In the present study we have analyzed the viability and metabolic competence of isolated rat hepatocytes subjected first, to subzero nonfreezing storage (up to 120 h at -4 °C) in modified University of Wisconsin (UW) solution with 8% 1,4-butanediol, and then to a normothermic rewarming step (KHR media, 37 °C, up to 120 min, carbogen atmosphere). Results were compared with hepatocytes stored up to 120 h at 0°C in modified UW solution and with freshly isolated hepatic cells. We have found that only cell suspensions stored in subzero nonfreezing conditions were able to finish the rewarming period with a viability comparable with the control group. Also, we have investigated the enzyme activities and the relative expression at messenger RNAs levels of two of the Urea cycle (UC) enzymes: Carbamyl phosphate synthetase I (CPSI) and ornithine transcarbamylase (OTC), during 60 min of rewarming. Results were compared with the ammonium removal efficiency of the three groups.
In conclusion: These data indicated that hepatocytes preserved under cold or subzero conditions up to 120 h followed by 60 min of rewarming, maintain UC enzymes at levels similar to freshly isolated hepatocytes, allowing their use in bioartificial liver devices.
In the last years, the demand of isolated hepatocytes for cellular transplant and bioartificial liver devices has increased. To meet these requirements, various preservation techniques have been investigated. Hypothermic cold preservation in University of Wisconsin (UW) solution1 is a widespread and well-accepted method for maintaining isolated rat hepatocytes, but only for a period of time shorter than 72 h. Cryopreservation has been suggested as the best technique for long term storage of liver cells, however, cell recovery and viability post-preservation is insufficient.2 We previously described a subzero nonfreezing storage protocol that uses UW solution and 8% (W/V) 1,4-butanediol (BDL) as cryoprotectant agent. With this method we could maintained viable and functional isolated rat hepatocytes at -4 °C for up to 120 h, without damage due to ice crystal formation.3
It is important to distinguish between responses which the cell establishes during cooling, and those which depend upon cold storage but can only be detected after return to normothermic temperatures during the rewarming process (KHR media, 37 °C, up to 120 min, carbogen atmosphere). Due to this fact it is necessary to evaluate the viability and metabolic functions of the cold subzero stored hepatocytes during this rewarming process.
The ammonium detoxification is a specific function of hepatocytes that has important influence on the prevention of hepatic encephalopathy in patients with acute or chronic liver failure.4 The urea cycle (UC) is the metabolic pathway responsible for the ammonium removal. Carbamyl phosphate synthetase I (CPSI; EC 6.3.4.16) and Ornithine transcarbamylase (OTC) catalyze the first and second committed steps of waste nitrogen metabolism in the urea cycle, respectively. In a previous work,5 we have investigated the protein activities and the relative messenger RNAs expression levels of CPSI and OTC, in isolated hepatocytes preserved up to 120 h in UW solution at 0 °C, and during the rewarming step.
There are only few reports on the effects of cold storage on gene expression.6 In the present study we evaluate whether the subzero nonfreezing storage protocol developed in our laboratory could -1-To sensitize the hepatocytes viability and metabolic functions during the rewarming period, and/or -2-To modify the gene expression of the urea enzymes CPSI and OTC during this step. The Urea cycle enzymes were chosen due to its importance in the maintenance of a transcendental liver function, such as ammonia control and the fact that is a function that it is necessary to maintain in any artificial liver device.
Experimental proceduresAnimals. Male Wistar rats weighing 250-300 g were used in all experiments. Rats were allowed free access to standard laboratory diet and water ad libitum prior to the experiment and received care in compliance with international regulations. The National Council Committee of Argentine approved animal protocols.
Hepatocyte isolation. Rat hepatocytes were isolated by collagenase perfusion as it was described previously.345 Cell viability was tested by the exclusion of 0.4% trypan blue (TBE) in phosphate-buffered saline. Preparations with a TBE greater than 85% were considered suitable for the experiments.
Hepatocyte cold storage and rewarming. Isolated hepatocytes were rinsed twice and resuspended in freshly prepared cold (0°C) modified UW solution. The composition of the modified UW solution was previously described.7 Hepatocytes (120.106 cells in 40 mL UW solution) were allowed to settle to the bottom of a 50 mL screw cup polycarbonate tubes, and left undisturbed at 0°C up to 120 h. After that, the suspensions were washed twice with a rinse solution and sedimented (50 g, 3 min) in warm Krebs-Henseleit resuspension (KHR) media. The hepatocytes were subsequently incubated (120 min, 37 °C, 2-3×7106 cells/mL) in KHR media under carbogen atmosphere in a Dubnoff metabolic shaker.
Hepatocyte subzero nonfreezing preservation and rewarming. Freshly isolated hepatocytes were subzero nonfreezing preserved up to 120 h in modified UW solution with 8 % 1,4-BDL (120.106 cells in 40 mL UW solution + 8% 1,4-BDL), as previously described.3 After that cell suspensions were warmed to 0°C and then were washed once with the rinse solution containing 4% of 1,4-BDL and later, twice with rinse solution with no additive. Then, the hepatocytes were incubated (120 min, 37 °C, 2-3×106 cells/mL) in KHR media under carbogen atmosphere in a Dubnoff metabolic shaker.
Experimental protocol. Hepatocytes from Wistar rats isolated by collagenase digestion were resuspended in UW solution and divided into the following two groups: 1-subzero nonfreezing stored group (Subzero Storage), and 2-cold stored group (Cold Storage). They were stored for up to 120 h at the temperatures indicated. After that, they were rewarmed for 120 min in KHS media. Suspensions of freshly isolated hepatocytes were used as control. Aliquots of the suspensions were removed at 0, 60, and 120 min to evaluate the viability and metabolic function.
Cell viability measurementsLDH retention. The capacity of the cells to retain macromolecules as Lactate Dehydrogenase (LDH) was determined by measuring the LDH activity in the cell suspension (total activity) and in the supernatant (extracellular LDH).8,9 Results were expressed as the percentage of LDH retention by the cells.
Cell viability % = 100 - [100 × (extracellular LDH, total activity)]
PIassay. The capacity of the cells to exclude the fluorescent marker propidium iodide (PI) was established as follows: the cells were incubated with the fluorescent marker and the fluorescence intensity is measured. As previously described,10 cell viability is expressed as the ratio between the fluorescence originated from non-viable cells that have membrane damage and the fluorescence originated from all cells in the sample.
Evolution of cellular metabolites during the rewarming periodGlutathione assay. Total glutathione-GSH plus GSSG-concentration (GSHt) was determined by the enzyme coupled spectrophotometric assay of Tietze,11 as previously described.7 Results were expressed as nmol of GSH/106 cells.
ATP assay. ATP concentration was determined by an HPLC technique, as previously described.12 Results were expressed as nmol of ATP/106 cells.
Glycogen assay. Intracellular glycogen concentration was assessed by the amyloglucosidase enzyme assay, followed by an enzymatic determination of released glucose, technique described by Carr and Neff.13 Results were expressed as glycosidic units/106 cells.
MTT assay. The tetrazolium dye 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) is widely used to assess the viability and/or the metabolic state of cells. This colorimetric assay is based on the conversion of the yellow tetrazolium bromide (MTT) to the red formazan derivative by mitochondrial succinate dehydrogenase in viable cells. MTT reduction capacity of the suspensions was assessed as was previously described.14 Results were expressed as 0D/106 cells.
Determination of the ammonium removal efficiency. After the preservation period, cells were rinsed and rewarmed in KHR in the presence of ammonium chloride overload (0.2 mM final concentration). Aliquots of the suspension were removed at 0 and 60 min of the rewarming period and centrifuged (13,400g, 15 s). The supernatants were kept under liquid N2 until the enzymatic determination. Ammonium was determined enzymatically according to the method by van Anken et al.15 Ammonium removal efficiency (ARE) was calculated from the measured values of ammonium concentration as follows: ARE=[(C0-Ct)/C0]×100, where C0 is the ammonium concentration of the medium at t=0, and Ct is the ammonium concentration of the medium after 60 min of incubation. A value of 100 represents the total load - ammonium removal efficiency.16
RNA isolation, reverse transcription, and quantitative PCR. Total RNA was extracted using the Tri ReagentTM (Sigma Chem. Co. S. Louis, USA) according to the manufacturer’s instructions. Reverse transcription and quantitative PCR was made as previously described.3Table I shows the primers designed for each gene expression analysis.
Primers utilized for gene expression analysis.
| CPSI: | sense, 5’-ATC TGA GGA AGG AGC TGT CT-3’ |
| antisense, 5’-AAA ACC ACT TGT CAA TGG AT -3 | |
| OTC: | sense, 5’-ATG ACA GAT GCA GTG TTA GC-3’ |
| antisense, 5’-CAG GAT CTG GAT AGG ATG AT-3’ | |
| • -ACTIN: | sense, 5’-CAC TAT CGG CAA TGA GCG GT -3’ |
| antisense, 5’-ATT TGC GGT GCA CGA TGG A -3’ |
Determination of CPSI and OTC activities. Activities of CPSI were determined using a rapid colorimetric assay described by Pierson.17 OTC activity was measured as the rate of citrulline formation from ornithine and carbamyl phosphate.18
Statistical analysis. Statistical significance of the differences between values was assessed by analysis of variance (ANOVA) followed by Scheffe’s multiple range tests. A p value less than 0.05 were considered statistically significant.
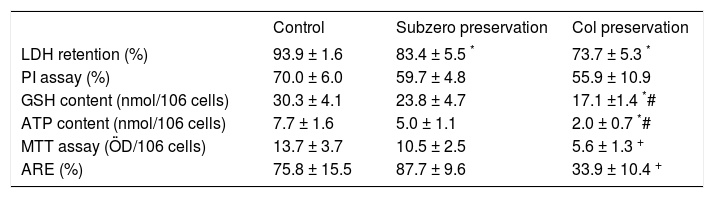
ResultsEvolution of cell viability during the rewarming periodTable II shows the cellular viability and the time course evolution of metabolites content after 120 min of the rewarming period, determined in freshly isolated and cold stored hepatocytes. We have found that at the end of rewarming period, cells suspensions subjected to subzero nonfreezing storage showed viabilities and metabolic functions comparable to freshly isolated hepatocytes. It is important to point out that, despite LDH and PI tests are based both on permeability properties of the plasma membrane; in all cases the viability levels estimated by LDH are greater than those valued from PI. We previously reported that this phenomenon could be due to the loss of extracellular LDH activity during the isolation and purification of the cell suspension.8
Cellular viability at the end of the rewarming step. Cells were stored and rewarmed as was described in Material and Methods. The cell viability was assessed by LDH, PI, GSH, ATP, MTT and ARE tests. Values are expressed as means ± SD of samples obtained from four preparations.
| Control | Subzero preservation | Col preservation | |
|---|---|---|---|
| LDH retention (%) | 93.9 ± 1.6 | 83.4 ± 5.5 * | 73.7 ± 5.3 * |
| PI assay (%) | 70.0 ± 6.0 | 59.7 ± 4.8 | 55.9 ± 10.9 |
| GSH content (nmol/106 cells) | 30.3 ± 4.1 | 23.8 ± 4.7 | 17.1 ±1.4 *# |
| ATP content (nmol/106 cells) | 7.7 ± 1.6 | 5.0 ± 1.1 | 2.0 ± 0.7 *# |
| MTT assay (ÖD/106 cells) | 13.7 ± 3.7 | 10.5 ± 2.5 | 5.6 ± 1.3 + |
| ARE (%) | 75.8 ± 15.5 | 87.7 ± 9.6 | 33.9 ± 10.4 + |
Regarding the glycogen content, we observed that freshly isolated hepatocytes began the rewarming period with the highest values (Controls: 2.19 ± 0.52, Subzero Stored: 0.15 ± 0.52, and Cold Stored: 0.09 ± 0.03 glycosidic units/106 cells). At the end of 90 min. of rewarming step, almost all the glycogen was consumed. However, the control group is the most glycogen consuming group; this behavior could be associated with a greater metabolic capacity of this suspension during the rewarming period.
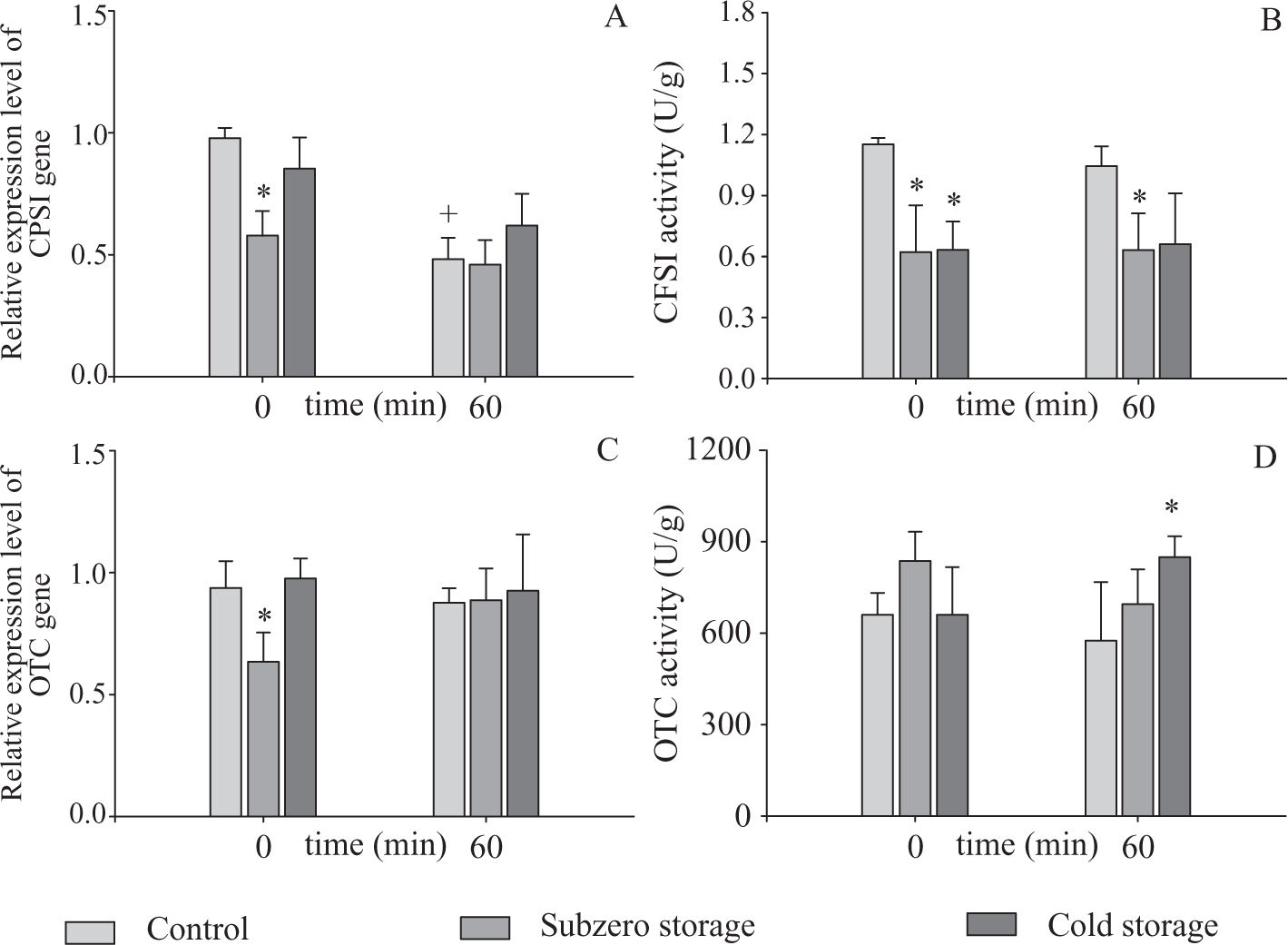
Gene expression and enzyme activity during rewarming periodWhen freshly isolated hepatocytes were rewarmed during 60 min, a diminution of approximately 50 % in CPSI transcript levels was observed. No differences were found for OTC gene (Figure 1A). These results suggest that the relative expression level of CPSI gene is susceptible to the normothermic reoxygenation period.
Relative gene expression levels and activities of CPSI (A and B) and OTC (C and D) during the rewarming step (0 and 60 min). The measurements were made in freshly isolated and subzero and cold stored hepatocytes (120 h).
Data are expressed as means ± SD for 3 hepatocyte preparations. * Statistically different from Control,+ statistically different from t = 0 min
No changes were found between control and cold preservation groups, neither for t = 0 min nor for t = 60 min (Figure 1A) and (Figure 1C). On the other hand, OTC activity was increased after 60 min of rewarming in cells that were cold preserved 120 h (Figure 1D). After 60 min of rewarming, we found differences in CPSI activity only between the control group and cell suspensions preserved in subzero conditions (Figure 1B). Despite the differences found, after 60 min of rewarming the ARE of preserved hepatocytes did not show statistical differences between them, and in comparison with control group (Controls: ARE = 41.8 ± 8.7%; Subzero Stored: ARE = 52.9 ± 18.2%; Cold Stored: ARE = 27.27 ± 7.88%, n = 4). These results show that after 120 h of preservation and 60 min of normothermic rewarming, cells preserved in both conditions maintain a capacity to detoxify ammonia similar to control hepatocytes.
DiscussionHypothermia decreases the rate of cellular energy-requiring and degradative reactions and is considered to play an important role during prolonged period of ischemia. However, the optimal level of hypothermia during hepatic preservation is controversial. Thus, as it was demonstrated in the joined paper19 subzero non-freezing temperatures could be reached with the addition of cryoprotectives agents, in this case with 1,4-butanediol.
The objective of the present study was to investigate the hypothesis that cold preservation or subzero nonfreezing storage of hepatocytes maintain the hepatocytes viability and metabolic functions during the rewarming period, and/or and if it is able to maintain the gene expression of the urea enzymes CPSI and OTC during this step.
The obtained results showed: Subzero nonfreezing storage conditions presents some incapacity to retain LDH content as compared with control studies. However, the entire metabolic and functional test (GSH, ATP and GLN content, MTT assay and the capacity to eliminate an overload of NH4+) and showed similar behaviour between controls and preserved under nonfreezing conditions. Meanwhile, “conventional” hypothermic preservation showed a wide range of results, meaning that in those conditions (120 h), we are in the limits of preservation time.
Another point that we wanted to probe in this manuscripts was if subzero nonfreezing preservation affects gene expression at mRNA level and the respective proteins activities. To probe this we choose the Urea cycle, the metabolic pathway responsible for the ammonium removal. Carbamyl phosphate synthetase I (CPSI) and Ornithine transcarbamylase (OTC) were analyzed at mRNA level and at protein level. Although hepatocytes suspension subjected to subzero nonfreezing preservation initiated the rewarming step with CPSI and OTC gene relative expression levels statistically reduced in comparison with the control group, they were able to finish this period with values comparable with freshly isolated cells.
Over the years it has been used multiple technologies to provide insights into freeze-responsive gene and protein expression by different animal species, like in example: wood frogs. Initial studies searched for freeze or thaw responsive proteins by using 35S-methionine to label proteins either in vivo after intraperitoneal injection or during in vitro translation of mRNA isolated from control versus frozen frogs. Both studies illustrated freezethaw and organ-specific differences in protein biosynthesis in wood frogs with prominent synthesis of some 15–20 kDa proteins, but both were limited the inability to identify individual proteins. Interestingly, major recent advances in proteomics technology including 2-dimensional electrophoresis coupled with LC-MS (liquid chromatography-mass spectrometry) of peptide fragments might now make a return to such studies fruitful.20
The results presented in the current study suggests that the cold subzero stored hepatocytes during 120 h. are able to maintain the viability and metabolic functions after 120 min of rewarming. Also, these data indicates that the hepatocytes cold subzero stored and rewarmed, maintain UC enzymes at levels similar to freshly isolated hepatocytes.