.01. N = 6. E, This is the schematic diagram of IL-6/STAT3 signaling pathway that stimulates PAR-4 to promote the malignant behavior of HCC cells.
Next, we analyzed the expression of PAR-4 and the tumor proliferation marker Ki-67 in the tumor tissues of nude mice. The results showed that the positive expression rate of PAR-4 in the shPAR-4 group was significantly lower than that in the NC group. The positive rate of immunohistochemistry was analyzed using Aipathwell software for statistical analysis, and the difference was statistically significant (P < 0.05) (Fig. 8C). We further detected the intensity and range of Ki-67 staining and found that the positive expression rate of Ki-67 in the shPAR-4 group was lower than that in the NC group. Statistical analysis of the positive rate of staining showed that P < 0.05, as shown in Fig. 8D, suggesting that after interfering with the expression of PAR-4 in liver cancer cells, the expression of Ki-67 may be downregulated, thereby affecting tumor proliferation. Fig. 8E shows that PAR-4 stimulates the malignant behaviors of HCC cells via the IL-6/STAT3 signaling pathway.
4DiscussionHCC is a highly malignant tumor that seriously threatens the lives and health of individuals worldwide. However, the exact etiology and molecular mechanisms underlying HCC remain unclear. Currently, HCC is believed to be a complex process involving multiple factors and steps, among which HBV [33] and HCV [34] infections are the main etiologies. Great achievements have been made in surgical treatment, targeted therapy, and immunotherapy. However, the desired therapeutic effect is still not achieved. It has been confirmed that the occurrence and development of HCC are closely related to IL-6/STAT3, PI3K/Akt/mTOR, MAPK, and other signaling pathways. Therefore, it is important to explore the pathogenesis of HCC.
There is little evidence on the relationship between PAR-4 and HCC progression, drug resistance, or malignant behaviors. The activated IL-6/STAT3 signaling pathway can stimulate the malignant behaviors of HCC cells. However, whether the IL-6/STAT3 signaling pathway mediates the role of PAR-4 remains unclear. Therefore, further study how IL-6/STAT3 affects the occurrence of HCC malignant behaviors by regulating the expression of PAR-4. This is the first study to analyze the expression and pathway enrichment of PAR-4 in HCC from biological information, observe the expression of PAR-4 in HCC patients from clinical specimens, analyze the correlation between PAR-4 expression and clinicopathological features and prognosis, and explore the regulation of PAR-4 expression in HCC cells via the IL-6/STAT3 signaling pathway.
In the present study, we found that PAR-4 was highly expressed in liver cancer cells, and that exogenous IL-6 up-regulated the expression of PAR-4 in liver cancer cells. It is speculated that the IL-6/STAT3 signaling pathway may regulate the expression of PAR-4, thus affecting the occurrence of malignant behavior in liver cancer, as shown in Fig. 2, which is contrary to the results of current mainstream studies. PAR-4 is a tumor suppressor that promotes cancer cell apoptosis [35]. Some studies have shown that intracellular PAR-4 plays a role in inhibiting the pro-survival pathway and activating Fas-mediated apoptosis [36–38]. However, extracellular (secreted PAR-4) activates the extracellular apoptosis pathway by binding to GRP78 on the cell surface [39,40]. Several studies have emphasized the importance of PAR-4 in the prevention of cancer development and recurrence, which has the potential to be used as a promising anticancer therapeutic drug [41]. Recent studies have not only found that PAR-4 is involved in the pro-apoptotic effect of tumors, but also that PAR-4 can reduce the invasion and migration of tumor cells by inhibiting epithelial-mesenchymal transition [42].
Currently, research on the role of PAR-4 in HCC is lacking. IL-6 is an important cytokine in the HCC microenvironment, is highly expressed in HCC, and plays an important role in the proliferation, invasion, metastasis, and drug resistance of HCC [43]. This study aimed to explore the role of IL-6 in the regulation of PAR-4 expression, thereby affecting the malignant behaviors of HCC. In this study, we investigated whether IL-6 plays a role in the development of HCC by inhibiting the expression of PAR-4. However, in the present study, exogenous recombinant human IL-6 promoted the expression of PAR-4 in HCC cells, which was contrary to the expected experimental results. To clarify the expression of PAR-4 in liver cancer, we first performed bioinformatics analysis. Using the ULCAN-TCGA database, the results indicated that the expression of PAR-4 was inconsistent between cancer and normal tissues of different tumors. Among them, the expression of PAR-4 in kidney cancer [44], breast cancer [45], endometrial cancer [46], bladder cancer [47], prostate cancer [48], and other tumor tissues is lower than that in normal tissues, which is consistent with classical mainstream research results (PAR-4 is poorly expressed in tumor tissues and is a cancer suppressor) [49]. According to the results of TCGA database, the expression of PAR-4 in tumor tissues was higher than that in normal liver tissues. The STRING database also revealed a protein-interaction relationship between PAR-4 and STAT3. KEGG pathway analysis of the interacting proteins indicated that PAR-4 might be related to the JAK/STAT signaling pathway.
To further understand the biological role of PAR-4, we detected the expression of PAR-4 in the tissues of 30 clinical HCC patients using immunohistochemistry and Western blotting. The results showed that the expression of PAR-4 in cancer tissues was significantly higher than that in the corresponding adjacent tissues, and the expression of IL-6 in HCC tissues was positively correlated with the expression of PAR-4. This is consistent with previous results from a part of a biological information database. Therefore, we hypothesized that IL-6 or IL-6/STAT3 signaling pathway regulates the expression of PAR-4 in HCC cells.
Ki-67, also known as MKI67, is generally considered a cell proliferation marker. The Ki-67 positive cell index was the highest in HCC and the lowest in normal liver tissue and was found to be correlated with tumor growth rate, histological stage, and tumor recurrence in HCC. The higher the Ki-67 index, the faster the progression of HCC and the worse prognosis [50,51]. The results of this study showed that PAR-4 expression correlated with tumor size, Ki-67 content, and histological grade in patients with HCC. Patients with HCC and high PAR-4 expression had larger tumors, lower histological differentiation, and a higher Ki-67 positive index. Survival curves were drawn according to the expression level of PAR-4 and the survival of HCC patients, and the results showed that the median overall survival of HCC patients with low PAR-4 expression was higher than that of patients with high PAR-4 expression. Based on these clinical findings, we hypothesized that high PAR-4 expression may be involved in stimulating the development of HCC.
Combined with the results from the biological information database, PAR-4 may be related to the JAK/STAT3 signaling pathway. The expression of IL-6 and PAR-4 in clinical specimens is positively correlated, and previous studies have shown that IL-6 can up-regulate the expression of PAR-4 in liver cancer cells. It is speculated that the IL-6/STAT3 signaling pathway can regulate the expression of PAR-4 in liver cancer cells. To explore how the IL-6/STAT3 signaling pathway regulates the expression and function of PAR-4 in tumor cells, we first examined the cellular localization of PAR-4 by confocal immunofluorescence laser and found that PAR-4 was localized in both the nucleus and cytosol, which was consistent with other research results [52]. As a stimulating factor, we screened for the concentration and duration of exogenous recombinant human IL-6 protein using the MTT assay. After treatment with IL-6 for 24 h, the proliferation rate of liver cancer cells increased significantly (P < 0.001) from 0 ng/mL to 20 ng/mL. Compared with IL-6 concentrations of 0, 2.5, 5, and 10 ng/mL, the difference was statistically significant. However, with further increase in concentration, the effect of IL-6 on promoting proliferation was not further enhanced, which may be due to receptor closure or the existence of a negative feedback mechanism. Therefore, an concentration (20 ng/mL) of IL-6 was selected for our subsequent experiments, which was similar to the screening results of other studies [53]. When stimulated with 20 ng/mL IL-6, the expression of PAR-4 was significantly increased in HCC cells. These results indicated that IL-6/STAT3 signaling pathway could up-regulate the expression of PAR-4. These results showed that IL-6 and PAR-4 expressions were positively correlated.
As a classical extracellular stimulator of the IL-6/STAT3 signaling pathway, IL-6 induces conformational changes after binding to its receptor and then activates GP130 on the cell membrane surface, triggering the formation of a homodimer of GP130, which leads to the activation of JAKs and STAT3. After phosphorylated STAT3 (p-STAT3) forms a homodimer, p-STAT3 translocates from the cytosol into the nucleus and then combines with the promoter regions of downstream effector target genes, thereby regulating tumor cell growth, differentiation, and development [54]. Many studies have shown that blocking STAT3 activation reduces the survival, proliferation, migration, and invasion abilities of tumor cells [55–57]. Current studies have shown that there are three approaches to target STAT3: modulation of upstream regulators, RNA interference, and direct targeting of the STAT3 protein [58]. STAT3 may not be completely blocked by upstream regulators because of the interactions between many molecular pathways, and RNA interference has a long way to go before it is approved for clinical use. Therefore, small-molecule inhibitors targeting STAT3 may be a better strategy for inhibiting STAT3. Stattic is one such inhibitor targeting the SH2-domain of STAT3 [59,60].
To explore the mechanism by which IL-6 up-regulates the expression of PAR-4 in HCC cells, HCC cell lines with interfering IL-6R were successfully constructed. When IL-6 was used to treat HCC cells while interfering with the expression of IL-6R, the expression levels of p-STAT3 and PAR-4 did not change significantly. The experimental results indicated that when the expression of IL-6R was inhibited in HCC cells, IL-6 could not bind to IL-6R, and the effect of IL-6/STAT3 signaling pathway upregulation on PAR-4 expression was blocked. In order to make this speculation more convincing, the study further treated liver cancer cells with the STAT3 inhibitor Stattic, and the results showed that the expression levels of PAR-4 and p-STAT3 decreased after STAT3 inhibitor treatment. When HCC cells were treated with IL-6 and Stattic simultaneously, the expression levels of PAR-4 and p-STAT3 decreased compared to those treated with IL-6 alone, and total STAT3 did not change significantly. These results indicate that the STAT3 inhibitor blocked the upregulation of PAR-4 by IL-6. The results showed that IL-6 activated the IL-6/STAT3 signaling pathway, up-regulated the expression of PAR-4, and inhibited the IL-6/STAT3 signaling pathway, which reversed the upregulation of PAR-4 expression induced by IL-6. These results prove that PAR-4 is highly expressed in HCC and that PAR-4 may be involved in the occurrence and development of HCC. The IL-6/STAT3 signaling pathway up-regulate the expression of PAR-4. Therefore, we investigated the biological function and role of PAR-4 in HCC at the cellular level in vitro and in vivo.
First, we used lentivirus transfection technology to construct stable HCC cells that interfered with PAR-4, and successful transfection was the basis for subsequent cell experiments. Western blotting and RT-CPR verified the interference effect of PAR-4. To explore the role of PAR-4 in HCC cells, we used cell scratch, Transwell, and cell clone formation assays to verify the role of PAR-4 in the proliferation and metastasis of HCC cells. The results showed that the proliferation and migration abilities of HCC cells were significantly increased after treatment with exogenous IL-6. This is consistent with the results of current studies on IL-6 in liver cancer [61]. After interference with PAR-4 expression, the healing and repair abilities, clone formation, and migration of HCC cells were weakened. When the HCC cell lines were interfered the expression of PAR-4 then treated with exogenous IL-6, the proliferation, clone formation and migration of HCC cell lines were significantly weakened compared with the NC group that treated with IL-6, indicating that after the interference the expression of PAR-4, the effect of exogenous IL-6 on the proliferation, clone formation and migration of HCC cells may be weakened.
With the continuous development of medicine, anticancer drugs related to HCC continue to emerge, including sorafenib [62,63], Atezolizumab [64], Lenvatinib [65], PD-1/PD-L1, and other drugs. The treatment of HCC patients has also reached a new step. Sorafenib is a multi-kinase inhibitor that targets RAF, VEGF, PDGF, and other tyrosine kinases. Before Lenvatinib was marketed, sorafenib was used as a standard targeted therapy for liver cancer, as it can inhibit tumor cell proliferation and the generation of vascular tumors [62]. Currently, the objective response rate of HCC treatment is only 13% and the disease control rate is 58.4%. Many patients do not benefit from the treatment [66]. In this study, the effect of sorafenib on the sensitivity of HCC cells in NC and PAR-4 stable cell lines was assessed using trypan blue staining and flow cytometry. According to the results of a literature review [67,68], the IC50 of Sorafenib on PLC and other liver cancer cells is 5–10 µg/mL, which is converted to 2.33–4.64 a concentration. In this study, a concentration of 5 µg/mL was used to treat liver cancer cells, and the apoptosis rate of liver cancer cells increased after interfering with the expression of PAR-4. These results indicated that PAR-4 may affect the sensitivity of HCC cells to Sorafenib treatment.
Ras protein is overexpressed in various tumors. The Ras protein acts as a molecular switch, participates in a variety of intracellular signal transduction pathways, and plays a key role in cell proliferation, differentiation, apoptosis, adhesion, and migration [69]. Overexpression of the Ras protein promotes the occurrence and development of HCC. An increasing number of studies have provided new insights and new targeted drugs for HCC treatment using Ras inhibitors [70]. Src is an oncogene that initiates the phosphorylation of Src in cell membranes [71], thereby inducing signaling pathways that promote cell proliferation, adhesion, and migration/invasion. Overexpression and/or elevated Src activity is associated with the progression of various tumors. Particularly in HCC, studies have found that the expression level of c-Src in HCC tissues is significantly increased and negatively correlated with patient survival [72,73], and downregulation of c-Src expression can inhibit the growth, invasion, and migration of HCC cells. Ras and Src are involved in the proliferation and metastasis of HCC cells, respectively. In the present study, the effects of PAR-4 on Ras and Src expression in HCC cells were studied. The results showed that compared to the NC group, the expression changes of Ras and Src in HCC cells were decreased after interfering with the expression of PAR-4. These results suggest that interference with the expression of PAR-4 may inhibit the expression of Ras and Src, thereby inhibiting the proliferation and development of HCC. When exogenous IL-6 was used to treat shPAR-4-hepatoma cells, the expression of Ras and Src proteins was significantly lower than that in the non-interfered group, indicating that the knockdown of PAR-4 may attenuate the proliferation and development of hepatoma induced by IL-6.
These results are based on in vitro studies at the cellular level. As the occurrence and development of tumors in vivo are biological processes regulated by multiple factors, simple in vitro experiments are far from sufficient to prove the role of PAR-4 in HCC. Therefore, in this study, we conducted in vivo experiments on subcutaneous tumorigenesis in nude mice. We observed an important effect of PAR-4 on HCC tumorigenesis in vivo. The results showed that compared with the control group, the tumor volumes and weigh of the shPAR-4 group was larger than that of the control group. These results indicate that interference with PAR-4 expression may inhibit tumor formation in nude mice.
Next, we prepared a microarray of tumor tissues for immunohistochemical detection and confirmed that the expression of PAR-4 in tumor tissues of shPAR-4 cells was lower than that in the control group. Ki-67 is generally regarded as a marker of cell proliferation, and Ki-67 is positively correlated with tumor growth rate and tumor recurrence in HCC. This study analyzed Ki-67 in tumor tissues and found that Ki-67 in shPAR-4 tumor tissues was lower than that in the NC group, which was consistent with the clinical results shown above, indicating that the expression of PAR-4 in HCC may affect the tumor proliferation rate. The expression of PAR-4 is low in other tumors and is recognised as a tumor suppressor. Why is the opposite the case in liver cancer? Further proteomic or modification omics studies are needed to determine whether PAR-4 is different in liver cancer and how the IL-6/STAT3 pathway affects the expression of PAR-4. The CHIP-SEQ database of transcription factors can be analyzed bioinformatically to identify transcription factors that bind to PAR-4 promoter and verify their binding to PAR-4 promoter by ChIP technology. The association of candidate transcription factors with PAR-4 expression can be further validated using liver cancer database data and validated using HCC tissue samples. By silencing candidate transcription factors, the expression changes of PAR-4 will be detected and the transcription factors that directly regulate PAR-4 expression will be verified to further elucidate the mechanism of PAR-4 overexpression in HCC and the oncogenic pathway involved.
5ConclusionsThe results of this study indicate that PAR-4 is highly expressed in patients with HCC and that high expression of PAR-4 is associated with poor prognosis. The IL-6 /STAT3 signaling pathway up-regulates the expression of PAR-4. In vivo and in vitro experiments showed that the proliferation, migration, and tumorigenicity of HCC cells decreased after the interference of PAR-4 expression, which may also affect the sensitivity of HCC cells to sorafenib. Meanwhile, interference with PAR-4 expression could significantly attenuate the proliferation and migratory abilities mediated by IL-6 in HCC cells, which may involve a variety of molecular mechanisms. PAR-4 is expressed at low levels in other tumors, and is recognized as a tumor suppressor. Although many studies have shown that PAR-4 is a tumor suppressor, the results of this study indicate that PAR-4 is an important molecule that promotes the malignant behaviors of HCC cells. Why the opposite results appear in liver cancer needs to be studied further using proteomics or modifiers to determine whether PAR-4 is different in liver cancer. PAR-4 is highly expressed in HCC and high PAR-4 expression is positively correlated with poor prognosis. IL-6/STAT3 signaling pathway up-regulated the expression of PAR-4, and PAR-4 has a function for promoting the malignant behaviors of HCC cells. Targeted inhibition of PAR-4 expression may be an effective new treatment strategy for HCC patients.
Authors contributionsML and MZ designed experiments. JX, KL, ZG, JL, HL, QZ, BL, and WL performed experiments. ML and HL supervised the study and analyzed the data. ML and MZ wrote the manuscript. All the authors have read and approved the final manuscript.
FundingThis work was supported by the National Natural Science Foundation of China(Nos. 82060514, 82460602), Natural Science Foundation of Hainan Province(Nos. 820RC634, 2019CXTD406, 2019CR204, 820QN403, and 824RC517), Hainan Health Industry Scientific Research Project Foundation (No. 20A200496), Education Department of Hainan Province(No. Hnky2022-34). And the Research Project of Take off the Proclamation and Leadership of the Hainan Medical College Natural Science Foundation (No. JBGS202106), and Hainan Provincial Science and Technology Special Fund(No. ZDYF2021SHF222), and Hainan Provincial Association for Science and Technology Program of Youth Science Talent and Academic Innovation(QCXM 201922).
Availability of data and materialsAll data generated or analyzed during this study are included in this manuscript. This manuscript does not contain data from any individual person (not applicable).
We thank Dr. Wenting Wang for assistance with the cellular migration assays.