Small RNA molecules such as microRNAs, for many years considered to be superfluous genomic material, are now known to play important regulatory roles in apoptosis, cell proliferation and differentiation, angiogenesis and thus in carcinogenesis. Primary liver carcinomas such as hepatocellular carcinomas, cholangiocarcinomas and mixed variants show a rising incidence with high mortality among affected patients but lack effective targeted therapies except the new multiple kinase inhibitor Sorafenib. This review elucidates the recent contributions of miRNA gene expression analyses to a better understanding of the complex molecular interactions in liver carcinogenesis and highlights their future promise to provide novel tools for improved diagnostics, more accurate prognostic assessment and tailored molecular therapies in liver cancer.
«A small rock holds back a great wave» [Homer, The Odyssey, 800 B.C.-700 B.C.]
IntroductionMicroRNAsMicroRNAs (miRNAs) have taken center stage in cancer research during the short span of the last 6 years since they have been shown to be functional in humans.1-7 They represent one of the most abundant classes of regulatory genes in mammals and likely influence over one third of all proteinencoding human genes.8-12 MiRNAs themselves do not encode proteins but rather function by targeting specific messenger RNAs for degradation or translational inhibition and thus decrease the expression of the resulting protein.13,14 Although only about 550 miRNAs have been identified thus far, estimates about their total number exceed 1,000, and they may comprise approximately 1-3% of the currently known genes in the human genome.12,15,16 Many miRNAs are expressed exclusively or preferentially in certain tissue types, e.g. miR-122 in liver parenchyma.16-21
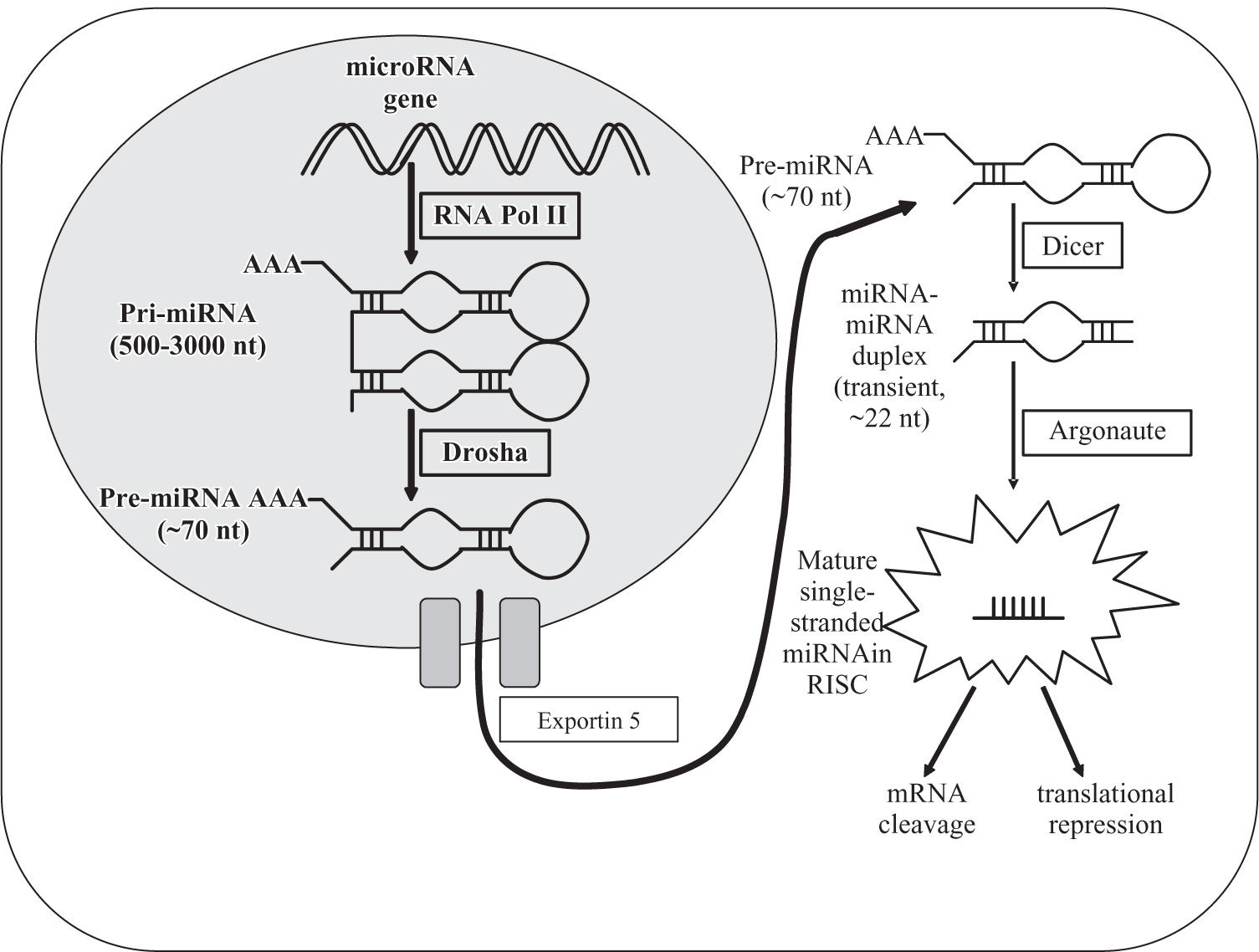
miRNA biosynthesis and regulationIn 1993, miRNAs were first identified through the discovery that the 22-nucleotide RNA lin-4 is important for the exact timing of post-embryonic development in the nematode Caenorhabditis elegans.22 MiRNAs are a class of small (~19-22 nt) endogenous RNAs that develop in a step-wise process through hairpin precursors cleaved by the dsRNA- specific endonucleases Drosha, Dicer and Argonaute (Figure 1).9 MiRNAs interact with target messenger RNA at specific sites to induce cleavage or inhibit translation.1-12 While complementarity with targets is often perfect in plants, it commonly involves bulges and loops in animals and humans, thus complicating the identification of putative targets.12 As a result, the specific function of most mammalian miRNAs is still unknown.12,13 A single miRNA can bind to and regulate many different target messenger RNAs, and conversely, many different miRNAs can influence each single messenger RNA.9,12 Newer data demonstrate that a large number of miRNAs are transcribed but are not processed to the mature miRNAs.23 MiRNA genes are frequently located at chromosomal fragile sites and regions of loss of heterozygosity.13 MiRNA expression might be regulated at multiple steps of RNA biogenesis, although it remains unknown how this control is achieved.10 In general, it is thought that miRNA function is adjusted by alteration of their processing enzymes, promoter hypermethylation or loss of miRNA binding sites of their target genes.24
MicroRNA biosynthesis in the hepatocyte. Precursor microRNA is initially cropped into the characteristic hairpin structure in the nucleus by the enzymes RNA Polymerase II and Drosha, then transferred to the cytoplasm by Exportin-5 and further processed by Dicer and Argonaute into the mature short single-stranded micro RNA, which exerts its regulating function on the appropriate messenger RNA after incorporation into the RISC (RNA-induced silencing complex).
Primary liver cancer consists in 80-90% of hepatocellular carcinoma (HCC) and much less frequently of cholangiocarcinoma (intrahepatic or hilar, so-called Klatskin tumor) and rarely of mixed hepatocellular-cholangiocarcinoma, which may derive from hepatic stem cells such as oval cells or hepatogones.25,26 HCC is the fifth most common cancer worldwide, accounts for approximately 500,000 deaths annually and continues to increase in incidence despite vaccination against Hepatitis B virus (HBV).26-29 HCC often develops in the setting of liver cirrhosis caused by its main risk factors, HBV or Hepatitis C Virus (HCV) infection, excessive alcohol consumption or hemochromatosis.26,30,31 It is thought that most HCCs develop through a progressive pathway from premalignant nodular lesions such as dysplastic nodules.32 Remarkably, even after studying thousands of HCCs, only limited knowledge has been gathered regarding genomic alterations during the development and progression of HCCs in humans.26,28 Overall, the molecular mechanisms of hepatocarcinogenesis are still poorly understood.28 Pathways and molecules that have been identified to play crucial roles in hepatic cancers are cell cycle regulatory proteins such as p53, c-Myc, Cyclin D1, the Wnt/***entity***catenin signalling pathway, and multiple tyrosine kinase growth factor ligands and receptors, including epidermal growth factor, hepatocyte growth factor, fibroblast growth factor and vascular endothelial growth factor.33,34 In contrast, cholangiocarcinomas often occur in the absence of liver cirrhosis.35-38 They have a poor prognosis with a median survival of only 13 months.39 Since they are highly chemoresistant, the need for improved treatment options is particularly urgent for cholangiocarcinomas.37 The mechanisms regulating cholangiocarcinoma growth and resistance to chemotherapy are poorly understood.37 Identification of new target molecules that are critically involved in the development of primary liver carcinomas and dysregulated specifically in tumors will be essential to understand the mechanisms and improve prognostication and therapeutic intervention of hepatic cancers.
microRNAs in human cancerWe are still in the early stages of understanding the intricacies of the miRNA puzzle that contributes to disease development in humans.40-42 Some miRNAs, similar to messenger RNAs, are expressed in a tissue-specific manner, and human adult tissues have unique miRNA profiles.43,44 Cancer is a complex genetic disease involving structural and expression abnormalities of both coding and noncoding genes.43,45 MiRNAs interact with classic oncogene and tumor suppressor networks and thereby contribute to the initiation and progression of many if not all human malignancies.41,43-46 MiRNAs that are downregulated in cancer and target oncogenes act as tumor suppressors, while miRNAs that are upregulated in cancer and target tumor suppressor genes act as oncogenes.41,43-47 Several individual miRNAs stand out and have been implicated in the development of human malignancies, for example miR-145 in carcinoma originating in the colon, breast, lung or prostate,4,48-52 miR-15a and miR-16-1 in chronic lymphocytic leukemia, mir-221 in papillary thyroid carcinoma,53 the miR-17-92 polycistronic cluster in lung carcinoma11 and miR-21 in glioblastoma.54 Several of the abnormally expressed miRNAs in human cancers target transcripts of protein-encoding genes well-known to be involved in carcinogenesis, such as the BCL-2 anti-apoptotic gene by the miR-15a/miR- 16-1 cluster, the Ras oncogenes by let-7 family members, the E2F1 transcription factor by the miR-17-92 cluster or the BCL-6 anti-apoptotic gene by miR-127.2,11,55 Although the analysis of single miRNAs provides a focussed understanding of their influence on known molecular pathways, the real power of miRNA research lies in large-scale miRNA expression fingerprints of many hundreds of miRNAs in tumors of varying etiologies. Recently, studies have emerged directly implicating miRNAs in cancer and thus giving rise to a new molecular taxonomy of human cancers based on miRNA profiling.5,44 A comprehensive analysis of the miRNA expression in diverse neoplasms showed an even higher accuracy for tumor diagnosis using the miRNA genetic fingerprint than using a profile of more than 16,000 messenger RNAs.56 Thus, miRNA profiles are excellently suited for the classification and diagnosis of human malignancies.4,43,47 In addition, the prognosis of patients with certain carcinomas can be determined using miRNA profiling.52,57 Further, miRNA signatures have been shown to correlate with the degree of histological tumor differentiation in HCCs58 or with specific pathologic features such as estrogen and progesterone receptor expression in breast carcinomas.50 In the future, miRNA profiles could potentially aid in determining the primary site of a tumor or metastasis of unknown origin, providing a unique opportunity for targeted therapy and eliminating the use of empiric chemotherapy.59 Since miRNA research has progressed rapidly over the past several years, a promising future for miRNAs in the realm of cancer diagnostics seems likely.4
microRNAs in hepatocellular carcinomamiRNAs in the non-neoplastic liver and in premalignant liver lesionsHepatocellular tumor development is thought to develop in a multi-step process requiring the accumulation of several structural and genomic alterations and affecting many different pathways.28,59 It has been suggested that many of the miRNA changes that occur during hepatocarcinogenesis do so early, so that many changes that predispose to HCC have already taken place in liver cirrhosis and other premalignant lesions.33 Subsequent changes in the miRNA expression in the transition from cirrhosis to HCC seem to be much less marked.33 A progressive downregulation of miR-145 and miR-198 from cirrhotic tissue to dysplastic nodules and further to HCCs of increasing histological grades has been observed.60 The fact that abnormal miRNA expression patterns are already present in premalignant lesions has also been shown for other organ sites such as miR-143 and miR-145, which are downregulated in colonic adenomas as well as adenocarcinomas51 and miR-221, which is upregulated in papillary thyroid carcinomas and also in peritumoral thyroid parenchyma.53 Changes of miRNA patterns have been demonstrated to occur before tumor formation in a HCC-model of rats exposed to tamoxifen.61 Therefore, it remains a tantalizing possibility that miRNAs could serve as early warning markers for cancer initiation or progression.4,43,44
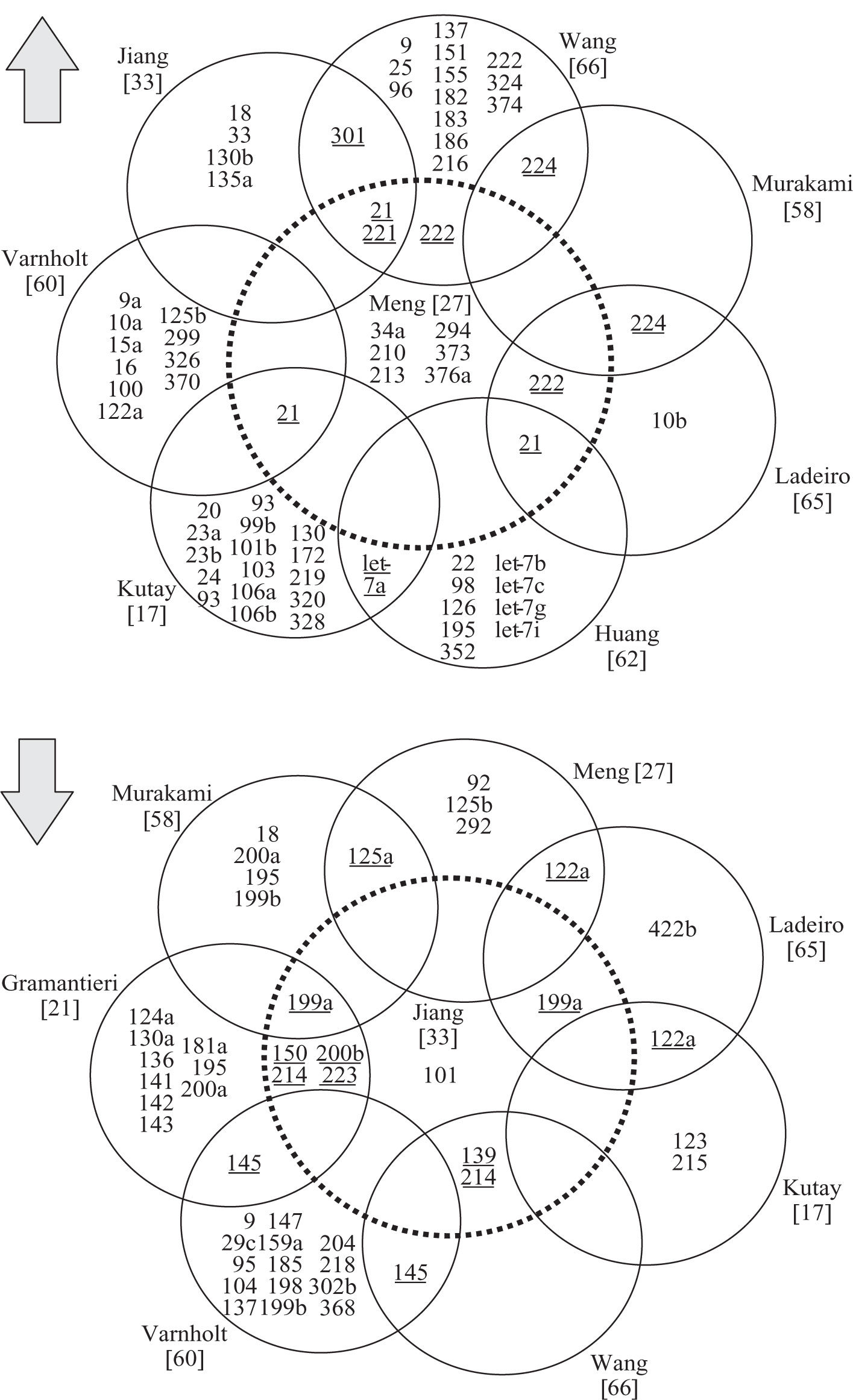
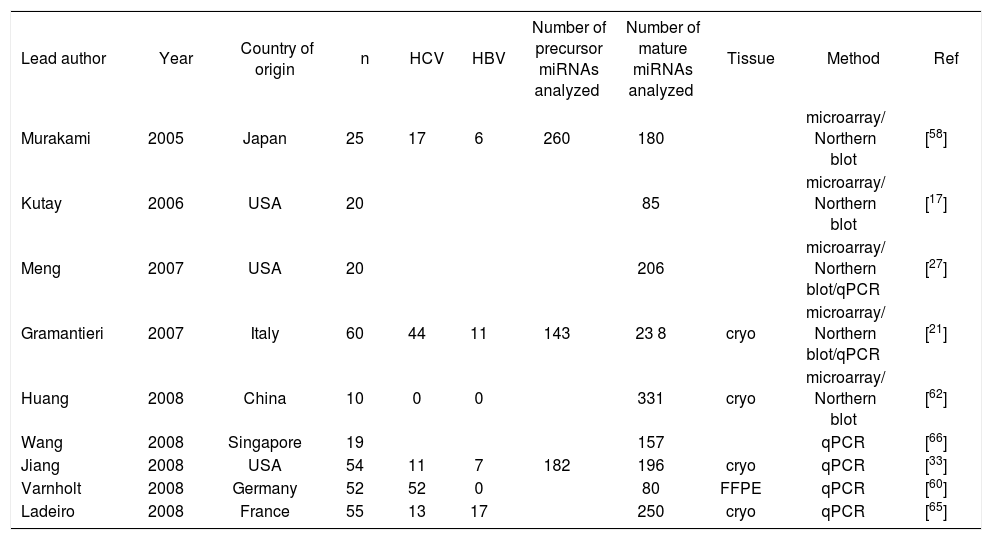
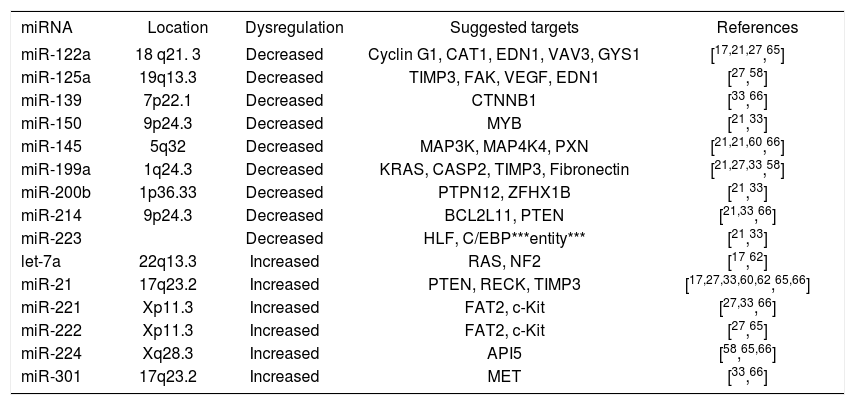
miRNAs in hepatocellular carcinomaSince the first publication of a miRNA gene expression profile by Murakami et al. in 2005, a total of 9 comprehensive miRNA analyses in HCCs have been reported thus far (Table I). Geographical origins of patients included those from the USA, Italy, France, Germany, Japan, Singapore and China. While the predisposing risk factors and etiologies of HCCs in these studies were quite inhomogenous, one study selectively included a large number of only HCV-infected patients60 and other authors limited their samples to those that were not infected with hepatotropic viruses.62 Over the past few years, a trend of methodologies from microarray platforms coupled with Northern blot confirmation toward widespread application of real-time quantitative PCR is evident (Table I). Although it has been shown that formalin-fixed paraffin-embedded (FFPE) tissue can be reliably be used for miRNA expression analyses,18,63 most studies collected data from snap-frozen liver tumor material with the exception of one study that utilized FFPE samples.60 Many hundred precursor and mature miRNAs have been examined, but only limited overlap exists between the results of upregulated (Figure 2a) and downregulated (Figure 2b) miRNAs in HCCs. Reasons for the diverse miRNA gene signatures could be variations in methodologies or in the sample origin spanning different geographical regions and ethnic groups. The overexpressed miRNAs let-7a, miR-21, miR-221, miR-222, miR-224, miR-301 and the underexpressed miR-122a, miR-125a, miR-139, miR- 145, miR-150, miR-199a, miR-200b, miR-214, miR-223 have been found to be dysregulated by more than one group of authors and are thus more likely to be of significance in hepatocellular carcinogenesis. These miRNAs are further characterized in Table II.
Characteristics of published microRNA gene expression analyses in hepatocellular carcinoma.
| Lead author | Year | Country of origin | n | HCV | HBV | Number of precursor miRNAs analyzed | Number of mature miRNAs analyzed | Tissue | Method | Ref |
|---|---|---|---|---|---|---|---|---|---|---|
| Murakami | 2005 | Japan | 25 | 17 | 6 | 260 | 180 | microarray/ Northern blot | [58] | |
| Kutay | 2006 | USA | 20 | 85 | microarray/ Northern blot | [17] | ||||
| Meng | 2007 | USA | 20 | 206 | microarray/ Northern blot/qPCR | [27] | ||||
| Gramantieri | 2007 | Italy | 60 | 44 | 11 | 143 | 23 8 | cryo | microarray/ Northern blot/qPCR | [21] |
| Huang | 2008 | China | 10 | 0 | 0 | 331 | cryo | microarray/ Northern blot | [62] | |
| Wang | 2008 | Singapore | 19 | 157 | qPCR | [66] | ||||
| Jiang | 2008 | USA | 54 | 11 | 7 | 182 | 196 | cryo | qPCR | [33] |
| Varnholt | 2008 | Germany | 52 | 52 | 0 | 80 | FFPE | qPCR | [60] | |
| Ladeiro | 2008 | France | 55 | 13 | 17 | 250 | cryo | qPCR | [65] |
cryo=snap-frozen; FFPE: formalin-fixed paraffin-embedded; qPCR: quantitative real-time PCR; Ref: Reference
Frequently dysregulated microRNAs in hepatocellular carcinomas.
| miRNA | Location | Dysregulation | Suggested targets | References |
|---|---|---|---|---|
| miR-122a | 18 q21. 3 | Decreased | Cyclin G1, CAT1, EDN1, VAV3, GYS1 | [17,21,27,65] |
| miR-125a | 19q13.3 | Decreased | TIMP3, FAK, VEGF, EDN1 | [27,58] |
| miR-139 | 7p22.1 | Decreased | CTNNB1 | [33,66] |
| miR-150 | 9p24.3 | Decreased | MYB | [21,33] |
| miR-145 | 5q32 | Decreased | MAP3K, MAP4K4, PXN | [21,21,60,66] |
| miR-199a | 1q24.3 | Decreased | KRAS, CASP2, TIMP3, Fibronectin | [21,27,33,58] |
| miR-200b | 1p36.33 | Decreased | PTPN12, ZFHX1B | [21,33] |
| miR-214 | 9p24.3 | Decreased | BCL2L11, PTEN | [21,33,66] |
| miR-223 | Decreased | HLF, C/EBP***entity*** | [21,33] | |
| let-7a | 22q13.3 | Increased | RAS, NF2 | [17,62] |
| miR-21 | 17q23.2 | Increased | PTEN, RECK, TIMP3 | [17,27,33,60,62,65,66] |
| miR-221 | Xp11.3 | Increased | FAT2, c-Kit | [27,33,66] |
| miR-222 | Xp11.3 | Increased | FAT2, c-Kit | [27,65] |
| miR-224 | Xq28.3 | Increased | API5 | [58,65,66] |
| miR-301 | 17q23.2 | Increased | MET | [33,66] |
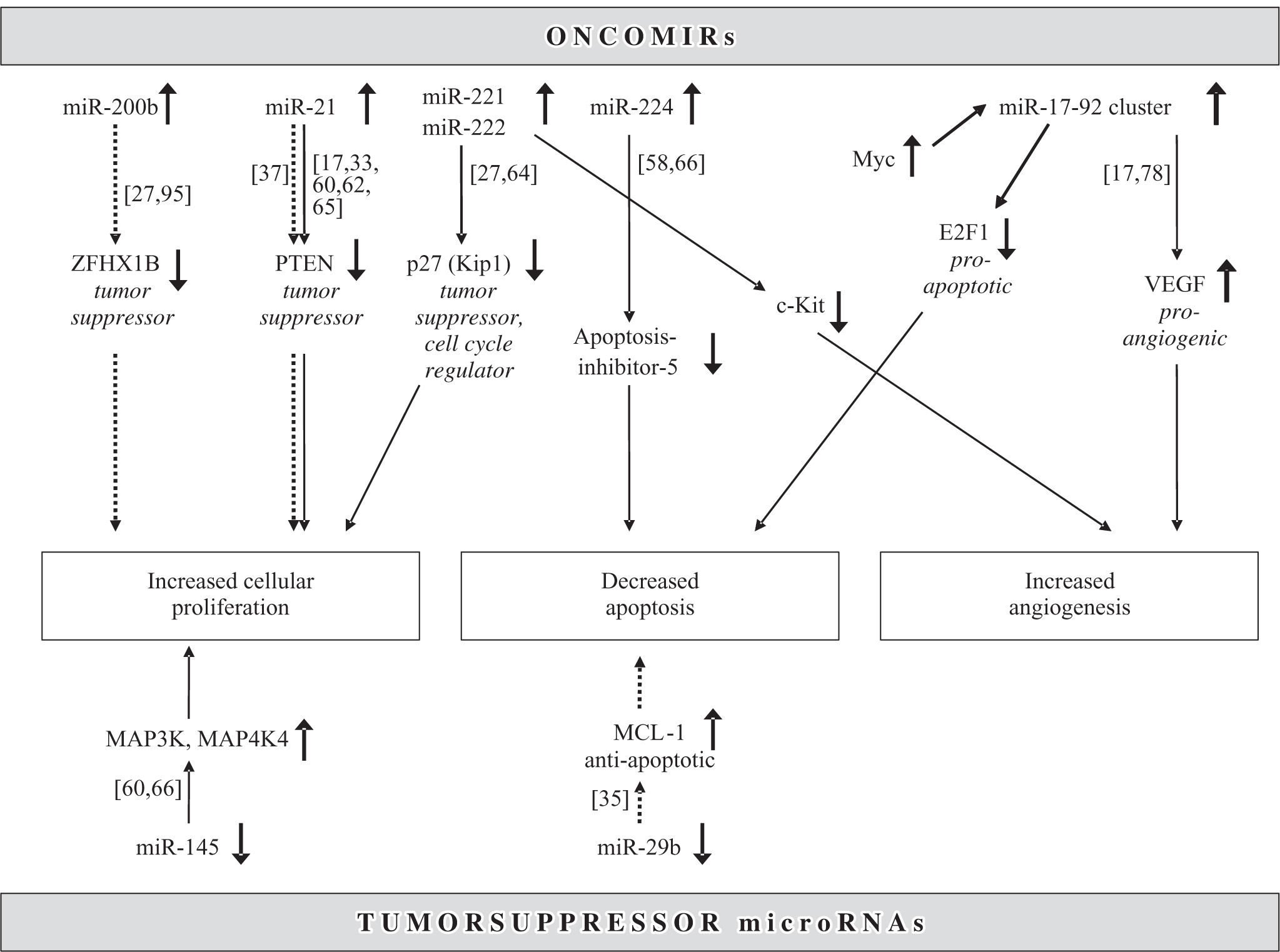
Mir-221 and miR-222 are encoded in tandem on the X-chromosome (Table II) and, since their overexpression directly results in upregulation of the tumor suppressor and cell cycle regulator p27(Kip1) (Figure 3), they can be viewed as a new family of oncogenes targeting p27(Kip1).64 Not only are both miR-221 and miR-222 significantly overexpressed in HCCs when compared to benign liver tumors and non-tumorous liver tissues,21,27,65 but miR-221 is also part of a gene signature that significantly correlates with HCC outcome.33 Another miRNA that constitutes this prognostically important miRNA gene signature is miR-100. It is likewise upregulated in HCCs and thus acts as an oncomiR.60 MiR-100 has also been found to be aberrantly expressed in breast, lung and ovarian cancer, is located on chromosome 11q23-q24-D, and has yet to be identified targets.13 Since Murakami et al. first showed that miR-224 is upregulated in HCCs,58 recent studies demonstrated that this miRNA is also upregulated in benign tumors such as liver adenomas and focal nodular hyperplasia, albeit to a lesser degree.65 MiR-224 targets apoptosis inhibitor-5 (API-5), and the expression of these two molecules are inversely proportional (Figure 3).66 Additional miRNAs overexpressed in HCCs are let7a17,62 and miR-301, which is also increased in pancreatic cancer.33,67 A frequently aberrant miRNA that was found to be upregulated in HCCs in seven different studies was miR-21.17,27,33,60,62,65,66 Both let-7 and miR-21 are also highly overexpressed in malignant cholangiocytes and the latter increases the sensitivity of human cholangiocarcinoma cell lines to the chemotherapeutic agent gemcitabine.37 In addition, mir-21 has been shown to be upregulated in glioblastoma54 and carcinoma from the pancreas,68,69 breast,50 stomach,68 thyroid,70 colon71 and prostate.68 In colonic adenocarcinoma, miR- 21 expression is significantly associated with lymph node positivity and distant metastases, and is a prognostic factor independent of TNM stage.72,73 Meng et al. have demonstrated that miR-21 targets the tumor suppressor gene PTEN (Figure 3), which is a key contributor to HCC pathogenesis and growth, and its protein product is frequently absent in HCCs.27 A polycistron named the miR-17-92 cluster, which comprises seven miRNAs and resides in the intron 2 of the C13orf25 gene at chromosome 13q31.3, is upregulated in rodent and human HCCs.11,17,33 Its overexpression results from transcription activation by c-Myc after direct binding to the genomic locus encoding the miR-17-92 cluster (Figure 3).74-76 It is known that miRNAs from the miR- 17-92 cluster act as oncogenes by influencing the translation of the E2F1 messenger RNA.77 In addition, since miR-17-92-transduced cells form larger, better perfused tumors and the Myc proto-oncogene sustains vascular endothelial growth factor (VEGF) production, it has been suggested that the miR-17-92 cluster exerts a proangiogenic effect.78 Mir-10b is another miRNA upregulated in HCCs compared to non-tumorous liver parenchyma.65 It promotes cell migration and invasion and has been found in breast carcinomas to be associated particularly with those that metastasize widely.79 Whether this pro-metastatic miRNA also leads to early or more frequent metastases in HCCs remains to be elucidated.65 Recently, first results have been published linking certain miRNAs to different etiologies and risk factors in HCCs. For example, the overexpression of miR-96 was associated with HCCs arising solely in the setting of HBV infection.65
Regulation of common molecular pathways in hepatocarcinogenesis by microRNAs. Upregulated (upper half) and downregulated (lower half) microRNAs act as oncomirs and tumor suppressors, respectively. Solid line: dysregulated in hepatocellular carcinoma, dotted line: dysregulated in cholangiocarcinoma.
Downregulation of miR-126 was strikingly associated with HCCs caused by excessive alcohol consumption but not those arising due to other hepatotoxic agents or hepatotropic viruses.65 This miRNA acts as a tumor suppressor not only in HCCs but also in breast cancers, particularly highly metastatic ones.80 Another miRNA that has been found to be dysregulated in HuH7 hepatic cancer cell lines,48 human HCCs21,60,66 as well as carcinomas from other organ sites such as the prostate, lung,52 colon,48,49,51 breast50 and thyroid70 is miR-145. In colorectal cancers, those more than 50 mm in maximal diameter are characterized by lower miR-145 expression than larger tumors.81 In addition, a progressive downregulation of miR-145 from normal breast parenchyma to mammary carcinomas with a high proliferation index was observed.50 Similarly, HCCs from low to high histological tumor grades show progressively decreasing expression of miR-145.60 Predicted target sites for miR-145 are MAPK transduction proteins such as MAP3K3, MAP4K4, MYCN, FOS, YES and FLI-1.48,50 Other downregulated miRNAs are miR-198,60 which is located in the 3´ untranslated region of the messenger RNA for human follistatin related protein,82 miR-199a21,27,33,58 and miR-200a, which is also underexpressed in colonic adenocarcinoma.4,58
miR-122MiR-122 plays a crucial role in the understanding of liver disease, because it is expressed exclusively in the liver where it constitutes 70% of the total miRNA content.17,20,83 While miR-122 is the most abundant miRNA in adult livers, other miRNAs such as miR-92a and miR-483 are highly expressed in fetal livers.84 The depletion of miR-122 compromises liver function and reduces cholesterol levels by targeting the expression of genes involved in cholesterol biosynthesis.85,86 In HCV infection, miR-122 facilitates viral replication by an unknown mechanism and seems to be required for efficient viral RNA expression.20 This is evident because HCV RNA can replicate in HuH7 liver carcinoma cells, which express miR-122, but not in HepG2 cells, which do not express miR-122.20 The first suggestion that miR-122 may play an important role in the development of HCCs was made by Etiemble et al. in 1989 in woodchucks.87 It took another 13 years until the human equivalent of miR-122 was discovered,88 and, since then, most studies have found miR-122 to be significantly downregulated in HCCs.17,21,27,65 However, one study reported miR-122 to be upregulated in a patient population being entirely HCV-infected.60 These differences may be related to the close interactions between miR-122 and the hepatitis C virus and remain to be elucidated in detail. Recently, Cyclin G1 has been identified as one of the targets of miR-122.21 Anti-miR-122 oligonucleotides have been shown to lead to specific, dose-dependent silencing of miR-122 without signficiant hepatotoxicity in mice and therefore promise to represent the first of a novel class of small molecules that could be used in molecular targeted therapy in liver diseases.85,89-91
miRNAs in prognosis and metastasis of hepatocellular carcinomaA miRNA gene signature consisting of 20 miRNAs that is significantly associated with venous metastases in HCCs has recently been reported.92 Determining the expression levels of these miRNAs may thus be a useful tool to classify patients with HCCs at an early stage and improve their clinical outcome.92 Dysregulation of selected miRNAs is associated with an altered response of tumors to commonly used chemotherapeutic agents. For example, miR-214 induces cell survival and cisplatin resistance through targeting the PTEN/Akt pathway93 while inhibition of miR-21 and miR-200b increases the sensitivity of cholangiocarcinoma cells to gemcitabine.37 MiRNA profiles may in the future provide information to guide oncologists in choosing a tailored therapy for individual patients.40,43,59 In combination with recently developed technologies that allow the systemic delivery of small RNA mimics or inhibitors to humans, these discoveries hold great promise for the development of innovative therapeutic strategies.85
microRNA in cholangiocarcinomaGene expression profiles of miRNAs are much less detailed in cholangiocarcinomas than in hepatocellular carcinomas. Cholangiocarcinomas are highly chemoresistant biliary malignancies with poorly understood mechanisms of growth regulation.35-39 Most studies thus far have utilized cell cultures or rodent models.35-38 In cholangiocarcinomas, upregulated oncomiRs are miR-141, miR-21, miR-23a, miR-27a, let-7a and miR-200b, while downregulated tumor suppressor miRNAs are miR-29b and miR-3 7 0.35-38,94 MiR-141 is highly overexpressed in malignant cholangiocytes and may target the CLOCK gene, which regulates circadian rhythms and can act as a tumor suppressor.37 Inhibition of miR-141 decreases cell growth of cholangiocarcinoma cells.37 Let-7a and miR-21 have been found to be overexpressed in both HCCs17,27,33,60,62,65,66 and cholangiocarcinomas.37,38 The underlying mechanism, at least in cholangiocytes, may be mediated by interleukin-6 and contribute to a constitutive phosphorylation of Stat-3 by NF2.38 The oncogenic miR-21 has already been characterized above in the section on HCCs and is likewise upregulated in cholangiocarcinoma.37,94 In summary, miR-21 targets PTEN and is an anti-apoptotic and pro-survival factor.54,94 Human cholangiocarcinoma cells are increasingly sensitive to the anti-tumor agent gemcitabine with inhibition of miR-21 and miR-200b. Interestingly, miR-200b shows inversely proportional expression levels in HCCs and cholangiocarcinomas since it is downregulated in the former21,33 and upregulated the latter.37,94 A suggested target gene for miR-200b is the PTPN12, which, if dysregulated, may contribute to tumor cell survival and carcinogenesis.37 MiR-200b has also been proposed to repress the expression of ZFHX1B, a transcription factor involved in the TGF***entity*** signalling pathway and in processes of epithelial to mesenchymal transition via regulation of E-Cadherin.95
MiR-29b expression is reduced in cholangiocarcinoma cell lines and thus acts as a tumor suppressor (Figure 3).35 It has also been found to be downregulated in chronic lymphocytic leukemia, colon cancer and breast cancer.4,8,50,52 Of note, a reduced expression of mir-29b was associated in particular with breast carcinomas that lacked estrogen and progesterone receptors and displayed an aggressive behavior.50 MiR-29b is present at a locus on chromosome 7q32, which coincides with the common fragile site FRA7H, thus possibly explaining the frequent miR-29b downregulation in a number of cancers.13 The suppression of miR-29b expression in cholangiocarcinoma cells leads to an overexpression of Mcl-1 and renders the cells resistant to cell death.35 The expression of miR-370 is reduced in malignant cholangiocytes compared to non-malignant cholangiocytes.36 Since miR-370 is also decreased in the early phase of hepatotoxicity by acetaminophen or carbon tetrachloride, it is speculated to regulate an oxidative-stress-related gene.96 MiR-370 is embedded in a CpG island and targets MAP3K8, which is consequently upregulated in cholangiocarcinoma cells lines as well as in tumor cell xenografts in vivo.36,94 Tight epigenetic regulation of miR-370 occurs by hypermethylation and through interleukin-6.36,94 Enhanced miR-370 expression suppresses growth of malignant human cholangiocytes and may therefore be suited as a novel target for molecular therapeutic strategies using small RNAs. Clearly, additional detailed investigations about the miRNA expression using human cholangiocarcinoma samples are needed in the future to elucidate the mechanisms involved in the oncogenesis of these enigmatic tumors.
Future outlookWe are now catching a first glimpse of a hopefully brighter future for patients with primary liver carcinomas than currently available. Small molecules such as microRNAs could potentially lead the way not only to early and accurate diagnoses but also to novel targeted therapeutic strategies for hepatic cancers.