Objective. We estimated the prevalence and identified the resistance pattern of HBV genotypes H and G in HBV monoinfected and HIV co-infected patients.
Material and methods. A cross-sectional prevalence and analytic study were performed in chronic hepatitis B patients at the Hospital de Infectologia, La Raza National Medical Center in Mexico City. Chronic HBV monoinfected and HIV co-infected patients were included. HBeAg, HBV viral load and genetic analysis of mutations were collected; CD4+ cells count from HIV co-infected patients and HIV RNA were measured. We calculated the prevalence and exact 95% binomial confidence interval and the Odds ratios (OR) with 95% confidence intervals to assess the relationship between the presence of risk factors and HBV genotypes H or G.
Results. We enrolled 77 patients, 67 men and 10 women with 37 HIV co-infected patients. The distribution of HBV genotypes was: HBV genotype H 55 (71% [95% CI 60% to 80%]), HBV genotype G 16 (20.7%), HBV genotype F 4 (5.1%) and HBV genotype A 2 (2.6%). The most frequent mutations presented in 8 HIV co-infected patients and one mono-infected patient with antiretroviral therapy (ART) experience were rtM204V and six of them showed genotype G (6/9). Mono-infected HBV patients exposed more probability to HBV genotype H than co-infected HIV patients OR 13.0 (CI 95% 3.40-49.79), p = 0.0001. In contrast co-infected patients presented less possibility to have genotype H, 0.56 (CI 95% 0.42-0.75).
Conclusions. This study confirms the high prevalence of HBV genotype H in Mexico; furthermore, our results suggest that HBV genotype G predominates in co-infected patients. As well, rtM204V and rtL180M mutations are common in HBV-HIV co-infected patients with genotype G and ART experience.
An estimated 350 million persons worldwide are chronically infected with hepatitis B virus (HBV).1 Carriers [defined as persons positive for hepatitis B surface antigen (HBsAg) for more than 6 months] of HBV are at increased risk of developing cirrhosis, hepatic decompensation, and hepatocellular carcinoma (HCC).2,3 Currently 9 HBV genotypes (A-I) with specific geographic distributions have been identified worldwide.4-6 These genotypes are based on a nucleotide divergence in the entire genome of at least 8%.7 These nine genotypes have a characteristic geographical distribution, but most have a worldwide prevalence because of human migration. As well, several studies have suggested that genotypes may correlate with disease activity and may affect the response to antiviral treatment.8-11
Among the estimated 36 million people living with HIV worldwide, 10% are chronically infected with HBV.12 Nucleosides analogues are associated with the emergence of mutations in the YMDD motif of HBV DNA polymerase domain C and with upstream compensatory mutations in polymerase domains A and B that, collectively, reduce treatment efficacy.13 The rate of emergence of HBV resistance mutations occur frequently and only one single mutation may invalidate the activity of some drugs (e.g., M204I for lamivudine (LAM), emtricitabine (FTC) and telbivudina (LdT)). Several changes are required to compromise the activity of others (e.g., L180M + M204V + T250V for entecavir (ETV).14 The development of resistance to one agent could lead and increase the cross-resistance and reduce or completely hamper the activity of other drugs. This is particularly true for LAM resistance mutations, which annul the activity of FTC and LdT, to a lesser extent of ETV, and occasionally of ADV adefovir (ADV), while tenofovir (TDF) remains active in most instances.15,16
In Mexico, genotypes H and G have been reported as the most common, although genotypes prevalence in HIV co-infected patients is unknown as mutations pattern on these patients.17,18 The purpose of this study was to estimate the prevalence and identify resistance pattern of HBV genotypes H and G, in HBV monoinfected and HIV co-infected patients.
Material and MethodsWe performed a cross-sectional prevalence and analytic study. This survey was carried out on chronic hepatitis B patients from May 2007 to September 2009 at the Hospital de Infectología, La Raza National Medical Center in Mexico City.
ParticipantsAdults were eligible if they had been positive for hepatitis B surface antigen (HBsAg) for at least 6 months, with anti-HBsAg (anti-HBs antibodies) negative; HBV DNA levels more than 500 IU per milliliter and increase in the aminotransferases serum levels.
During the enrollment period, 77 patients were admitted for evaluation if they presented a diagnosis of chronic hepatitis B. We recruited 37 HIV/HBV infected patients with fulfilled criteria of chronic hepatitis B from a total of 422 HIV patients in our cohort. Clinical and laboratory data were collected: general attributes, risk factors, family history, HIV or HCV co-infections, antiretroviral (LAM or FTC) experience, blood cells count, liver function tests and HBeAg as well as HBV viral load and mutations genetic analysis. When patients presented HIV co-infection, CD4+ cells count and HIV RNA were measured.
Amplification of HBV DNA and mutations detectionHepatitis B virus genotype and BCP/Precore mutations was performed by Quest Diagnostics Nichols Institute, San Juan Capistrano, CA., USA, according its methodology. HBV DNA was detected in EDTA plasma isolates by polymerase chain reaction (PCR) with HBV-specific primers. The sequence analysis was done for mutations at codons 173, 180, 202, 204, 207, in the reverse transcriptase region of the polymerase gene; nucleotides 1858 and 1896 of the pre-C region; and nucleotides 1762 and 1764 of the BCP region. The HBV genotype was determined via computer-aided alignment and phylogenetic analysis of the amplified portion of the S gene.
EthicsProcedures were approved by the local Ethical Committee of the hospital with register number R2009-3506-4 (Comité Local de Investigación en Salud 3506; Instituto Mexicano del Seguro Social), in accordance with the Helsinki Declaration of 1975. Informed consent was obtained in order to take the blood samples for the analysis.
Statistical methodsThe prevalence and extract 95% binomial confidence interval were calculated. Odds ratios and 95% confidence intervals were analyzed to assess the relationship between risk factors and the presence of HBV genotypes H or G.
ResultsPatients characteristicsWe enrolled 77 Mexican patients, 67 men and 10 women. Chronic HBV was confirmed in all cases.
- •
The mean (± SD) age of these subjects was 34.2 ± 12.3 years.
- •
AST 75.2 ± 113 IU/mL.
- •
ALT 93.6 ± 130 IU/mL.
- •
The mean (± SD) HBsAg was 238,400 ± 50,100 IU/mL.
- •
HBeAg 876.6 ± 778.2 IU/mL.
- •
HBV DNA 20,200,000 IU/mL (IQR 155,500225,000,000).
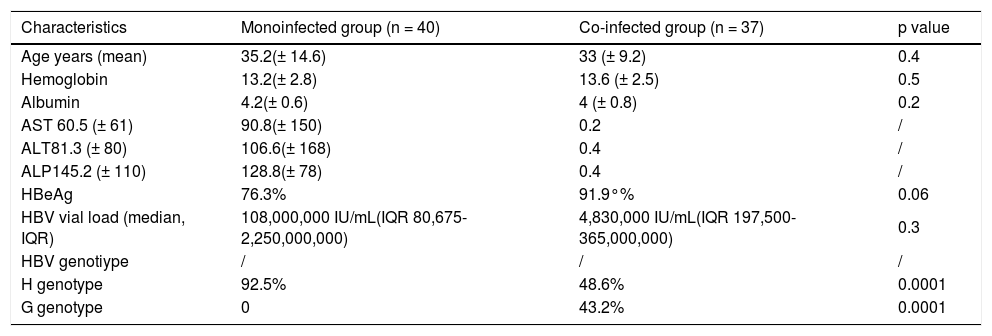
Twelve (15.6 %) subjects were HBeAg negative. Thirty seven were HIV co-infected patients, with CD4+ cells count of 260.9 ± 226 cells/mm3 and HIV RNA of 35,784 copies/mL (IQR 2,537-165,479). Eight patients (21.6%) showed a history of receiving LAM (all of them were co-infected HIV/HBV patients) and one mono-infected patient received LAM therapy for 2 years (Table 1).
Basal characteristics.
| Characteristics | Monoinfected group (n = 40) | Co-infected group (n = 37) | p value |
|---|---|---|---|
| Age years (mean) | 35.2(± 14.6) | 33 (± 9.2) | 0.4 |
| Hemoglobin | 13.2(± 2.8) | 13.6 (± 2.5) | 0.5 |
| Albumin | 4.2(± 0.6) | 4 (± 0.8) | 0.2 |
| AST 60.5 (± 61) | 90.8(± 150) | 0.2 | / |
| ALT81.3 (± 80) | 106.6(± 168) | 0.4 | / |
| ALP145.2 (± 110) | 128.8(± 78) | 0.4 | / |
| HBeAg | 76.3% | 91.9°% | 0.06 |
| HBV vial load (median, IQR) | 108,000,000 IU/mL(IQR 80,675-2,250,000,000) | 4,830,000 IU/mL(IQR 197,500-365,000,000) | 0.3 |
| HBV genotiype | / | / | / |
| H genotype | 92.5% | 48.6% | 0.0001 |
| G genotype | 0 | 43.2% | 0.0001 |
IQR: intequartili range 25th-75th.
The major transmission mechanism identified in mono-infected patients was hemodialysis, and sexual transmission in co-infected HIV/HBV subjects.
Finally from the 422 HIV infected patients in our hospital, 37 presented chronic hepatitis B 8.7% (95% CI 6.4% to 11.8%).
HBV genotypingHBV genotypes from 77 patients showed a distribution of 55 HBV genotype H patients (71% [95% CI 60% to 80%]), 16 HBV genotype G patients (20.7% [95% CI 13% to 31%]), 4 HBV genotype F patients (5.1%) and 2 HBV genotype A patients (2.6%). We did not find mixed HBV genotypes. Mono-infected patients 37/40 (91%) vs. co-infected HIV patients 18/37 (50%) had HBV genotype H. Mono-infected HBV patients had more probability to present HBV genotype H, OR 13.0 (CI 95% 3.4049.79), p = 0.0001, than co-infected HBV patients with HIV.
Resistance mutationsMutations were detected in 8 HIV co-infected patients and one mono-infected patient with ART experience where LAM was part of the regimen during a period between 12 to 35 months and detectable HBV DNA. Although, not all patients had resistance mutations, six showed genotype G, two genotype H and one genotype A with the following mutations:
- •
rtL180M.
- •
rtM204V.
- •
rtV207V/I.
- •
rtV173L.
- •
ntG1896A/T.
- •
ntG1896T, and
- •
ntC1858A.
Most frequent mutations were (Table 2):
Characteristics of pattern mutations.*
| Patients | Genotype | Pattern mutation |
|---|---|---|
| 1 | G | rtL180M, rtM204V, rtV207V/I, rtV173V/L, ntG1896A |
| 2 | G | ntG1896A |
| 3 | G | ntG1896A |
| 4 | G | rtL180M, rtM204V, ntG1896A |
| 5 | G | rtL180M, rtM204V, rtV173L, ntG1896A/T, ntC1858A |
| 6 | G | rtL180M, rtM204V, rtV173L, ntG1896A |
| 7 | H | rtM204V/I, ntG 1896A |
| 8 | A | rtL180M, rtM204V |
| 9† | H | rtL180M, rtM204V, rtM202G |
- •
ntG1896A/T (7/9).
- •
rtM204V (7/9), and
- •
rtL180M (6/9).
This study demonstrated the high prevalence of HBV genotype H in Mexico and the differences between HBV genotype H in mono-infected patients and HIV co-infected patients which showed a prevalence of 8.7% of chronic HVB in a cohort of HIV patients. A difference among the prevalence of HBV genotype H and G in mono-infected and co-infected patients was identified, suggesting differences in the skill transmission mechanism. As well, we observed the presence of rtM204V and rtL180M mutations in HVB/HIV co-infected patients with genotype G and lamivudine treatment experience.
We had two patients with HBV genotype A, one of them with infection acquired in Spain; the other patient referred to have sex with foreigner men.
Our findings are consistent with those of ArauzRuiz and Alvarado-Esquivel,16,17 who found HBV genotype H in Mexican population, it was considered more an Amerindian genotype which has been rarely found in other countries,3,4,19 but more found in HBV isolates from Nicaragua, Mexico, USA and Japan.5,9 In our study, this genotype was predominant in mono-infected patients, usually under hemodialysis.
Livingston, et al. found in Alaska, a predominance genotype D, they associated too genotype F with hepatocellular carcinoma and T1762/A1764 associated mutations which could not be determine in our study.10
Other associated mutations as T1762/A1764 associated with genotype F and hepatocelluar carcinoma could not be determined in our study.10
Studies of long-term treatment with lamivudine in patients with HBeAg positive chronic hepatitis B reveal 11 and 14% with lamivudine resistance in 1 year, and 6 to 27% in HBeAg negative chronic hepatitis B.20,21 All our patients who developed resistance mutations had more than one year with lamivudine experience. As well, we found similar pattern mutations that Lai et al., with lamivudine resistance based on the signature mutations rtM204I and rtM204V which in the case of rtM204V was frequently accompanied by the rtL180M secondary mutation.22
We found 7 patients with ntG1896A which is a hepatitis B virus precore (PC) mutation that read an open reading frame that creates a stop codon causing premature termination of the PC protein, similar to reported by Lin CL, et al. We found similar data that suggest a high prevalence of precore stop codon and basal core promoter mutation in Mexican patients with chronic hepatitis B genotype G. Even thought, we did not find association between this mutation and HBeAg-negative, only two of 8 patients presented HBeAg-negative, maybe we did not detect mixed genotypes.23,24
Hence Chen, et al. described that during lamivudine treatment, drug resistance develops a similar rate in HBeAg positive and HBeAg negative, as well CHB and Lamivudine-resistant HBV mutants have been shown to replicate inefficiently in vitro in the absence of PC mutations. In addition, the PC stop codon mutation appears to increase the replication efficacy of lamivudine-resistant virus but does not affect in vitro drug sensitivity.24
In the case of rtV207V mutation that been described by Margeridon-Thermet, et al., in hepatitis B virus genotype A, we found it in only one patient with G genotype.25,26
A previous study reported by Chudy, et al., showed in a reported case of an HBV infection was caused exclusively by genotype G; in fact, we found some HBV genotype G in monoinfected patients associated to transfusion and hemodialysis, however, we had more HIV/HBV co-infected patients with this genotype.27
The study has a number of limitations; no conclusion about the course of HBV/HIV coinfection in H and G genotype could be done because the main complications of HBV infection (i.e., cirrhosis and hepatocellular carcinoma) are observed many years after infection. Importantly this is the first study that describes mutations pattern in these genotypes.
These results emphasize the value of screening for hepatitis B virus in HIV co-infected and hemodialysis patients and confirmed the high prevalence of HBV genotype H and HBV genotype G in Mexico.
Finally, in our study we found an important resistance pattern that confers resistance to lamivudine in patients who had antiretroviral treatment experience with lamivudine as a monotherapy to chronic hepatitis B. As well a difference among the prevalence of HBV genotypes H and G in monoinfected and co-infected patients, could suggest different transmission mechanisms. We consider that future research with a higher sample size should be developed to confirm these data.
AcknowledgementsWe thank the study participants, study investigators, and study site staff.
We thank to Helen Elizabeth Curtis-Twomey for her contribution for copyedit the paper.
Competing InterestsThere are no financial conflicts of interest.