Background and aim. Acetylsalicylic acid (ASA) has been shown to downregulate HCV expression; however, the involved mechanisms are unknown. We used proteomic analysis to compare protein expression profiles between human hepatocarcinoma cells (Huh7) and Huh7-HCV cells harboring expression of non-structural HCV proteins, to elucidate the mechanism(s) involved in ASA-mediated downregulation of HCV replication.
Material and methods. Both cell lines were treated or untreated with 4 mM ASA and harvested at 0, 24, 48 and 72 h to isolate total proteins, which were resolved by two-dimensional gel electrophoresis (2DE) to separate them by isoelectric point (pI), followed by fractionation by molecular weight (MW). Gels were scanned and analyzed with PD-Quest software V8.0.1, and proteins were elucidated by the specific pI and MW using TAGIDENT software. Statistics analysis included the t-test.
Results and Discussion. Different protein patterns among hepatocytes expressing HCV-proteins in ASA treated and untreated cells were found. Among proteins differentially expressed in Huh7-HCV cells, we found proteins related to cell proliferation (MTMR6, FAM22, HDGF and HCF–1) after 24 h of ASA treatment; and upregulation of angiostatin, PI4KA and STAT–1 after 48 h of treatment. Finally, at 72 h of ASA exposure, we identified overexpression of adenyl-succinate synthase, 2’–3’-di-deoxyadenosine, ubiquitin-protein-ligase E6A, adenylosuccinate-lyase and ni-brin (NBN).
Conclusion. We found that ASA induces different protein patterns in Huh7-HCV cells promoting activation of proteins involved in cell progression, repair of double strand breaks, proliferation, inhibition of apoptosis and growth stimulation at the same time that it decreased HCV expression.
Chronic HCV infection can lead to serious liver disease, such as cirrhosis and hepatocellular carcinoma (HCC). Although considerable progress has been made in recent years, the mechanism of replication and pathogenesis of HCV infection are still elusive. Hepatitis C virus (HCV), a member of the family Flaviviridae, is an enveloped virus with a single-stranded, 9.6-kb RNA genome of positive polarity. HCV affects 3% of the world population and is considered a serious public health problem. Despite notable advances in research, the mechanism responsible for the development of chronic HCV infection is not yet known.1–4
Standard treatment for HCV-infected patients consists of the combination of ribavirin and pegylated interferon alpha, and recently the use of a protease inhibitor (boceprevir or telaprevir) has been suggested, but not all treated patients have a sustained virological response.5,6 The development of new compounds against HCV has been difficult due to inappropriate culture systems and a lack of animal models. However, with the knowledge generated through models of genomic and subgenomic replicons, a rational design of agents that specifically block virus replication has become possible.3,7–10
In this regard, Trujillo, et al., employing a subge-nomic HCV-replicon system, reported that ASA decreases HCV-RNA and viral protein levels (60%) in cells harboring viral protein expression;11 however, the mechanisms involved were poorly defined. Evaluation of genes associated to HCV-disease has centered on microarray expression profiles and more recently, the use of proteomic approaches.12 Proteomic technology development has provided a powerful tool for the study of biological processes that result in the overall alteration of protein expression.13–15 By comprehensively examining differential protein expression profiles between normal, diseased, or drug-treated samples using 2-dimensional electro-phoresis (2DE) or protein chips, proteomics has been able to provide information on new biomarkers, disease-associated targets and the process of pathogenesis. Proteomic analysis has been used to determine changes in host cell protein expression related to different viral infections, cytokine treatment, and carcinogenesis.16
Based on this, we examined the differential expression of proteins in the Huh7 cell line that stably expresses nonstructural HCV proteins (Huh7 HCV-replicon cells) and the parental cell line exposed to ASA in order to elucidate the mechanism involved in ASA-mediated downregulation of HCV expression. We observed that the presence of ASA induces a differential protein pattern in ASA-treated cells promoting activation of proteins involved in cell progression, proliferation, inhibition of apoptosis, and growth stimulation at the same time that it decreased HCV expression. This study allowed us to increase our understanding of the mechanisms of HCV gene regulation.
Material and MethodsReagentsReadyStrip™ IPG strips (pH 3–10, linear, 17 cm), rehydration buffer, iodoacetamide and mineral oil were purchased from Bio-Rad (Hercules, CA). To isolate total protein we used protease inhibitors (Roche Applied Science, Mannheim, Germany). A standard calibration curve was made with bovine serum albumin (BSA) (Amresco, Solon, OH). Acrylamide and N-N’-metilbisacrilamide were purchased from Invitrogen (Life Technologies Corporation, Carlsbad, CA). Molecular weight marker and isoelectric point marker were purchased from Bio-Rad. N,N,N’N’- tetramethylethylenediamine TEMED (Bio-Rad), PSA (Sigma-Aldrich Co., St Louis, MO), acetic acid (Avantor Performance Materials, Inc., Center Valley, PA), urea (Sigma-Aldrich Co.), di-thiothreitol (DTT) (Sigma-Aldrich Co.) and methanol (Sigma-Aldrich Co.) were reagent grade.
Cell cultureWe used a genotype 1b HCV subgenomic replicon cell culture system (Huh7 HCV-replicon) described previously and Huh–7 parental cells, which allow studying molecular mechanisms involved in HCV replication.9 Cells were maintained in Advanced Dulbecco’s Modified Eagle Medium (ADMEM) (GIB-CO-BRL, Grand Island, NY) supplemented with 2% heat-inactivated Fetal Bovine Serum (FBS) (GIBCO-BRL), 1% nonessential amino acids, 100 U of penicillin G and 100 of streptomycin per mL at 37 °C in a humidified atmosphere with 5% CO2. Cells were maintained in culture in the presence of 500 of G418/mL (Geneticin) (Life Technologies Corporation, Carlsbad, CA), which was removed two days before the experiments. For the different treatments, Huh7 HCV-replicon cells were plated one day before and then media was changed and cells were treated with 4 mM ASA considering this moment as time zero and incubating cells for up to 72 h. As we previously reported, viability assays demonstrated that there are no cytotoxic effects of aspirin at the concentrations used on parental and HCV replicon-containing cells.11
Extraction of total protein and sample preparationHuh7 parental and HCV-replicon cells were treated with 4 mM ASA (Sigma-Aldrich) and incubated at 0, 24, 48 and 72 h. At the end of each time, cells were harvested and total proteins were extracted using the MicroRotofor Lysis kit (Bio-Rad) followed by protein purification using the Ready Prep 2-D Cleanup kit (Bio-Rad), according to the manufacturer’s instructions. Total protein quantification was carried out by Bradford’s method (BioRad). Protein extracts were verified by SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electro-phoresis) before two-dimensional separation. Extracts were stored at –80 °C until used.
Second dimension electrophoresis (2DE) and proteomic analysisFor 2DE analysis, equal amounts of protein were used (120 μg). Samples were passively rehydrated into 17-cm pH 3–10 lineal IPG strips (Bio-Rad) at room temperature for 16 h. After this step, the first dimension separation was made in an IEF Protean Cell (Bio-Rad) using a gradient of 10,000 V in 2.5 h and 40,000 V/h. The IPG strips were then equilibrated in buffer (6 M urea, 2% SDS, 50 mM Tris-Cl, pH 8.8, 30% glycerol) supplemented with 0.5% DTT for 15 min at room temperature followed by 4.5% iodoacetamide in equilibration buffer for another 15-min incubation at room temperature. Separation in second dimension was performed using Protean II xi cell system (Bio-Rad) and 12% polyacrylamide gels using 150V and a temperature of 4 °C. A protein molecular weight (MW) and isoelectric point (pI) of 7 known proteins was added to establish reference points in the subsequent estimation of molecular weight and isoelectric point of the proteins resolved for each sample. All gels were prepared in triplicate.
Proteomic analysis and identification of overexpressed proteinsProteins were revealed using the Silver Stain Plus kit (Bio-Rad). Protein profiles displayed in 20 × 20 cm and 1.5 mm thickness gels were digitalized using the GS-800 densitometer (Bio-Rad). Analysis of 2DE was performed using PDQuest V8.0.1 software (Bio-Rad). Furthermore, we identified differentially expressed proteins in accordance with Gasteiger, et al.,17 where a bioinformatic protein tool calculates the estimated pI and MW of a specified Swiss-Prot/TrEMBL entry or a user-entered aa sequence. Briefly, proteins were identified and elucidated by individual pI and MW using Tagldent (Uniprot consortium, 2002–2012) (www.expasy.org/tools/pi_tool.html). TagIdent can identify proteins by matching sequence tags against proteins in Swiss-Prot from one or more species within a specified pI and MW range. These parameters are useful to know the approximate region of a 2DE gel where a protein may be found.
In addition, results of 2DE were evaluated using principal component analysis (PCA) in order to evaluate proteins with major relevance to the studied conditions by using the following PCA mathematical algorithm:
Xij = ai1.Z1f + aik ˙Zkj = Σ ais ˙Zsk
X= factor matrix.
a = coefficients.
Z = standardized matrix.
To determine factorial coefficients (level of correlation and principal component) with Unscrambler 9.8 software. Data reported are mean values ± standard deviations of the mean of at least three independent experiments. Statistical analysis was performed by Student’s t test (p < 0.05).
ResultsWe collected the cell proteome after each incubation period (0, 24, 48 and 72 h) with either 4mM ASA or mock treatment (treated with vehicle), focusing on maximal inhibition of HCV-gene expression in Huh7 HCV-replicon cells (48 and 72 hours post-treatment) (about a 60% decrease; *P < 0.05).11
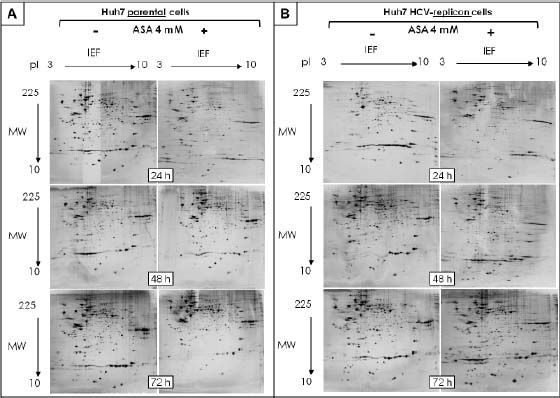
The protein expression patterns of parental and HCV-replicon Huh7 cell lines were examined using 2DE-based proteomics, PCA analysis and further bioinformatic in silico protein identification. The amount of analyzed gels included: 18 separated replicates of analyzed proteomes from ASA-treated and untreated Huh7 cells and the same number of gels to analyze proteomes from Huh7 HCV-replicon cells. In order to identify the pI and MW, analysis of proteins migration pattern vs. reference proteins pattern were performed using PDQuest software. Two-dimensional patterns showed between 674 and 1,416 proteins per gel, with most being located in the high molecular weight zone and with a slightly acid pI, distributed in a range of 10 to 140 kDa (Figures 1A and 1B). The same internal standard was used for all samples to allow better technical quality and less variance between gels.
Bidimensional separation of proteins from Huh7 parental and HCV-replicon cells obtained after 2DE. Proteins were separated on pH 3–10L IPG strips, with 120 µ3 sample loaded, followed by SDS-PAGE on a 12% gel (20 cm × 20 cm × 1.5 mm). Gels were stained with silver nitrate (Bio-Rad). A. 2DE analysis of protein extracted from Huh7 cells treated with 4 mM ASA at different times. B. 2-DE analysis of protein extracted from Huh7 HCV-replicon cells treated with 4 mM ASA at different times.
The average number of spots representing either individual or similarly migrating proteins obtained with parental and HCV-replicon cells were 923 and 816, respectively.
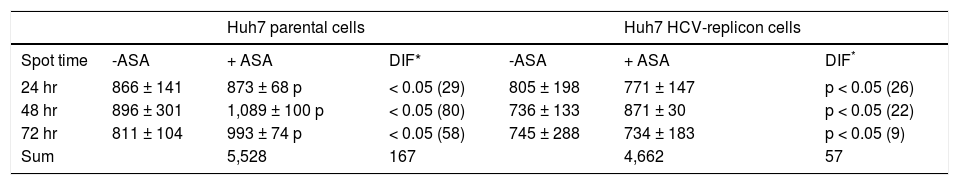
After spot comparisons and normalization with basal conditions, we identified numerous spots expressed differently (up and down regulated) between both cell lines and upon ASA treatment. We detected 37 protein spots expressed qualitatively different and 343 spots expressed quantitatively different. Comparison of gels from both cell lines (parental and HCV-replicon) showed 24 protein spots significantly upregulated in Huh7 replicon cells (more than threefold, Student’s t test, p < 0.05). Figures 1A and 1B show representative protein expression profiles of both cell lines in the absence or presence of 4 mM ASA at different times of exposure. We found differentially expressed proteins, either qualitatively and/or quantitatively at 24, 48 and 72 h with ASA treatment. At 24 and 48 h, we observed the highest number of statistical significant differentially expressed proteins as shown in table 1.
Differential proteins found in Huh7 parental and Huh7 HCV-replicon cells after acetylsalicylic acid (ASA) treatment.
| Huh7 parental cells | Huh7 HCV-replicon cells | |||||
|---|---|---|---|---|---|---|
| Spot time | -ASA | + ASA | DIF* | -ASA | + ASA | DIF* |
| 24 hr | 866 ± 141 | 873 ± 68 p | < 0.05 (29) | 805 ± 198 | 771 ± 147 | p < 0.05 (26) |
| 48 hr | 896 ± 301 | 1,089 ± 100 p | < 0.05 (80) | 736 ± 133 | 871 ± 30 | p < 0.05 (22) |
| 72 hr | 811 ± 104 | 993 ± 74 p | < 0.05 (58) | 745 ± 288 | 734 ± 183 | p < 0.05 (9) |
| Sum | 5,528 | 167 | 4,662 | 57 |
A principal component analysis (PCA) of the protein spot maps demonstrate that the differences in protein expression were found mainly between ASA-treated and untreated cells. The explained variance (EV = 1/n●XtX) with two principal components (CP1 and CP2) for each time were as follows: at 24 h we observed an EV of 86% and 12%; at 48 h the EV was 57% and 32%; and at 72 h it was 64% and 28%, for ASA-treated and untreated cells respectively.
Based on our previous results, it was expected that the different physiological properties of parental and HCV-replicon cells, could lead to the induction of different protein expression profiles following exposure to ASA. In both cell types the differences in protein expression in response to aspirin treatment were analyzed for the 1,416 spots detected in 2DE analysis, using an independent t test. Interestingly, we observed that parental cells have a different response to ASA exposure, because the total and the sum of protein differentially expressed at the three times are higher than the proteins differentially expressed by HCV-replicon cells upon ASA exposure (167 against 57 proteins, respectively) (Table 1). Furthermore, HCV-replicon cells seem to have a decreased number of differentially expressed proteins after 72 h of ASA treatment (Table 1). Among them, there are some spots that keep the differential expression in more than one evaluated time.
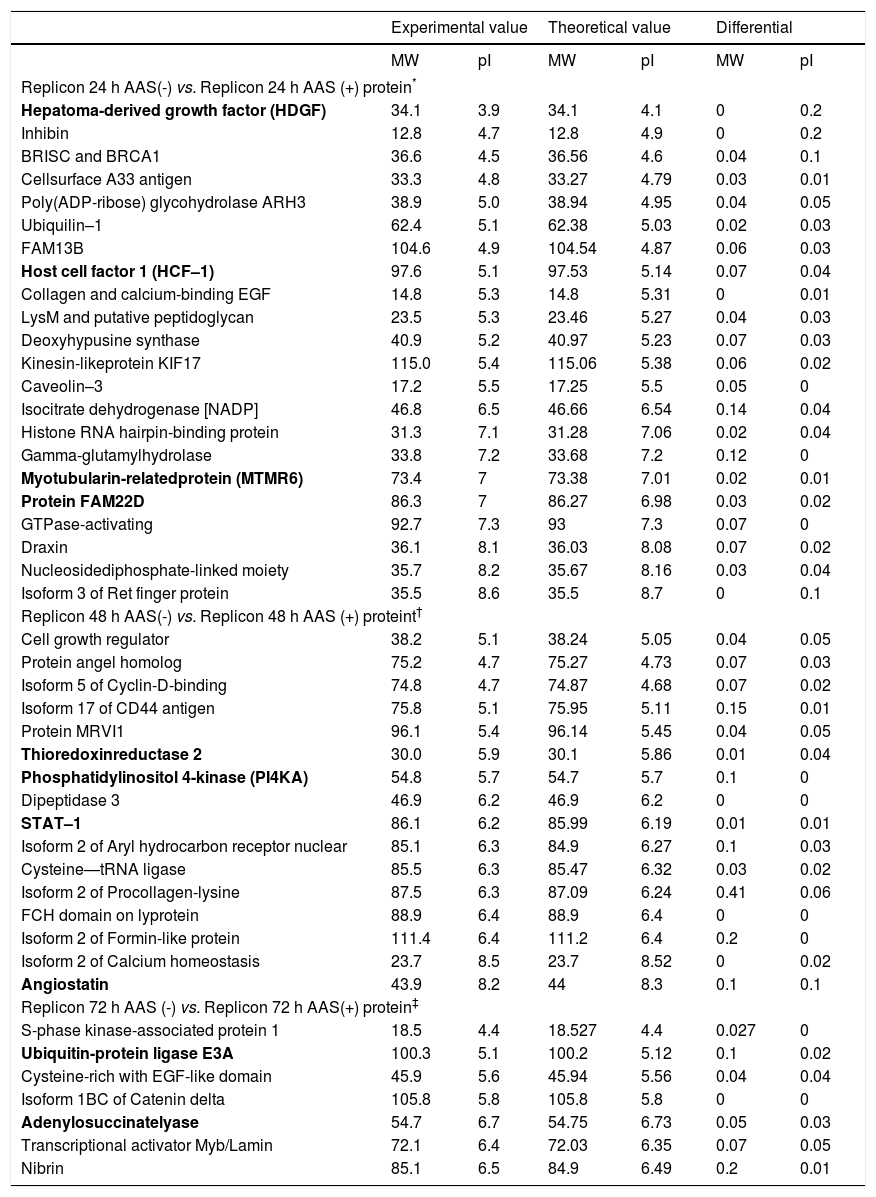
Using proteomic analysis, 289 proteins were shown to be differentially expressed between Huh7-HCV replicon cells in the presence or absence of ASA. Regarding the identification of proteins involved in the down regulation of HCV–1b replication mediated by acetylsalicylic acid, we specifically found that most of the proteins over expressed at 24 h with ASA treatment were related to cell proliferation processes, showing the expression of proteins MTMR6, FAM22, HDGF and HCF–1. After 48 h of ASA exposure, we observed the up regulation of angiostatin, PI4KA, and STAT-1. Finally, at 72 h of ASA treatment over expression of adenylosuccinate synthase (a protein involved in purine synthesis in the liver), 2’, 3’-di-deoxyadenosine, ubiquitin-protein ligase E6A, adenylosuccinate lyase and nibrin was observed (Table 2).
Comparison of the identified proteins found between exposed and unexposed HCV replicon cells to ASA.
| Experimental value | Theoretical value | Differential | ||||
|---|---|---|---|---|---|---|
| MW | pI | MW | pI | MW | pI | |
| Replicon 24 h AAS(-) vs. Replicon 24 h AAS (+) protein* | ||||||
| Hepatoma-derived growth factor (HDGF) | 34.1 | 3.9 | 34.1 | 4.1 | 0 | 0.2 |
| Inhibin | 12.8 | 4.7 | 12.8 | 4.9 | 0 | 0.2 |
| BRISC and BRCA1 | 36.6 | 4.5 | 36.56 | 4.6 | 0.04 | 0.1 |
| Cellsurface A33 antigen | 33.3 | 4.8 | 33.27 | 4.79 | 0.03 | 0.01 |
| Poly(ADP-ribose) glycohydrolase ARH3 | 38.9 | 5.0 | 38.94 | 4.95 | 0.04 | 0.05 |
| Ubiquilin–1 | 62.4 | 5.1 | 62.38 | 5.03 | 0.02 | 0.03 |
| FAM13B | 104.6 | 4.9 | 104.54 | 4.87 | 0.06 | 0.03 |
| Host cell factor 1 (HCF–1) | 97.6 | 5.1 | 97.53 | 5.14 | 0.07 | 0.04 |
| Collagen and calcium-binding EGF | 14.8 | 5.3 | 14.8 | 5.31 | 0 | 0.01 |
| LysM and putative peptidoglycan | 23.5 | 5.3 | 23.46 | 5.27 | 0.04 | 0.03 |
| Deoxyhypusine synthase | 40.9 | 5.2 | 40.97 | 5.23 | 0.07 | 0.03 |
| Kinesin-likeprotein KIF17 | 115.0 | 5.4 | 115.06 | 5.38 | 0.06 | 0.02 |
| Caveolin–3 | 17.2 | 5.5 | 17.25 | 5.5 | 0.05 | 0 |
| Isocitrate dehydrogenase [NADP] | 46.8 | 6.5 | 46.66 | 6.54 | 0.14 | 0.04 |
| Histone RNA hairpin-binding protein | 31.3 | 7.1 | 31.28 | 7.06 | 0.02 | 0.04 |
| Gamma-glutamylhydrolase | 33.8 | 7.2 | 33.68 | 7.2 | 0.12 | 0 |
| Myotubularin-relatedprotein (MTMR6) | 73.4 | 7 | 73.38 | 7.01 | 0.02 | 0.01 |
| Protein FAM22D | 86.3 | 7 | 86.27 | 6.98 | 0.03 | 0.02 |
| GTPase-activating | 92.7 | 7.3 | 93 | 7.3 | 0.07 | 0 |
| Draxin | 36.1 | 8.1 | 36.03 | 8.08 | 0.07 | 0.02 |
| Nucleosidediphosphate-linked moiety | 35.7 | 8.2 | 35.67 | 8.16 | 0.03 | 0.04 |
| Isoform 3 of Ret finger protein | 35.5 | 8.6 | 35.5 | 8.7 | 0 | 0.1 |
| Replicon 48 h AAS(-) vs. Replicon 48 h AAS (+) proteint† | ||||||
| Cell growth regulator | 38.2 | 5.1 | 38.24 | 5.05 | 0.04 | 0.05 |
| Protein angel homolog | 75.2 | 4.7 | 75.27 | 4.73 | 0.07 | 0.03 |
| Isoform 5 of Cyclin-D-binding | 74.8 | 4.7 | 74.87 | 4.68 | 0.07 | 0.02 |
| Isoform 17 of CD44 antigen | 75.8 | 5.1 | 75.95 | 5.11 | 0.15 | 0.01 |
| Protein MRVI1 | 96.1 | 5.4 | 96.14 | 5.45 | 0.04 | 0.05 |
| Thioredoxinreductase 2 | 30.0 | 5.9 | 30.1 | 5.86 | 0.01 | 0.04 |
| Phosphatidylinositol 4-kinase (PI4KA) | 54.8 | 5.7 | 54.7 | 5.7 | 0.1 | 0 |
| Dipeptidase 3 | 46.9 | 6.2 | 46.9 | 6.2 | 0 | 0 |
| STAT–1 | 86.1 | 6.2 | 85.99 | 6.19 | 0.01 | 0.01 |
| Isoform 2 of Aryl hydrocarbon receptor nuclear | 85.1 | 6.3 | 84.9 | 6.27 | 0.1 | 0.03 |
| Cysteine—tRNA ligase | 85.5 | 6.3 | 85.47 | 6.32 | 0.03 | 0.02 |
| Isoform 2 of Procollagen-lysine | 87.5 | 6.3 | 87.09 | 6.24 | 0.41 | 0.06 |
| FCH domain on lyprotein | 88.9 | 6.4 | 88.9 | 6.4 | 0 | 0 |
| Isoform 2 of Formin-like protein | 111.4 | 6.4 | 111.2 | 6.4 | 0.2 | 0 |
| Isoform 2 of Calcium homeostasis | 23.7 | 8.5 | 23.7 | 8.52 | 0 | 0.02 |
| Angiostatin | 43.9 | 8.2 | 44 | 8.3 | 0.1 | 0.1 |
| Replicon 72 h AAS (-) vs. Replicon 72 h AAS(+) protein‡ | ||||||
| S-phase kinase-associated protein 1 | 18.5 | 4.4 | 18.527 | 4.4 | 0.027 | 0 |
| Ubiquitin-protein ligase E3A | 100.3 | 5.1 | 100.2 | 5.12 | 0.1 | 0.02 |
| Cysteine-rich with EGF-like domain | 45.9 | 5.6 | 45.94 | 5.56 | 0.04 | 0.04 |
| Isoform 1BC of Catenin delta | 105.8 | 5.8 | 105.8 | 5.8 | 0 | 0 |
| Adenylosuccinatelyase | 54.7 | 6.7 | 54.75 | 6.73 | 0.05 | 0.03 |
| Transcriptional activator Myb/Lamin | 72.1 | 6.4 | 72.03 | 6.35 | 0.07 | 0.05 |
| Nibrin | 85.1 | 6.5 | 84.9 | 6.49 | 0.2 | 0.01 |
The most relevant proteins are labeled in bold.
After 24 h post-treatment with ASA, HCV promotes the activation of proteins involved in cell proliferation and cell cycle progression (MTMR6, FAM22, HDGF and HCF–1).
Recently it has been reported that salicylates induce antiviral activity against some flavivirus18 and exert an influence on the replication of other virus as the influenza virus, HCMV and varicella-zoster virus.19,20 Furthermore, Spier, et al.19 have shown that aspirin attenuates HCMV infectivity and gene expression mediated by COX-2 in coronary artery smooth muscle cells. Despite extensive studies of NSAIDs, little is known about their effects on viral infection. We have previously demonstrated that the antiviral effect of ASA is partially dependent on the inhibition of COX-2 activity and prostaglandin synthesis in the HCV replicon cell system.11 In this work we wanted to get more information about the complete mechanism. We have applied proteomic techniques and bioinformatic tools to
globally analyze the protein expression profiles of a human liver carcinoma cell line Huh7 in the absence and presence of HCV replication and ASA treatment, to elucidate the components of HCV replication and the cellular mechanisms involved in responses to ASA treatment. Many proteins were shown to be differentially expressed between both cell lines and upon ASA exposure. It was possible to obtain a protein expression pattern of hepatocytes that express the HCV proteins treated with ASA, which represents the first step in identifying differential protein profiles involved in the downregulation of HCV expression mediated by ASA. We employed proteomic techniques to elucidate the mechanism of interaction between HCV and host cell response to ASA treatment. Many results were coincident with already reported studies that use a different full-length/subgenomic replicon HCV cell culture system.21–23
These expression differences between studied conditions are in coincidence with previous reports on the differential expression of the Huh7 cells and cells harboring HCV-non-structural proteins;24 however, in addition, different protein patterns among hepatocytes expressing HCV-proteins and cells treated with ASA were found.
We also used a bioinformatic tool, TagIdent, to list proteins in a defined pI and/or MW region. The Tagldent tool (http://www.expasy.org/tools/ tagident.html) serves two main purposes. First, it can create lists of proteins from one or more organisms that are within a user-specified pI or MW range. This is useful to find proteins from the database that may be in a region of interest on a 2D gel. Secondly, the program can identify proteins from 2D gels by virtue of their estimated pI and MW, and a short protein “sequence tag” of up to six amino acids. The sequence tag can be derived from protein N-termini, C-termini, or internally, and generated by chemical or mass spectrometric sequencing techniques. As sequence tags are highly specific (e.g., there are 160,000 different combinations of four amino acid sequence tags), they represent a form of protein identification that is useful for organisms that are molecularly well defined and have a relatively small number of proteins (e.g., Escherichia coli or Saccharomyces cerevisiae).
Infection with HCV has been shown to affect the biology of host cell systems, including cell cycles and signaling pathways, contributing to viral patho-genesis. Based on this, it could explain the results that we obtained comparing the proteins differential ly expressed from parental and HCV-replicon cells in response to ASA exposure (Table 1). In addition, ASA treatment affected host cell protein expression, but at the same time, the protein profile is affected by the presence of HCV proteins. Additional experiments should be performed in order to specifically identify and validate the proteins involved in this response. Further identification of protein spots (by using trypsin digestion, followed by MALDI-TOF MS measurement) induced by ASA treatment could allow us to later identify new candidates that could be used as antiviral therapy targets. Additionally, an important contribution will be the generation of a protein signature for the definition of therapeutic targets, and diagnostic and prognostic markers.
We found that HCV promotes the activation of proteins involved in cell proliferation and cell cycle progression. MTMR6, FAM22, HDGF, and HCF-1 have previously been associated with inhibition of apoptosis, binding oncoproteins, stimulation of cell growth and cell cycle progression, respectively. After 48 h post-treatment with ASA, the virus continues promoting proliferation and progression in the host cell by inducing protein PI4KA and angiostatin. However, at the same time, proteins related to cellular arrest and apoptosis were identified such as STAT1 and thioredoxin reductase. Previously, Hartman, et al. reported that STAT1 in the presence of IFN promoted cell cycle arrest and apoptosis.25 While, Hinze, et al. and Ganther, et al. suggested that thioredoxin reductase participated as an antioxidant, reducing the viral process.26,27 Our results showed that at 72 h of ASA exposure, proteins such as ubiquitin-protein ligase E6A, adenylosuccinate lyase and nibrin, are overexpressed and may participate in the inhibition of HCV replication. In this respect, Shirakura, et al. mentioned that ubiquitin-protein ligase E6A is involved in the HCV core protein degradation.28 Also, adenylosuccinate lyase is required to activate an antiviral pathway involving the 2’, 3’ di-deoxiadenosine protein. Another protein upregulated was nibrin, which has been reported to have oncogene activity and repairs double strand breaks. The proteins identified may suggest that in the early hours of viral infection, the virus promotes proliferation and cell cycle deregulation in benefit of HCV, and that after treatment with ASA, the infected cell begins a process of defense against the virus. Further experiments should be performed in order to confirm these proteomic and in silico findings. The proteomic analysis carried out in this study provides global information of proteomic alteration of Huh7 HCV replicon cells in the presence of antiviral activity mediated by ASA and the clues for further understanding the mechanism of HCV replication and pathogenesis, and host gene regulation.
Abbreviations- •
ASA: acetylsalicylic acid.
- •
HCV: hepatitis C virus.
- •
2DE: two dimensional gel electrophoresis.
Financial support was provided by the CONACYT, grant numbers CB-2006-1-58781 and SALUD-2008-C01–86996 to AMRE.
AcknowledgementsWe thank Sergio Lozano-Rodriguez, M.D. of the “Dr. Jose E. Gonzalez” University Hospital (UANL) for his assistance in reviewing the manuscript.