MiR-122 has been regarded as a tumor suppressor. Toll-like receptor 4 (TLR4) has been found to be closely related to metastasis and immune escape of hepatocellular carcinoma (HCC). In the study, we sought to investigate the effect of miR-122 on HCC and the expression of TLR4.
Patients or Materials and methodsReal-time PCR and Western blot were performed to detect the expressions of target factors. CCK-8 and flow cytometry analysis were employed to evaluate cell viability and apoptosis, respectively. Luciferase reporter assay was used to determine whether miR-122 could directly regulate the expression of TLR4. Enzyme-linked Immuno Sorbent Assay was adopted to detect the secretion of inflammatory cytokines.
ResultsBoth down-regulation of miR-122 and up-regulation of TLR4 were found to be correlated with low overall survival rate of HCC patients. TLR4 may be a direct target gene of miR-122. Over-expression of miR-122 induced apoptosis and inhibited cell viability of HCC by down-regulating TLR4, enhanced the expression of pro-apoptotic genes and suppressed the expression of anti-apoptotic genes. MiR-122 inhibited expressions and activities of inflammatory cytokines, including vascular endothelial growth factor (VEGF), interleukin 6 (IL-6), cyclooxygenase-2 (Cox-2) and prostaglandin E2 (PGE2) and also reduced the expression of matrix metallopeptidase 9 (MMP-9). Furthermore, activities of phosphatidylinositide 3-kinases (PI3K), Akt and nuclear factor-kappa B (NF-κB) were suppressed by miR-122.
ConclusionsDown-regulation of miR-122 facilitated the immune escape of HCC by targeting TLR4, which was related to PI3K/Akt/NF-κB signaling pathways. Our study may provide a possible strategy for the treatment of HCC.
Hepatocellular carcinoma (HCC) is the most common primary liver cancer that mainly develops from chronic inflammatory liver diseases such as liver fibrosis and liver cirrhosis [1–3]. The incidence of HCC in China is 50% over the world's cases. The progression of HCC is regulated by various factors, to be specific, immune system plays a central role in the response to tumors and cell-mediated immunity is responsible for identifying and eliminating the genetically altered cells after transformation, namely immune surveillance [4]. However, some tumor cells could resist supervision from immune system. In case that abnormal antigen was recognized on the cell surface, transformed cells would be destructed and down-regulation of tumor antigens would therefore directly lead to the spread of cancer cells [5,6]. This suggested that immune escape might also exert an effect in HCC patients [7,8].
To the best of our knowledge, attack of intestinal bacteria, can aggravate liver fibrosis and cirrhosis [9], which is closely associated with the recognition of pathogen-associated molecular patterns (PAMPs). Toll-like receptors (TLRs) belong to the critical pattern recognition receptors (PRRs) family, which govern both innate and adaptive immune responses [10,11]. Toll-like receptor 4 (TLR4) is an important member of TLRs as a ligand of lipopolysaccharide (LPS) [12]. Moreover, except for the immune cells, expression of TLR4 has been detected in many cancer cells including HCC. Activation of TLR4 promotes tumor cells proliferation and metastasis and suppresses tumor cells apoptosis [13]. Furthermore, the activation of TLR4 could activate phosphoinositide 3-kinase (PI3K)/protein kinase B (Akt) and NF-κB pathways, which will trigger the secretion of inflammatory cytokines [14,15]. Thus, TLR4-based immune therapeutic strategies may be used for the treatment of cancers including HCC.
MicroRNA (miRNA) is a non-coding small RNA that takes charge of regulating the expression of targeted messenger RNA (mRNA) at the post-transcriptional level [16]. Extensive studies have reported the implicated roles of miRNAs in cancer [17–19]. MiR-122 was first identified in mouse [20] and the expression of miR-122 is extremely abundant in adult mouse liver [21]. Previous studies have suggested that miR-122 serves as a tumor suppressor in HCC [22–24]. However, the relationship between miR-122 and TLR4 in HCC is still little known. Based on these investigations, it is likely that some uncovered connections between miR-122 and TLR4 may exert pivotal roles in the immune surveillance of HCC. Therefore, the study was mainly to explore the potential associations between miR-122 and TLR4 in the immune escape of HCC and illustrate the possible underlying mechanisms.
2Materials and methods2.1Tissue samplesThe written informed consent was received from 37 patients with HCC. According to diagnosis reports, the size of tumor that larger than 5cm existed on 13 patients while that smaller than 5cm existed on the rest 24 ones; for the alteration of the tumor, high differentiation occurred to 8 patients while moderate or low differentiation occurred to 29 patients. This study was approved by the Institutional Review Board of Shenzhen University General Hospital. All the patients didn’t receive radiotherapy or chemotherapy before surgery. Cancer tissues and normal tissue samples (>5cm distance from the cancer location) were collected for determining the expressions of miRNAs, mRNAs, and proteins.
2.2Cell culture and transfectionHCC cell lines (Hep3B and MHCC97H) and non-tumor liver cell line (HL-7702) were purchased from the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco's-modified Eagle's medium (DMEM) with 10% fetal bovine serum under a humidified atmosphere of 5% CO2 at 37°C. MiR-122 vector and the negative miRNA control were obtained from Genecopoeia (Guangzhou, China). When the confluence reached 65%, the cells were cultured in serum-free medium overnight. MiR-122 or negative miRNA control was transfected with Lipofectamine 3000 following the manufacturer's instructions. Then the cells were incubated at 37°C for 48h prior to the subsequent experiments. Such experiments were undertaken at least three times independently.
2.3Cell proliferation assayThe cell viability was detected by Cell Counting Kit-8 (CCK-8) (CA1210, Solarbio, China) following the manufacturer's instruction. In brief, the cells were seeded at a density of 1×103 per well in 96-well plates. Afterwards, CCK-8 solutions were added in each well and the plates were maintained at 37°C for 4h. The absorbance at 450nm was measured by a spectrophotometer (Bio-Rad, USA).
2.4Flow cytometry for apoptosis evaluationAnnexin V-FITC/PI fluorescence dye (V13241, Invitrogen, USA) was adopted to evaluate the apoptosis. The treated cells were mixed with 5μl Annexin V-FITC and maintained for 10min at room temperature. The cells were washed with PBS buffer and then added with 10μl PI. Flow cytometry was performed using CytoFLEX flow cytometer (Beckman Coulter, Brea, CA, USA) after 10min incubation in the dark. The results were analyzed by CELLQuest software (BD).
2.5Luciferase reporter assayThe potential binding site of miR-122 in the 3′UTR of TLR4 was predicted by the available online database (http://www.microrna.org/microrna/home.do). The wild type (wt)-3′UTR-TLR4 firefly luciferase reporter vector (named pmirGLO-TLR4, CDS-H13244-14) was purchased from Axybio Biotechnology Co., Ltd. The mutant (mut)-3′UTR-TLR4 was generated with site-directed mutagenesis kit (KM131204, Tiangen, China). The pRL-TK plasmid (Promega) and negative miRNA control were used as the luciferase control or negative control, respectively. The luciferase activity was examined with Dual-Glo Luciferase Assay System (E2920, Promega) according to the manufacturer's protocols.
2.6Enzyme linked immunosorbent (ELISA) assayBefore detection, cells were incubated in serum-free DMEM medium for 24h. Then, the supernatants were collected to determine the levels of VEGF (DVE00), IL-6 (D6050), MMP-9 (DM900) and PGE2 (KGE004B) by ELISA (R&D, USA) according to the manufacturer's instructions. The optical density at 450nm was detected by a Microplate Reader (Bio-Rad, USA).
2.7Real-time PCRTotal RNA was extracted using Trizol regent (15596018, Invitrogen, USA) according to the manufacturer's protocols. Two-Step Stemaim-it miR qRT-PCR Quantitation Kit (Shanghai Novland Co., Ltd, China) was purchased to detect the expression of miRNA. The RNA was reversely transcribed with M-MLV reverse transcriptase (M1705, Promega, USA) and oligodT. The amplification of cDNA was performed using AceQTM qPCR SYBR Green Master Mix (Nanjing Vazyme Biotech) to quantify the expression of TLR4. The real time PCR was carried on iCycleriQ real-time PCR system (Bio-Rad, Hercules, CA). The U6 or GAPDH served as the interval control. The relative expression levels were calculated using the 2−ΔΔCt method [25]. The sequences of primers are listed in Table 2.
The sequences of primers. The catalog number and dilution of primary antibody.
| Primer name | Sequence (5′-3′) |
|---|---|
| Bax | Forward: 5′-GTGCCGGAACTGATCAGAAC-3′ |
| Reverse: 5′-CCAAAGTAGGAGAGGAGGCC-3′ | |
| Bcl-2 | Forward: 5′-GCCTTCTTTGAGTTCGGTGG-3′ |
| Reverse: 5′-GAAATCAAACAGAGGCCGCA-3′ | |
| Survivin | Forward: 5′-AGCCAGATGACGACCCCATA-3′ |
| Reverse: 5′-CAAGGGTTAATTCTTCAAACTGCTT-3′ | |
| TLR4 | Forward: 5′-TCTGGGGAGGCACATCTTCT-3′ |
| Reverse: 5′-AGGTCCAAGTTGCCGTTTCT-3′ | |
| miR-122 | Forward: 5′-ACACTCCAGCTGGGTGGAGTGTGACAATCC-3′ |
| Reverse: 5′-TGGTGTCGTGGAGTCG-3′ | |
| U6 | Forward: 5′-ATTGGAACGATACAGAGAAGATT-3′ |
| Reverse: 5′-GGAACGCTTCACGAATTTG-3′ | |
| GAPDH | Forward: 5′-AGAACATCATCCCTGCATCC-3′ |
| Reverse: 5′-CACATTGGGGGTAGGAACAC-3′ |
| Primary antibody name | Catalog number | Dilution | Manufacturer |
|---|---|---|---|
| anti-TLR4 | ab13556 | 1:500 | Abcam |
| anti-survivin | ab76424 | 1:5000 | Abcam |
| anti-Bcl-2 | ab59348 | 1:700 | Abcam |
| anti-Bax | ab53154 | 1:1000 | Abcam |
| anti-AKT1/2 | ab182729 | 1:5000 | Abcam |
| anti-p-AKT (ser473) | ab81283 | 1:5000 | Abcam |
| anti-NF-κB | ab32360 | 1:2500 | Abcam |
| anti-VEGF | ab106580 | 1:1000 | Abcam |
| anti-PGE2 | ab167171 | 1:3000 | Abcam |
| anti-Cox-2 | ab15191 | 1:500 | Abcam |
| anti-IL-6 | 12153 | 1:1000 | Cell signaling technology |
| anti-MMP-9 | 13667 | 1:1000 | Cell signaling technology |
| anti-p-PI3K Kinase Class III (Ser249) | 13857 | 1:1000 | Cell signaling technology |
| anti-PI3 Kinase Class III | 4263 | 1:1000 | Cell signaling technology |
| anti-GAPDH | 5174 | 1:1000 | Cell signaling technology |
Total proteins were isolated using Total protein extraction kit (BC3640, Solarbio, China). The concentrations were examined using BCA Protein Assay Kit (CW0014, Cwbio, China). The proteins were loaded on 10% SDS-PAGE gel and electrophoresis to separate the target proteins. Having transferred onto PVDF membranes, the proteins were blocked using non-skimmed milk at room temperature for 2h. Primary antibodies were added to the membranes and incubated at 4°C overnight and the details were listed in Table 2. HRP-conjuncted secondary antibodies (sc-2963, 1:200 diluted, Santa Cruz, USA) were added and incubated for 1h at room temperature. Enhanced Chemiluminescence (ECL) detection regent (RPN2232, Amersham Life Sciences) was used to visualize the blots and exposure to X-ray film (Kodak, Amersham Life Sciences, UK). The optical density was read by Quantity One software version 4.6.9 (Bio-Rad Laboratories).
2.9Statistical analysisDifferences between tumor group and normal one were analyzed using paired t test. Kaplan–Meier methods were used to draw the survival curves; the best cut-off value was based on the ROC analysis. The correlations between the two groups were evaluated using Pearson's rank correlation coefficient. Chi-squared test was used for univariate analysis. Comparisons between the two groups were carried out with a two tailed Student's t-test. All statistical analyses were performed using SPSS V.14.0 for Windows (SPSS Inc, Chicago, Illinois, USA) and Prism 6.0 (GraphPad Software Inc.). P<0.05 was considered as statistically significant difference.
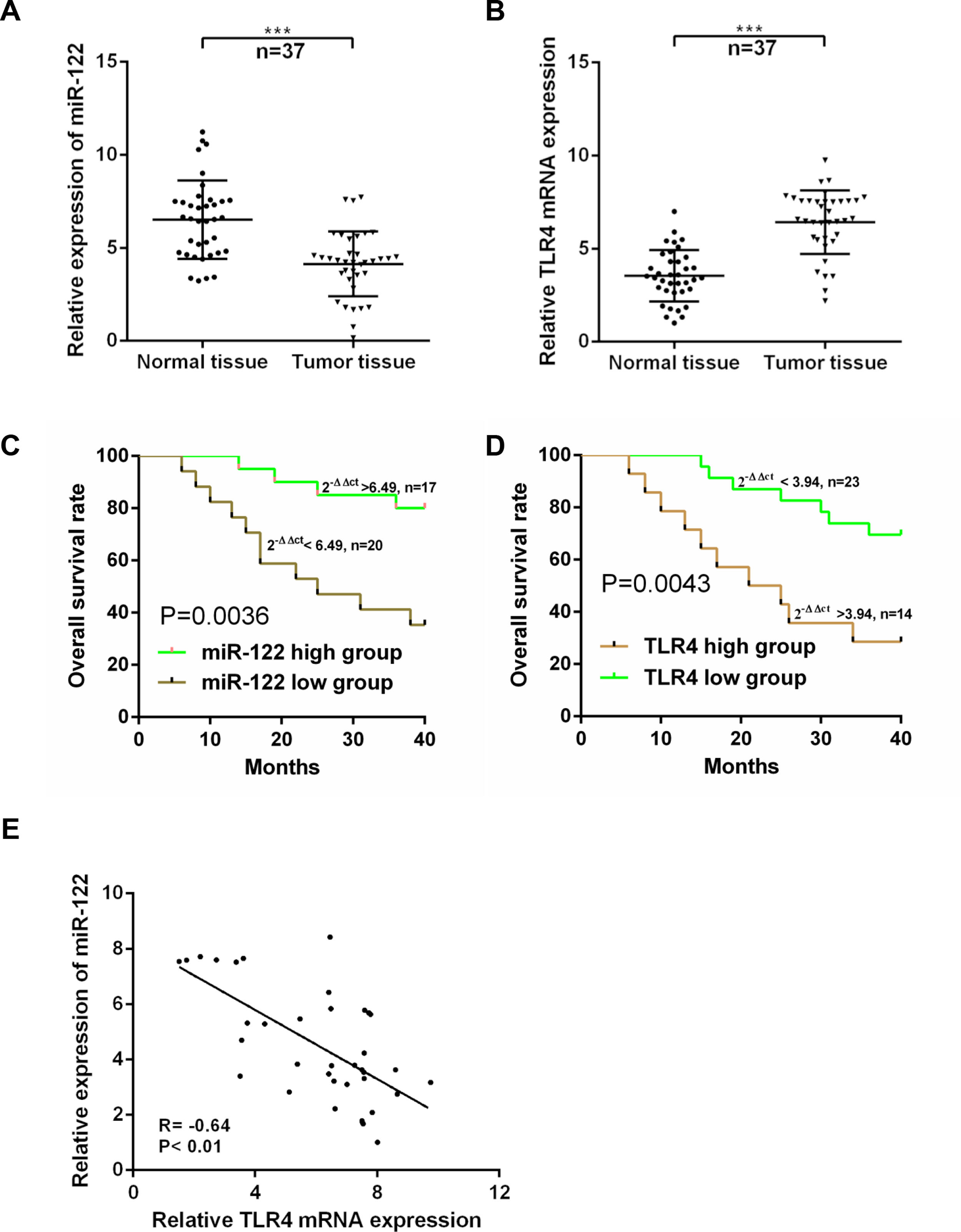
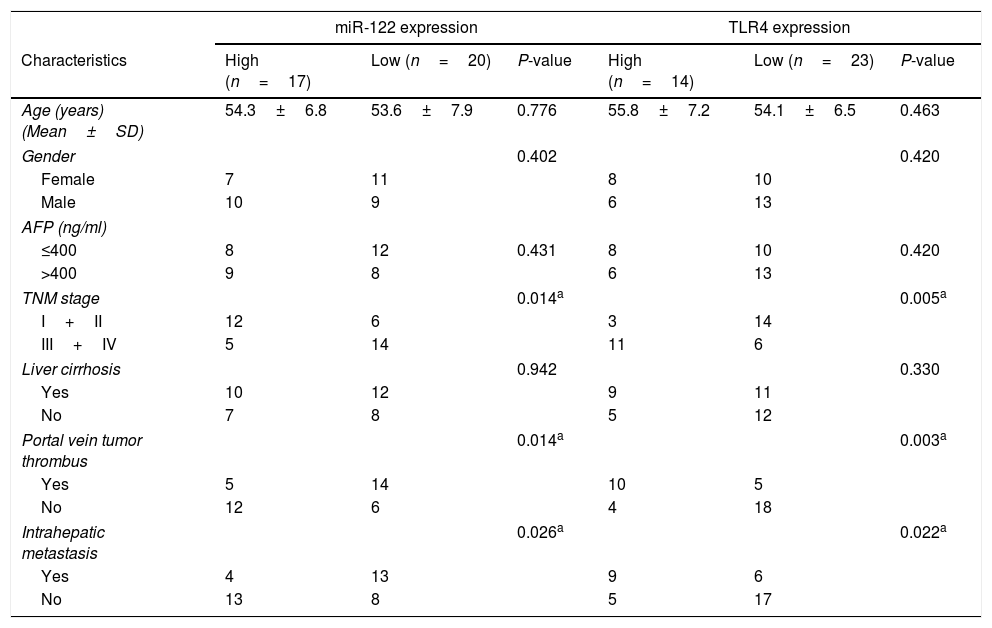
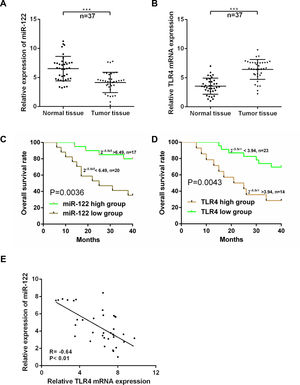
3Results3.1Expressions of miR-122 and TLR4 in HCC patientsExpressions of miR-122 and TLR4 in HCC tumor and normal tissues were examined using real time-PCR. As shown in Fig. 1A and B, compared to the normal tissues, the expression of miR-122 was dropped significantly (P<0.001); while that of TLR4 was elevated markedly in tumor tissues (n=37) (P<0.001). Based on the ROC analysis, 6.49-fold expression (2−ΔΔCt) was analyzed as the cut-off value to define the low (<6.49-fold) and high (>6.49-fold) miR-122 gene expressions, while 3.94-fold expression (2−ΔΔCt) was identified to define the expression of TLR4. According to the 40-month follow-up visit, overall survival of the HCC patients was rather poor in both low-expressed miR-122 group and high-expressed TLR4 group (P<0.05) (Fig. 1C and D). Meanwhile, expressions of miR-122 and TLR4 were strongly negative correlated in tumor tissues (r=−0.64, P<0.01) (Fig. 1E). Furthermore, the relationship among the expressions of miR-122 and TLR4 and clinical pathological features was investigated and summarized in Table 1. Notably, both low-expressed miR-122 and high-expressed TLR4 expressions were significantly associated with the advanced TNM stage (III+IV), portal vein tumor thrombus and intrahepatic metastasis.
Relationship of clinical pathological features and the expression of miR-122 and TLR4.
| miR-122 expression | TLR4 expression | |||||
|---|---|---|---|---|---|---|
| Characteristics | High (n=17) | Low (n=20) | P-value | High (n=14) | Low (n=23) | P-value |
| Age (years) (Mean±SD) | 54.3±6.8 | 53.6±7.9 | 0.776 | 55.8±7.2 | 54.1±6.5 | 0.463 |
| Gender | 0.402 | 0.420 | ||||
| Female | 7 | 11 | 8 | 10 | ||
| Male | 10 | 9 | 6 | 13 | ||
| AFP (ng/ml) | ||||||
| ≤400 | 8 | 12 | 0.431 | 8 | 10 | 0.420 |
| >400 | 9 | 8 | 6 | 13 | ||
| TNM stage | 0.014a | 0.005a | ||||
| I+II | 12 | 6 | 3 | 14 | ||
| III+IV | 5 | 14 | 11 | 6 | ||
| Liver cirrhosis | 0.942 | 0.330 | ||||
| Yes | 10 | 12 | 9 | 11 | ||
| No | 7 | 8 | 5 | 12 | ||
| Portal vein tumor thrombus | 0.014a | 0.003a | ||||
| Yes | 5 | 14 | 10 | 5 | ||
| No | 12 | 6 | 4 | 18 | ||
| Intrahepatic metastasis | 0.026a | 0.022a | ||||
| Yes | 4 | 13 | 9 | 6 | ||
| No | 13 | 8 | 5 | 17 | ||
AFP, alpha-fetoprotein; TNM, tumor-node-metastasis.
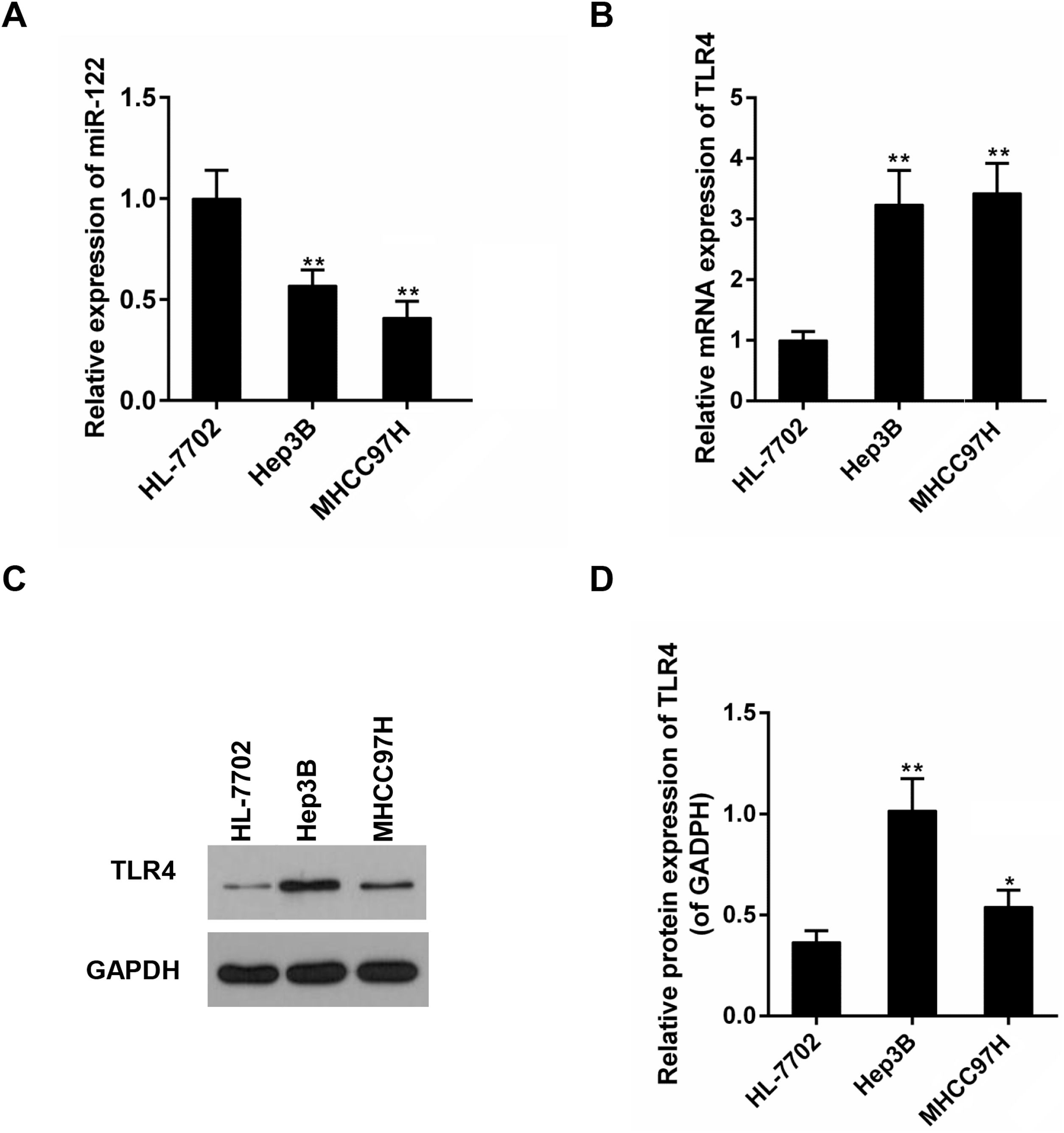
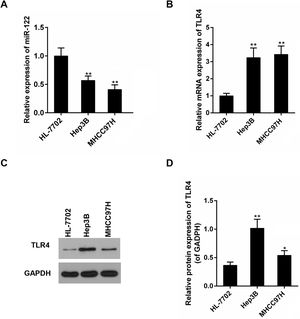
Subsequently, expressions of miR-122 and TLR4 were detected in a panel of HCC cell lines with different metastatic potentials (Hep3B and MHCC97H). It was displayed that, compared to the non-tumor liver cell line (HL-7702), mRNA expressions of miR-122 and TLR4 were declined and elevated in all HCC cell lines, respectively (Fig. 2A and B). The expression changes of miR-122 and TLR4 were bigger in MHCC97H. In protein level, an obvious expression change of TLR4 was observed in Hep3B (Fig. 2C and D).
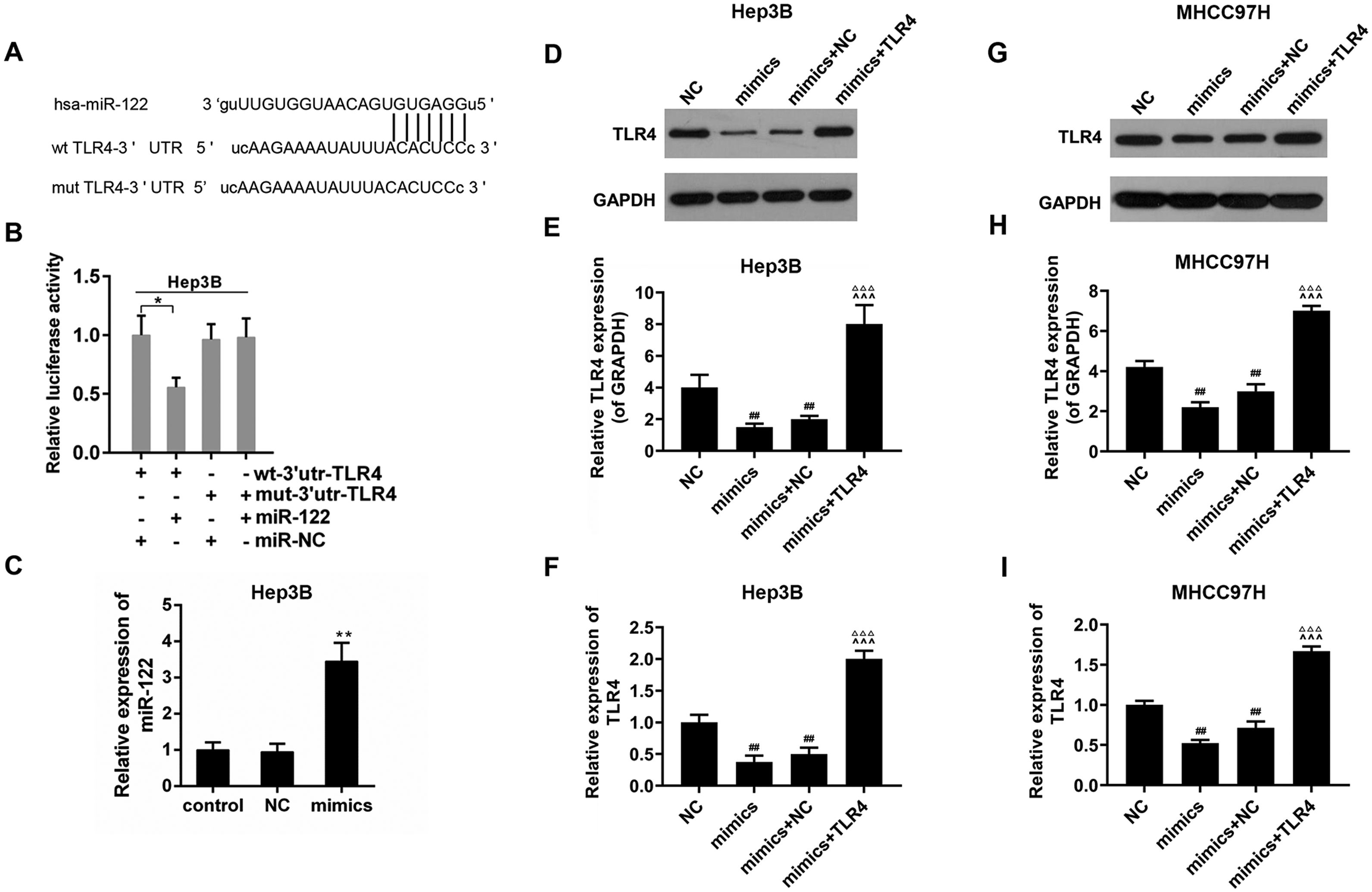
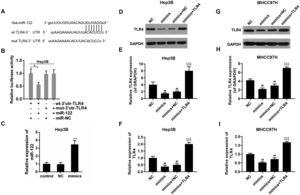
3.3TLR4 may be a direct target gene of miR-122The bioinformatics analysis showed that there was a potential binding sequence in the 3′UTR region of TLR4 (Fig. 3A). The luciferase activity of wt-3′UTR-TLR4 was inhibited in the presence of miR-122; while that of mut-3′UTR-TLR4 was not influenced in Hep3B (Fig. 3B). The transfect efficiency of miR-122 mimics was presented in Fig. 3C. To confirm the effect of miR-122, expression of TLR4 was also determined. The results revealed that the expression of TLR4 was down-regulated by miR-122 in the Hep3B and MHCC97H cells (Fig. 3D–I).
(A) The potential binding sites of miR-122 in TLR4 mRNA 3′-UTR. wt: wild-type; mut: mutant-type. (B) The effect of miR-122 on the luciferase activity of TLR4 mRNA 3′-UTR in Hep3B. *P<0.05. (C) The expression of miR-122 after transfection. mimics: miR-122 mimics. **P<0.01 vs. control. (D–I) The effect of miR-122 on the protein expression of TLR4 in Hep3B and MHCC97H cells, western blot was repeated three times. mimics: miR-122 mimics. ##P<0.01 vs. NC; ^^^P<0.001 vs. mimics; ΔΔΔP<0.001 vs. mimics+NC.
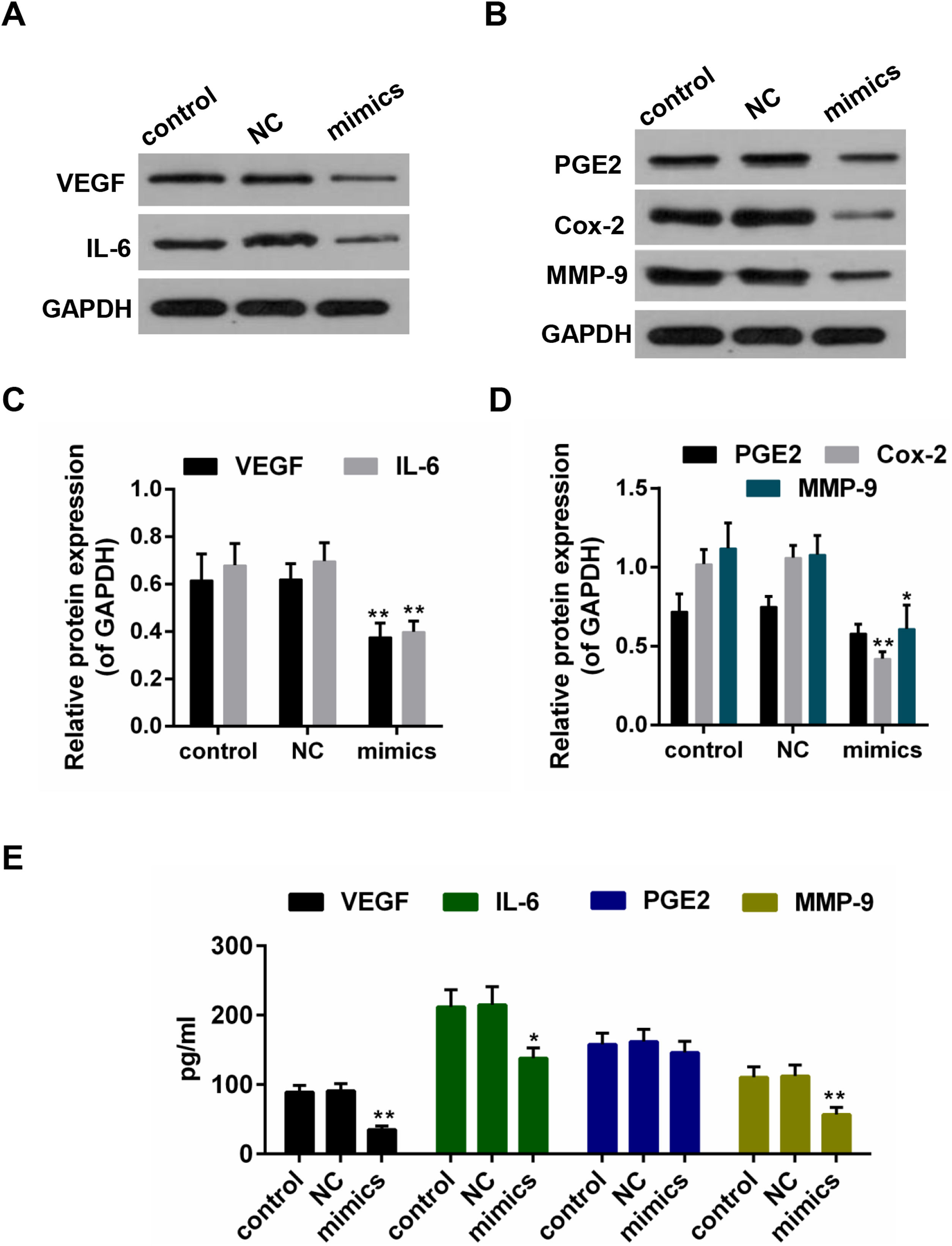
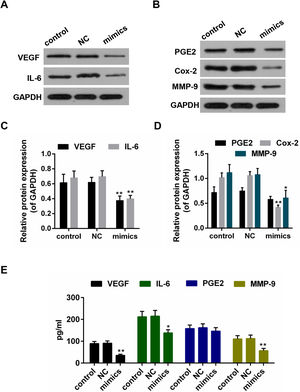
Results from the Western blot showed that the protein levels of VEGF, IL-6, PEG2, Cox-2 and MMP-9 were decreased in miR-122 mimics group (Fig. 4A–D). Furthermore, according to the ELISA assay, a significant declining trend was also shown in the expressions of VEGF, IL-6 and MMP-9 in miR-122 mimics group (Fig. 4E).
(A–D) The effect of miR-122 on the protein expressions of VEGF, IL-6, PGE2, Cox-2 and MMP-9 in Hep3B, western blot was repeated three times. *P<0.05 and **P<0.01 vs. control. (E) ELISA assay for determining the activity of VEGF, IL-6, MMP-9 and PGE2. *P<0.05 and **P<0.01 vs. control.
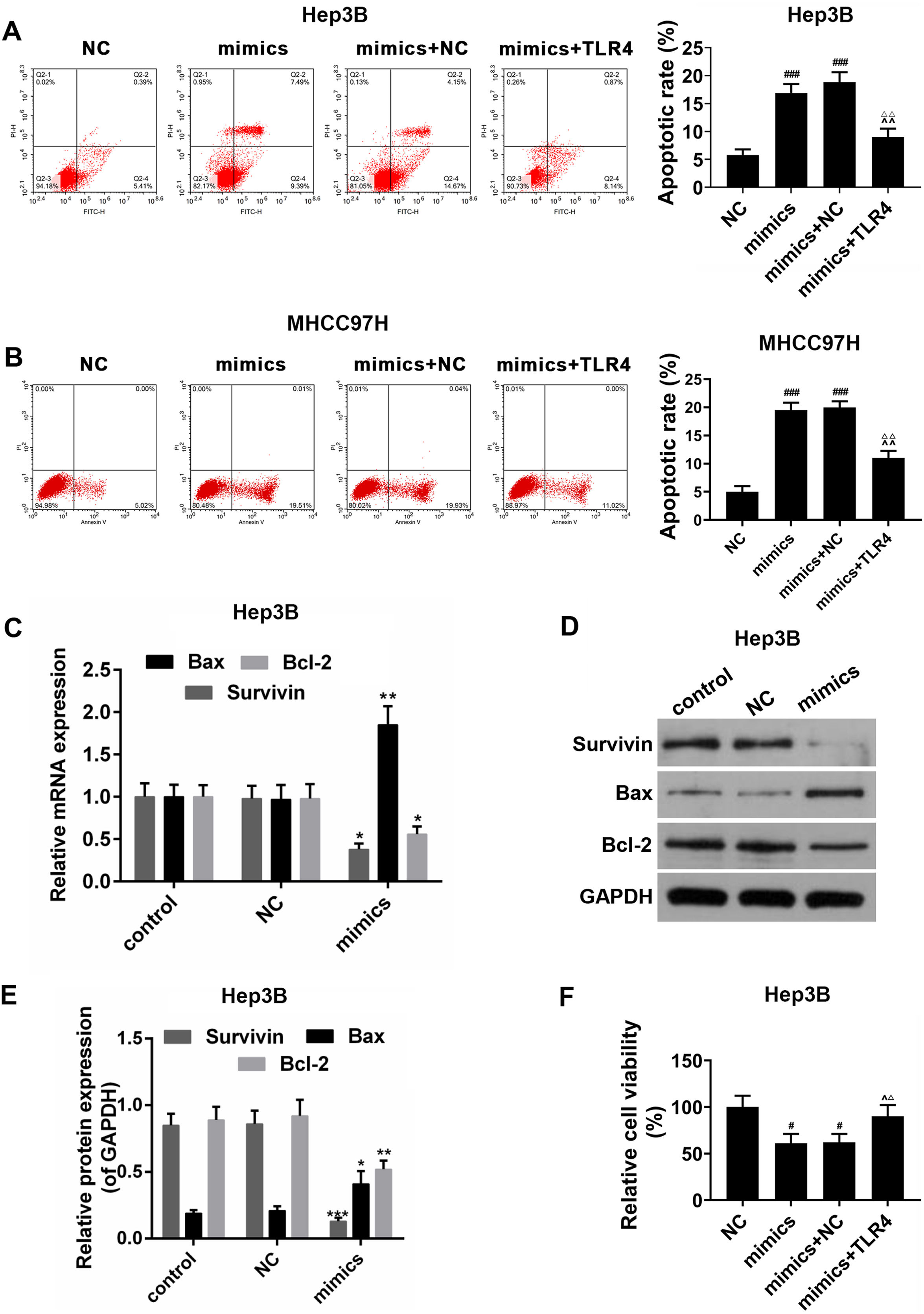
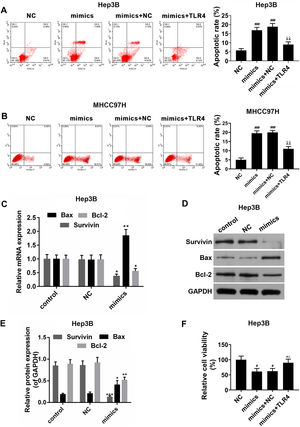
The effects of miR-122 on apoptosis and viability of HCC cells were further detected. In the Hep3B and MHCC97H cells, it was showed that apoptosis rate was increased in miR-122 mimics group, by comparison, a significantly lower apoptosis rate was observed in miR-122 mimics+TLR4 group (Fig. 5A and B). Meanwhile, as shown in Fig. 5C–E, the expression of pro-apoptotic protein (Bax) was abundantly up-regulated in miR-122 mimics group; while the expressions of anti-apoptotic proteins (survivin and Bcl-2) were down-regulated by miR-122. In addition, miR-122 inhibited cell proliferation, while TLR4 partially produced the opposite effect (Fig. 5F).
(A and B) The effect of miR-122 on apoptosis of Hep3B and MHCC97H cells. mimics: miR-122 mimics. ###P<0.001 vs. NC; ^^P <0.01 vs. mimics; ΔΔP<0.01 vs. mimics+NC. (C) RT-PCR for the expression of Bax, Bcl-2 and survivin. mimics: miR-122 mimics. *P<0.05 and **P<0.01 vs. control. (D and E) Western blot for the protein levels of Bax, Bcl-2 and survivin, western blot was repeated three times. mimics: miR-122 mimics. *P<0.05, **P<0.01 and ***P<0.001 vs. control. (F) The effect of miR-122 on viability of HCC cells. mimics: miR-122 mimics. #P<0.05 vs. NC; ^P<0.05 vs. mimics; ΔP<0.05 vs. mimics+NC.
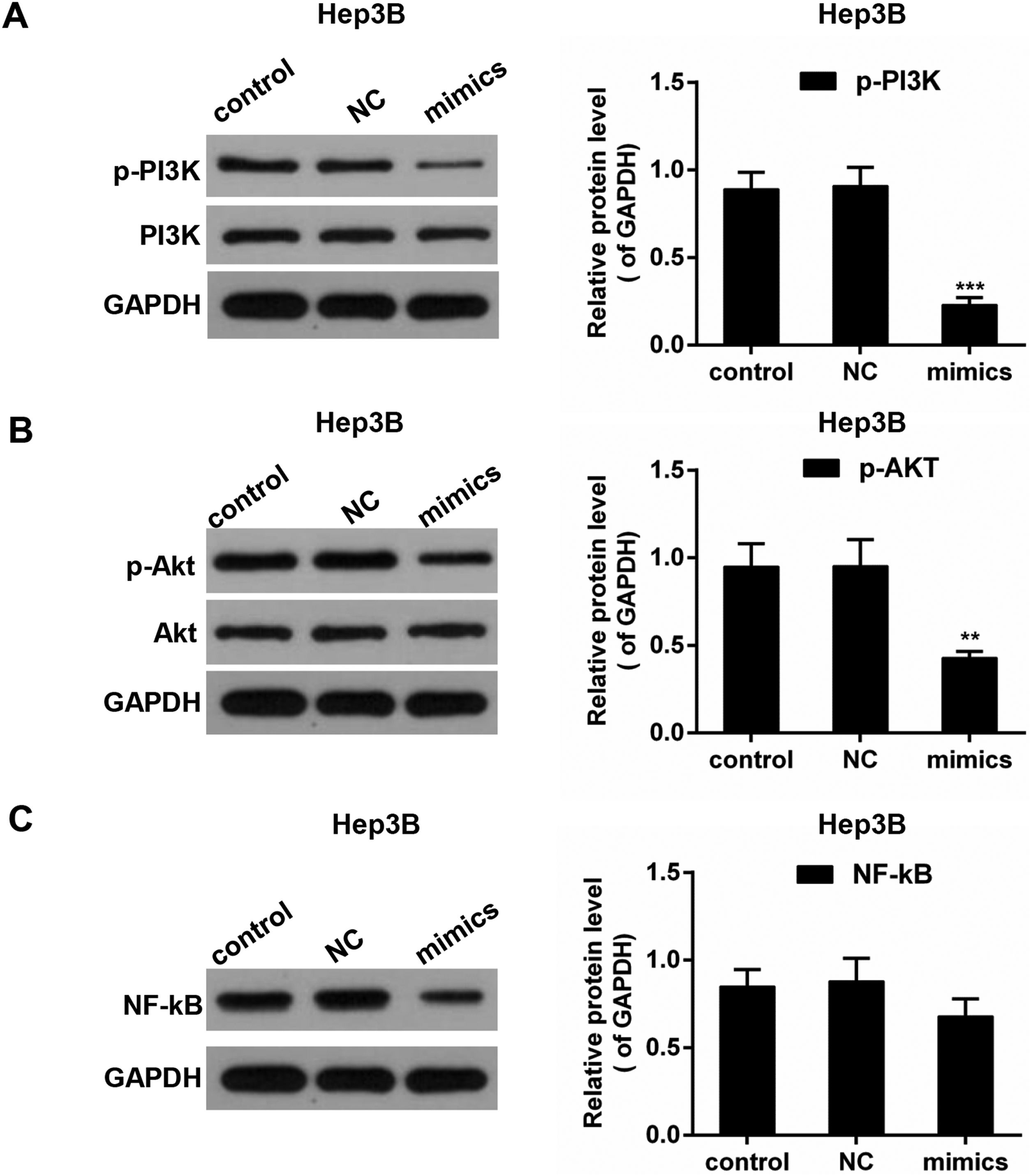
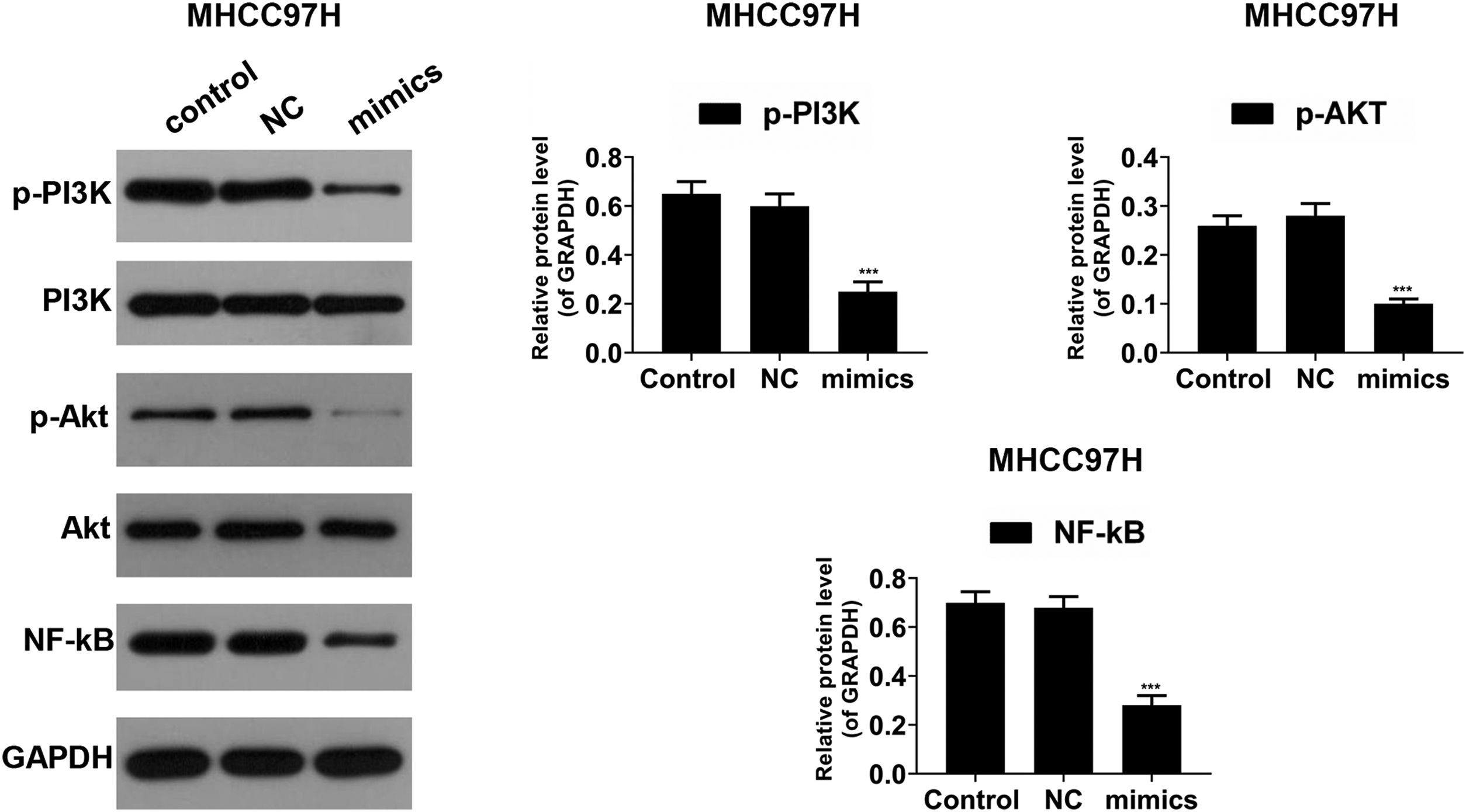
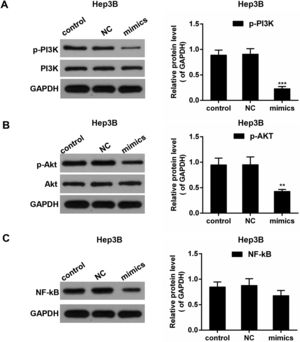
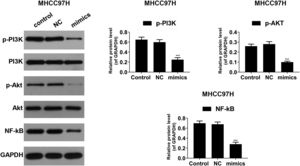
PI3K/AKT pathway and the transcription factor NF-κB play implicated roles in the regulation of the inflammation mediators [26]. To investigate the underlying mechanisms, Western blot was performed to detect the activities of PI3K, AKT and NF-κB. As shown in Fig. 6A–C, in Hep3B cells, the phosphorylated levels of PI3K (p-PI3K) and AKT (p-AKT) were dropped in miR-122 mimics group compared to the control group; the expression of NF-κB was reduced in the presence of miR-122. In MHCC97H cells, the phosphorylated levels of PI3K (p-PI3K) and AKT (p-AKT) was decreased in miR-122 mimics group compared to the control group, and the protein expression of NF-κB was decreased in the miR-122 mimics group (Fig. 7).
The current study proved that up-regulation of miR-122 promoted apoptosis and inhibited the escape of HCC from immune surveillance. This progress may be associated with activation of PI3K/AKT/NF-κB signaling pathways. It was widely accepted that attenuated immune surveillance and inflammation were close related to the tumor progression [27]. TLRs were important receptors of PAMPs, through which PAMPs could trigger inflammatory responses [28]. Liver is the first line of defense against gut-derived microbiota [29]. TLR4, as a member of TLRs, was activated in liver cells by recognizing LPS, which facilitated the progression of HCC [13]. Many mi-RNAs are intensively involved in cancer and immune system [30]. The abnormal expression of miR-122 is common in HCC. Nevertheless, the effect of miR-122 on immunosuppression of HCC is largely unknown.
In this study, the expressions of TLR4 and miR-122 were negatively correlated in patients with HCC, whose poor overall survival rate was both found in low miR-122 expression group and high TLR4 expression group. The inversed expression patterns of miR-122 and TLR4 were confirmed in a panel of HCC cell lines. MicroRNAs are generally regarded as regulators of mRNA translation; the corresponding mRNA level may even be up-regulated in an attempt to compensate protein inhibition. In this study, we found that TLR4 was a direct target gene of miR-122. The elevated miR-122 expression was associated with the decreased level of TLR4, a potential protein target. Moreover, HCC is derived from chronic inflammatory liver diseases. It has been proved that the growth and survival of liver cancer cells are often influenced by inflammatory cytokines [23,24]. The generated inflammatory cytokines are accompanied by inflammatory reactions, including VEGF, IL-6, Cox-2 and PGE2, which help cancer cells escape from host immune surveillance [31]. To investigate the effect of miR-122 on the immune escape of HCC, secretion of these cytokines was detected. The result showed that miR-122 decreased inflammation of HCC by reducing secretion of these cytokines, indicating that the immune escape of HCC may be prevented by miR-122. Consistently, it has been described that the activation of TLR4 facilitated the inflammation-driven HCC promotion and boosted the activation of Cox-2/PEG2 loop [32]. In addition, MMP-9 is linked to the metastasis and invasion of HCC, which is responsible for the degradation of extracellular matrix [33]. A previous study reported that the blockade of MMP-9 could inhibit HCC invasion [34]. The result in this study revealed that the secretion of MMP-9 was suppressed by miR-122. It was implied that miR-122 might be capable of preventing the metastasis and invasion of HCC.
Malignant proliferation and resistance to apoptosis are the major characteristics of tumor cells. There are many factors involving in the progress of apoptosis. Survivin belongs to inhibitor of apoptosis (IAP) family. In addition, Bcl-2 and Bcl-2-assocaited X proteins (Bax) are recognized as an anti-apoptotic protein and a pro-apoptotic protein, respectively [35]. In the current study, over-expression of miR-122 up-regulated expression of BAX whereas it down-regulated expressions of Bcl-2 and survivin. Meanwhile, viability of HCC cells was declined by miR-122, and overexpression of miR-122 promoted apoptosis. TLR4 reversed the effect of miR-122 on HCC cells. This suggested that miR-122 might induce apoptosis and suppress the growth of HCC cells by inhibiting TLR4 expression, which was consistent with previous studies in HCC [36,37].
PI3K/Akt pathway is important for regulating innate immune and TLR-mediated inflammatory reactions [38]. It has been documented that NF-κB is involved in various cellular events including the inflammatory responses [39]. To further investigate the potential mechanism of miR-122 on liver cancer cells, we detected the expressions of PI3K, Akt and NF-κB. The results displayed that both phosphorylation level of PI3K/Akt and expression of NF-κB were depressed by miR-122. It was suggested that miR-122 prevented the immune escape of HCC by inhibiting the PI3K/Akt/NF-κB signaling pathways. It was compatible to one previous study, in which the specific inhibition of PI3K/Akt mitigated the MMP-9-mediated invasion of HCC [34]. On the contrary, another study reported that the inflammatory responses can be attenuated by icariin through the activation of PI3K/Akt and the inhibition of NF-κB, which was a bit of contradictory to the findings in this study [26]. The conflict results may be attributed to the complexity of biological events, in which progress various signaling pathways may be involved. For example, the activity of NF-κB was reported to be influenced by the blocking of PI3K/Akt [40]. The relationship of these two factors in this study was still to be illustrated in in-depth studies. In addition, it has been reported that IGF1R was a target gene of miR-122, and Akt was a downstream IGF1R pathway [41]. The activation of Akt may be the result of miR-122 re-expression and not due to down-regulation of TLR4. Therefore, whether miR-122 regulates Akt through TLR4 remains to be further explored. Another limitation was that the direct binding of TLR4-3′UTR and miR-122 needed more solid evidences. In this study, we preliminarily analyzed the relationship among the expressions of miR-122 and TLR4 and clinical pathological features. In future studies, we will increase the sample size for further analysis.
In summary, expressions of miR-122 and TLR4 were negatively correlated in HCC patients. MiR-122 and TLR4 expressions were decreased and increased respectively in HCC patients and HCC cell lines. Moreover, over-expression of miR-122 promoted apoptosis and inhibited the immune escape of HCC by regulating the PI3K/Akt/NF-κB signal pathways. It was indicated that miR-122 might be a biomarker for the diagnosis of HCC, and TLR4 might be a promising target for preventing the immune escape of HCC.
Author contributionsXiaolin Wei conceived and designed the experiments and is responsible for the integrity of the work as a whole.
Hui Liu performed the experiments and wrote the manuscript;
Xiaowu Li analyzed the data and assist the manuscript writing;
Xiangde Liu helped perform the analysis with constructive discussions.
FundingThis research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of interestThe authors declare no conflicts of interest.