Background and aim. DNA methylation plays a critical role in the control of important cellular processes. The present study assessed the impact of promoter methylation (PM) of some genes on the antiviral response to antiviral therapy and it’s relation to the presence of fibrosis in HCV-4 infected patients from Egypt.
Material and methods. Clinical, laboratory and histopathological data of 53 HCV-4 infected patients who were subjected to combined antiviral therapy were collected; patients were classified according to their response to treatment and the fibrosis status. The methylation profiles of the studied groups were determined using the following genes: APC, P14ARF, P73, DAPK, RASSF1A, and O6MGMT in patients’ plasma.
Results.O6MGMT and P73 showed the highest methylation frequencies (64.2 and 50.9%) while P14 showed the lowest frequency (34%). Sustained virological response (SVR) was 54.7%with no significant difference in clinico-pathological or laboratory features between the studied groups. PM of O6MGM was significantly higher in non-responders (p value 0.045) while DAPK showed high methylation levels in responders with no significance (p value: 0.09) andPM of RASSF1A was significantly related to mild fibrosis (p value: 0.019). No significant relations were reported between PM of any of the studied genes and patients’ features.
Conclusion. PM of some Tumor Suppressor genes increases in chronic active HCV-4. However, only 06MGMT can be used as a predictor of antiviral response and RASSF1A as a marker of marked fibrosis in this small set of patients. An extended study, including more patients is required to validate the results of this preliminary study.
Hepatitis C virus (HCV) is the cause of a significant proportion of cases of chronic liver disease, hepatocellular carcinoma (HCC) and deaths from liver disease. Egypt has the highest prevalence of HCV worldwide (15%) and the highest prevalence of HCV-4, which accounts for almost 90% of the cases.1 Moreover a consistent increase of seropositivity for HCV antibodies with age was observed, with a peak level of 54.9% in all individuals for the age group 45-49 years.2 The goal of treatment is to prevent complications of HCV infection, which is mainly achieved by elimination of the virus, predicted by a sustained virological response (SVR).3 The main predictors of SVR are the IL28B (IFN k3) polymorphism, the HCV genotype, and the stage of fibrosis. Other predictors of response include baseline HCV RNA levels, the dose and duration of therapy, host factors e.g. body mass index, age, insulin resistance, gender, and the characteristics of liver disease e.g. the levels of alanine aminotransferase (ALT), gamma glutamyltransferase (GGT), and the stage of fibrosis or co-infection with HIV or other hepatotropic virus.4
Epigenetic changes, including promoter methylation (PM) of several genes play a critical role in the control of cellular processes through switching genes on or off leading to differential expression of the genes, which determines the expression of proteins.5 Some studies have shown that the presence of hepatitis viruses, especially HCV, could play a role in accelerating the methylation process which is involved in HCC development, potentiate the progression of HCV related liver disease and affect its response to treatment.6
We have previously reported, in a case control study, a high frequency of PM of the APC, FHIT, p15, p16, and E-cadherin in tumor tissues and plasma obtained from 28 HCC patients from Egypt. The Promotor methylation frequency (PMF) ranged from 67.9% for p16 to 89.2% for p15 with a high concordance rate between plasma and tissue samples.7 In another study, we found that PM of a group of genes increases with disease progression from CH to HCC.8 In this study the PMF of p14, p73, RASSF1A, CDH1 and O6MGMT was significantly higher in HCC and their ANT whereas PMF of APC was higher in CH and we were able to suggest a panel of 4 genes (APC, p73, p14, O6MGMT) that can independently classify cases into HCC and CH with high sensitivity and specificity.9 Then, it may be valuable to assess whether PM of our previously tested genes contribute to fibrogenesis and response to antiviral treatment in chronic HCV-4 related liver disease.
Therefore the current study was conducted to clarify the contribution of PM to: the development of fibrosis and the response to antiviral therapy using some genes that proved to be significant in our previously studies (APC, P14ARF, P73, DAPK, RASSF1A and MGMT).
Material and MethodsPopulation samplesThe study was conducted on 53 consecutive chronic HCV-4 patients from Egypt who were eligible for treatment with pegylated interferon α and ribavirin. All patients fulfilled the standards of care, inclusion and exclusion criteria for interferon therapy, which are applied on the national wide program controlled by the National Committee for Treatment of Viral Hepatitis. Informed consents were obtained from all the participants enrolled in the study, which was performed in accordance with the declaration of Helsinki, local and national laws (clinical trial NCT01758939).
For all patients height and weight were determined at baseline and body mass index (BMI) was calculated (weight in kilograms divided by height in meters squared). Laboratory investigations including complete liver profile, kidney function, Alfa-Fetoprotein (AFP), INR, and CBC were also done to justify suitability for therapy. HCV RNA was quantified in all patients’ sera using quantitative real time PCR at baseline, after 12, 24, 48 and 72 week of anti-viral therapy. Histological examination was done on core needle biopsies to determine the grade of necro-inflammation and the stage of fibrosis according to the Metavir scoring system prior to treatment. Clinical and laboratory follow up were done for every patient to report any possible adverse side effects and the response to treatment according to IFN treatment guidelines.
Detection of promoter methylationHigh molecular weight DNA was extracted from patient’s plasma samples collected before treatment, according to our previously published protocol.7 Briefly, an equal volume of equilibrated phenol (pH 7.0-7.5) was added to samples and vortexed. The upper aqueous layer was removed and an equal volume of phenol/chloroform (1:1) was added and vortexed. The upper aqueous layer was removed again and an equal volume of chloroform/isoamyl alcohol (24:1) was added and vortexed. This was followed by the addition of 3 M Sodium acetate (pH 4.7-5.2), DNA precipitation by ice-cold ethanol and overnight incubation at -80 °C. The fluid was decanted and the DNA pellet was dissolved in sterile water.7
The extracted DNA was subjected to bisulfite treatment using EZ DNA methylation kit which uses 300 ng of the extracted Nucleic acid. This was followed by MSP using the primer sequences and the methylation- specific PCR conditions illustrated in table 1. DNA methylation of CpG islands for p14, p73, APC, DAPK, RASSF1A and O6MGMT genes was determined using specific primers for methylated (M) and unmethylated (UM) DNA as previously described by.8 Negative control samples (without DNA) were included in each PCR set. PCR products were analyzed on 4% ethidium bromide-stained agarose gel and visualized under ultraviolet illumination. The methylation index (MI), defined as the ratio between the number of methylated genes and the total number of the studied genes for each sample was calculated for all patients.8
Primers sequences and conditions of the methylation specific PCR (MSP).
| Gene | Primer | Annealing temperature °C |
|---|---|---|
| DAPK (M) | GGATAGTCGGATCGAGTTAACGTC CCCTCCCAAACGCCGA | 60 |
| DAPK (U) | GGAGGATAGTTGGATTGAGTTAATGTT CAAATCCCTCCCAAACACCAA | 57 |
| p73 (M) | GGACGTAGCGAAATCGGGGTTC ACCCCGAACATCGACGTCCG | 59 |
| p73 (U) | AGGGGATGTAGTGAAATTGGGGTTT ATCACAACCCCAAACATCAACATCCA | 60 |
| O6-MGMT (M) | TTTCGACGTTCGTAGGTTTTCGC GCACTCTTCCGAAAACGAAACG | 55.2 |
| O6-MGMT (U) | TTTGTGTTTTGATGTTTGTAGGTTTTTGT AACTCCACACTCTTCCAAAAACAAAACA | 57 |
| p14 (M) | GTGTTAAAGGGCGGCGTAGC AAAACCCTCACTCGCGACGA | 54.9 |
| p14 (U) | TTTTTGGTGTTAAAGGGTGGTGTAGT CACAAAAACCCTCACTCACAACAA | 57.2 |
| APC (M) | TATTGCGGAGTGCGGGTC TCAACGAACTCCCGACGA | 62 |
| APC (U) | GTGTTTTATTGTGGAGTGTGGGTT CCAATCAACAAACTCCCAACAA | 59.2 |
| RASSF1A (M) | TTCGTCGTTTAGTTTGGATTTTG CCGATTAAACCCGTACTTCG | 54.4 |
| RASSF1A (U) | TGTTGTTTAGTTTGGATTTTGG TACAACCCTTCCCAACACAC | 52 |
This was done using Statistical Package for Social Sciences, Version 17.0 (SPSS, Inc., Chicago, III., USA) for Windows. Continuous variables were analyzed as mean values ± standard deviation (SD) or median (range) as appropriate. Percentages were calculated for categorical data. For categorical variables, differences were analyzed with χ2 (chi square) tests and Fisher’s exact test when appropriate. Differences among continuous variables with normal distribution were analyzed by Student’s T-test; comparison between three groups was done using Kruskelswallis test (non parametric analogue for ANOVA). P value of < 0.05 was considered statistically significant.
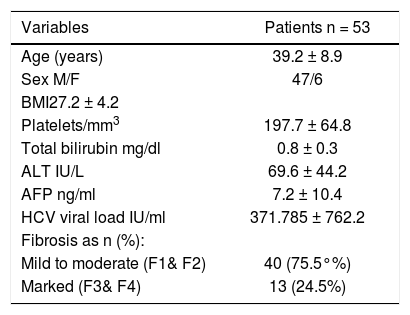
ResultsClinical and epidemiological data: Baseline demographic and laboratory features of all patients enrolled in the study in addition to the stage of fibrosis are illustrated in table 2. Out of the 53 patients assessed, 47 (89%) were males and 6 (11%) were females. Their ages ranged from 35 to 45 with a mean of 39.2. Forty patients (75.5%) showed mild to moderate fibrosis (F1/F2) and 13 (24.5%) showed marked fibrosis.
Clinico-pathological features of the studied patients.
| Variables | Patients n = 53 |
|---|---|
| Age (years) | 39.2 ± 8.9 |
| Sex M/F | 47/6 |
| BMI27.2 ± 4.2 | |
| Platelets/mm3 | 197.7 ± 64.8 |
| Total bilirubin mg/dl | 0.8 ± 0.3 |
| ALT IU/L | 69.6 ± 44.2 |
| AFP ng/ml | 7.2 ± 10.4 |
| HCV viral load IU/ml | 371.785 ± 762.2 |
| Fibrosis as n (%): | |
| Mild to moderate (F1& F2) | 40 (75.5°%) |
| Marked (F3& F4) | 13 (24.5%) |
Data are represented by Mean ± SD. AFP: α-fetoprotein. ALT: alanine aminotransferase. BMI: body mass index. F: female. HCV: hepatitis C virus. M: male.
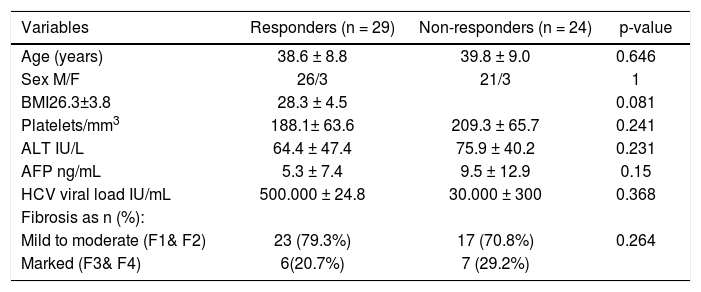
Out of 53 patients studied, 29 showed SVR (54.7%) and 24 showed either no response or relapse (45.3%). None of patients was excluded from the treatment due to emergence of any side effects and no patient received < 80% of the therapeutic schedule. The demographic, laboratory and histopathological parameters of those patients are illustrated in table 3. No significant difference was observed between the two groups (responders and non-responders) regarding the age and sex, the hematological parameters, liver profile, HCV viral load or different fibrosis stages.
Clinico-pathological features of the responder and non- responder patients
| Variables | Responders (n = 29) | Non-responders (n = 24) | p-value |
|---|---|---|---|
| Age (years) | 38.6 ± 8.8 | 39.8 ± 9.0 | 0.646 |
| Sex M/F | 26/3 | 21/3 | 1 |
| BMI26.3±3.8 | 28.3 ± 4.5 | 0.081 | |
| Platelets/mm3 | 188.1± 63.6 | 209.3 ± 65.7 | 0.241 |
| ALT IU/L | 64.4 ± 47.4 | 75.9 ± 40.2 | 0.231 |
| AFP ng/mL | 5.3 ± 7.4 | 9.5 ± 12.9 | 0.15 |
| HCV viral load IU/mL | 500.000 ± 24.8 | 30.000 ± 300 | 0.368 |
| Fibrosis as n (%): | |||
| Mild to moderate (F1& F2) | 23 (79.3%) | 17 (70.8%) | 0.264 |
| Marked (F3& F4) | 6(20.7%) | 7 (29.2%) |
Data are represented by Mean ± SD. P-value > 0.05 is not significant. AFP: α-fetoprotein. ALT: alanine aminotransferase. BMI: body mass index. F: female. HCV: hepatitis C virus. M: male.
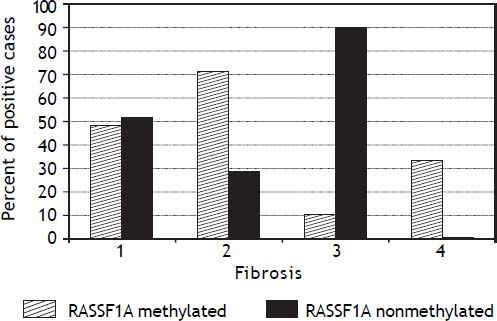
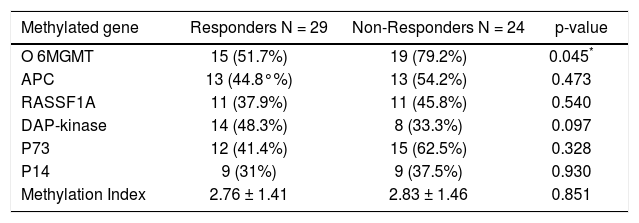
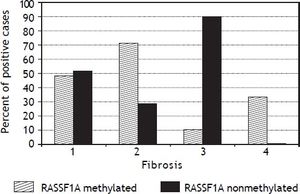
The PMFs of all studied genes are illustrated in table 4. The 06MGMT showed the highest PMF (64.2%) followed by P73 (50.9%) and APC (49.1%) whereas P14 showed the lowest PMF (34%). Out of the six genes assessed for PM, only RASSF1Agene methylation was significantly related to the presence of mild fibrosis (p value: 0.019, and 06MGMT was significantly correlated with patient’s response to therapy (p value: 0.045). On the other hand, PM of DAPK was higher in responders than in non responders, however, the difference between the two groups did not reach a statistically insignificant level (p value: 0.097) (Tables 5 and 6)(Figure 1).
Promotor Methylation Frequency of in the studied genes.
| Studied genes | Methylated genes n (%) | Un-methylateds gene n (%) |
|---|---|---|
| O6MGMT | 34 (64.2) | 19 (35.8) |
| APC | 26 (49.1) | 27 (50.9) |
| RASSF1A | 22 (41.5) | 31 (58.5) |
| DAP-kinase | 22 (41.5) | 31 (58.5) |
| P73 | 27 (50.9) | 26 (49.1) |
| P14 | 18 (34) | 35 (66) |
Numbers are represented as n (%).
The methylation status of the studied group in relation to IFN response.
| Methylated gene | Responders N = 29 | Non-Responders N = 24 | p-value |
|---|---|---|---|
| O 6MGMT | 15 (51.7%) | 19 (79.2%) | 0.045* |
| APC | 13 (44.8°%) | 13 (54.2%) | 0.473 |
| RASSF1A | 11 (37.9%) | 11 (45.8%) | 0.540 |
| DAP-kinase | 14 (48.3%) | 8 (33.3%) | 0.097 |
| P73 | 12 (41.4%) | 15 (62.5%) | 0.328 |
| P14 | 9 (31%) | 9 (37.5%) | 0.930 |
| Methylation Index | 2.76 ± 1.41 | 2.83 ± 1.46 | 0.851 |
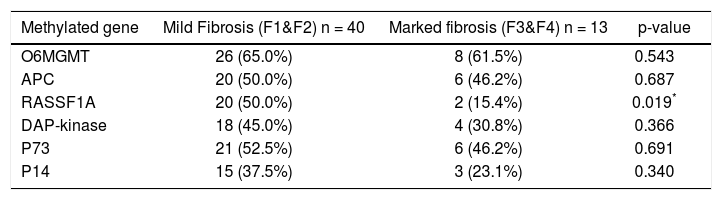
Correlation between promoter methylation of the studied genes and degree of fibrosis.
| Methylated gene | Mild Fibrosis (F1&F2) n = 40 | Marked fibrosis (F3&F4) n = 13 | p-value |
|---|---|---|---|
| O6MGMT | 26 (65.0%) | 8 (61.5%) | 0.543 |
| APC | 20 (50.0%) | 6 (46.2%) | 0.687 |
| RASSF1A | 20 (50.0%) | 2 (15.4%) | 0.019* |
| DAP-kinase | 18 (45.0%) | 4 (30.8%) | 0.366 |
| P73 | 21 (52.5%) | 6 (46.2%) | 0.691 |
| P14 | 15 (37.5%) | 3 (23.1%) | 0.340 |
P-value > 0.05 is not significant.
There is no significant difference between IFN responders and IFN non-responders regarding the methylation index (2.76 ± 1.4 and 2.83 ± 1.46; p = 0.851) respectively as well as no significant difference in methylation index between mild fibrosis (F1 and F2) and marked fibrosis (F3 and F4) (Table 5).
DiscussionAberrant methylation in the promoter regions of tumor suppressor genes (TSGs) is a crucial epigenetic alterations that contribute to deregulation of many cellular processes leading finally to the initiation and progression of human cancers.9,10 Several studies revealed that different types of cancer, including HCC, show distinct DNA methylation profiles; suggesting the existence of cancer- type specific methylation signatures.11,12 Other studies have mentioned a possible HCV- induced HCC methylation profile.6,8 Such epigenetic defects have also been observed in non-cancerous liver tissues of HCC patients, which are usually show evidence of chronic inflammation.13,14
Though Egypt has the highest prevalence of HCV infection in the world, DNA methylation profiles for HCV has not been well studied yet and there are only few studies in this context. In an early case control study by our group,7 we were able to detect a high frequency of APC, FHIT, p15, p16 and E-cadherin-PM (range 67.9-89.2%) in the plasma and tissues of 28 chronic HCV and/or HBV- associated HCC patients, with a high concordance for all studied genes. However, no significant association was found, in this study, between the methylation status of any gene and the presence of hepatitis virus infection. This was partially attributed to the small sample size in this study. Then, we assessed the contribution of methylation status to the development and progression of HCV- associated HCC and CH in Egyptian patients using a specific panel of genes (APC, FHIT, p15, p73, p14, p16, DAPK1, CDH1, RARb, RASSF1A, O6MGMT).8 We found that HCV infection may contribute to hepatocarcinogenesis through enhancing PM of certain genes. A panel of 4 genes (APC, p73, p14, O6MGMT) out of 11 tested genes successfully classified cases into HCC or CH with high accuracy (89.9%), sensitivity (83.9%) and specificity (94.7%).
A more extended confirmatory study, including 516 Egyptian patients with HCV-related liver disease (208 HCC, 108 liver cirrhosis, 100 CHC and 100 controls), was then performed to detect PM of P14, P15, P73 and Mismatch repair gene (O6MGMT) in patient’s plasma by using EpiTect Methyl qPCR Array technology.15 This study provided evidence that PM of the studied genes is an early event in hepatocarcinogenesis and showed specific DNA methylation signatures associated with the potential clinical applications in diagnosis and prognosis of HCC.15
The current study was then conducted to determine the impact of PM of our specified panel of genes on the degree of fibrosis in chronic HCV infected patients and their response to combined antiviral therapy.
Our results regarding the correlation between PM of the tested genes and patients’ response to antiviral therapy differ from previously published data in Western countries or USA. We found that only O6MGMT PM significantly affected patients’ response to antiviral therapy (p value, 0.045), being significantly higher in non-responders than in responders. This is explainable since some previous studies have shown that; 06MGM plays an important role in cytoprotection through preventing DNA damage and triggering DNA repair mechanisms. Therefore, PM of 06MGM is frequently detected in chronic hepatitis patients.16
On the other hand, PM of DAPK showed a tendency to affect patients’ response to treatment though this did not reach a statistically significant level (p = 0.097). This could be attributed, at least partially, to the small sample size. None of the other tested genes was significantly associated with response to antiviral treatment.
In the current study 06MGMT has the highest pretreatment PM frequency among HCV infected patients (64.2%) followed by P73 (50.9%), APC (49.1%), RASSF1A/DAP-kinase (41.5%) and P14 (34%). Our data in this regard is different from what has been previously reported by our group in chronic HCV and/or HBV-associated HCC)8 or by Gioia, et al.17 These two studies showed that PM of the RASSF1A gene was the most frequently detected with progression from regenerative conditions to cirrhosis. The variability in results of different studies regarding the frequency of PM of the assessed genes could be attributed to several factors including: the differences in the CpG sites tested, environmental factors, HCV genotype present as well as geographical and racial differences. However, some previous reports have also shown significant association between O6MGMT-PM and HCV infection including the study of Matsukura, et al.18
The p73 is the second most frequently methylated gene in our tested group. Data regarding PM of the p73 gene in chronic active hepatitis patients are still immature and the few available reports in literature show a low PM frequency in CH patients.19 This contradicts with our results since PM of the p73 was detected in 50.9% of the studied patients.
Data regarding PM of the APC gene varied significantly in different studies. In the current study, APC-PM was relatively high in CH patients (49.1%), confirming the data obtained from our previous study on the Egyptian population.8 However, Nomoto, et al.20reported a much lower frequency of APC- PM (21.6%) in CH patients with cirrhosis. One possible explanation for the difference between the results of the two studies could be attributed either to different HCV genotypes, the presence of fibrosis and/or environmental factors in Egyptian population.
Within our studied panel of genes, p14 showed the lowest frequency of PM among HCV patients (34%). Similar findings were reported by Anzola,21 who showed that p14 PM was associated with the pathogenesis of HCC and suggested that inactivation of p14 through PM could be an important mechanism for HCV-induced HCC.8,21 To the best of our knowledge, data regarding the impact of PM on the response to antiviral therapy in HCV-associated CH patients are still preliminary. Thus, further studies including larger number of patients are still needed to evaluate and validate the already available small studies including ours.
Only few studies have addressed the association between gene methylation status and the development of fibrosis in hepatitis patients. Murphy, et al.22 confirmed in their study the implication of methylation status of some genes in the progression of mild NAFLD (Non-alcoholic fatty liver disease) into advanced NAFLD, steato-hepatitis, fibrosis and carcinogenesis. Their study confirmed the presence of functionally relevant differences in the methylation patterns, which can distinguish a mild from an advanced disease. However no similar studies have been done in chronic hepatitis patients, except for the current study, which shows that only PM of the RASSF1A gene was significantly associated with mild fibrosis in the studied patients (p = 0.0.019). This provides an evidence for the role of an intact RASSF1A gene in the induction of fibrogenesis in chronic HCV patients.
We conclude that, promoter methylation of some genes increases in HCV genotype 4-associated chronic active hepatitis. However, only O6MGMT methylation can significantly affect patients’ response to antiviral treatment, whereas RASSF1A is involved in the regulating the process of fibrogenesis and therefore, it could be helpful in predicting the stage of fibrosis or in the differentiation between mild and marked fibrosis in those patients.
AcknowledgmentWe would like to thank Prof. Waleed M. Sief, professor of Virology and Immunology, Cancer Biology Department, National Cancer Institute for helping in statistical analyses of the data.
Abbreviations- •
AFP: Alfa-Fetoprotein.
- •
ALT: Alanine aminotransferase.
- •
BMI: body mass index.
- •
GGT: gamma glutamyltransferase.
- •
HCC: hepatocellular carcinoma.
- •
HCV: hepatitis C virus.
- •
HCVG4: hepatitis C virus genotype 4.
- •
M: methylated DNA.
- •
MI: methylation index.
- •
PMF: The Promotor methylation frequency.
- •
SVR: Sustained virological response.
- •
TSCs: tumor suppressor genes.
- •
UM: unmethylated DNA.