Relapse occurs in approximately 20% of Mexican patients with childhood acute lymphoblastic leukemia (ALL). In this group, chemoresistance may be one of the biggest challenges. An overview of complex cellular processes like drug tolerance can be achieved with proteomic studies.
MethodsThe B-lineage pediatric ALL cell line CCRF-SB was gradually exposed to the chemotherapeutic vincristine until proliferation was observed at 6nM, control cells were cultured in the absence of vincristine. The proteome from each group was analyzed by nanoHPLC coupled to an ESI-ion trap mass spectrometer. The identified proteins were grouped into overrepresented functional categories with the PANTHER classification system.
ResultsWe found 135 proteins exclusively expressed in the presence of vincristine. The most represented functional categories were: Toll receptor signaling pathway, Ras Pathway, B and T cell activation, CCKR signaling map, cytokine-mediated signaling pathway, and oxidative phosphorylation.
ConclusionsOur study indicates that signal transduction and mitochondrial ATP production are essential during adaptation of leukemic cells to vincristine, these processes represent potential therapeutic targets.
Aproximadamente el 20% de los pacientes mexicanos con leucemia linfoblástica aguda (LLA) infantil presentan recaídas. En este grupo, la quimiorresistencia es uno de los principales desafíos. Los estudios proteómicos pueden dar un panorama general de procesos celulares complejos como la tolerancia a fármacos.
MétodosLa línea celular de LLA de linaje B, CCRF-SB, fue expuesta de manera gradual al fármaco quimioterapéutico vincristina hasta observar proliferación celular en presencia de 6nM, como control se cultivaron células en ausencia del fármaco. Se analizó el proteoma de cada grupo mediante nanoHPLC acoplado a un espectrómetro de masas de tipo trampa de iones. Las proteínas identificadas se agruparon en categorías funcionales sobre-representadas con el sistema de clasificación PANTHER.
ResultadosEncontramos 135 proteínas expresadas exclusivamente en presencia de vincristina. Las categorías funcionales más representadas fueron la señalización asociada a los receptores tipo Toll, señalización dependiente de Ras, activación de células B y T, mapa de señalización CCKR, señalización mediada por citoquinas y la fosforilación oxidativa.
ConclusionesNuestro estudio indica que la transducción de señales y la producción de ATP mitocondrial son procesos esenciales durante la adaptación de células leucémicas a vincristina por lo que estos procesos representan potenciales blancos terapéuticos.
Childhood acute lymphoblastic leukemia (ALL) is the most common type of cancer and the second leading cause of mortality among Mexican children.1 Overall cure rates of newly diagnosed ALL patients is around 80%, while chemoresistance is one of the main challenges in the relapsed population.2–4 Although much progress has been achieved in the detailed characterization of the molecular processes directly involved in drug tolerance of cancer cells,5,6 more research is needed to develop effective therapeutic strategies to resensitize chemoresistant cells.6
Vincristine is a vinca alkaloid, which interacts with tubulin disrupting microtubule polymerization and favoring the cell cycle arrest in the M phase which is followed by induction of apoptosis.7 It is used in several stages of the ALL treatment.3 It has been reported that the P-glycoprotein MDR1 actively pumps vincristine outside the cell reducing its therapeutic effect.8 Inactivation of intracellular vincristine by the myeloperoxidase has also been reported to contribute to resistance in some types of leukemia.9 Moreover, resistant leukemia, and other types of cancer cells, commonly show deregulated apoptosis4,6,10 and signaling pathways involved in survival.5,6,11 The complex state of resistance is the result of the concerted action of multiple interacting genes, proteins, and metabolites; and this scenario is well suited for characterization with the omic technologies. Proteomics studies provide an overview of the changes in the relative abundance of the proteins of a cell or tissue under different conditions.12 The characterization of proteins, as the final executors of cellular activities, may lead to identification of putative therapeutic targets and a better understanding of the pathological states.13 In the present work, we describe the changes in the proteome of a B-ALL cell line after adaptation to vincristine. Our results allowed the identification of targetable signaling and metabolic steps which may represent potential targets to resensitize leukemic cells to vincristine.
2Methods2.1Growth conditionsThe B-lineage pediatric ALL cell line CCRF-SB (ATCC CCL-120) was grown in RPMI-1640 with 10% (v/v) fetal bovine serum, 100 U/ml penicillin, 0.1mg/ml streptomycin, 1% sodium pyruvate at 37°C under an atmosphere with 5% CO2.
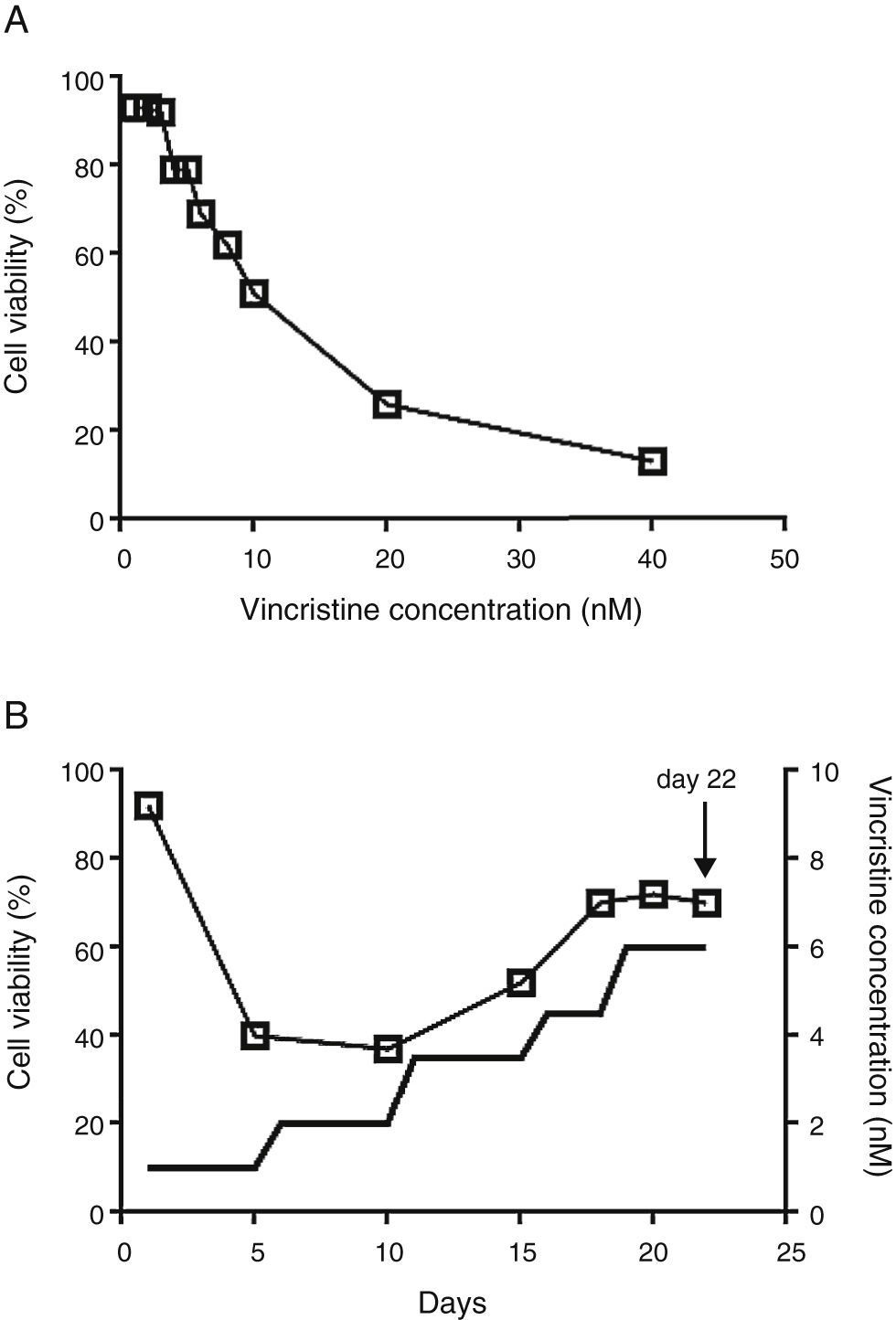
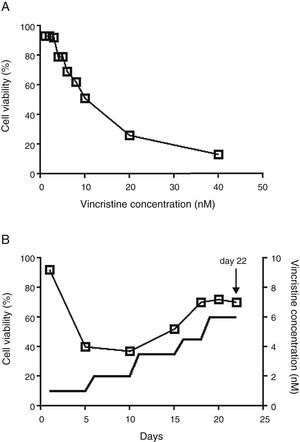
2.2Vincristine expositionCell viability was estimated with the MTT assay in 96 well plates.14 To determine the IC50, vincristine was added to 7 x104 cells in 100μL of media per well at 0, 1, 2, 3, 4, 5, 6, 8, 10, 20 and 40nM in triplicate for 48hours (Figure 1A). Gradual exposition was made as follows: 3 x106 cells were cultured in 5ml of media in the presence of 1nM vincristine for five days, and then were exposed to 2nM for the next five days, 3.5nM for five days, 4.5nM for three days, and 6nM for four days (Figure 1B). In every step viable cells were enriched by centrifugation at 800rpm 5min, only the pelleted cells were transferred to the subsequent drug concentration. After 22 days of gradual exposition, cells proliferated in the presence of 6nM vincristine, and cell viability was higher than 70%. Control cells were subjected to the same manipulations but cultured in the absence of vincristine.
The CCRF-SB cell line is sensitive to vincristine. A. To obtain the IC50 (10nM), cells were exposed to growing concentrations of vincristine for 48h. B. Cell viability (□) and vincristine concentration (—) during the gradual adaptation protocol. The arrow shows the time point at which cells were harvested for proteomic studies.
Cells were washed with cold PBS and resuspended in 500μL lysis buffer (4% SDS, 100mM DTT, 100mM TRIS pH 8.6 and a protease/phosphatase inhibitor cocktail, Thermo Scientific). Complete cell disruption was achieved by 20 cycles of 2seconds sonication at 50% amplitude in a sonics 130-Watt Ultrasonic Processor and 5seconds ice. After reduction (30min at 40°C) and alkylation (200mM iodoacetamide, 30min at room temperature in the darkness), 50μg of protein were precipitated overnight with nine volumes of ethanol at -20°C and washed twice with 90% ethanol. The dried pellet was dissolved in 50mM guanidinium chloride, 20mM TRIS and digested with mass spectrometry grade trypsin (1:50, Promega) overnight at 37°C. The resulting peptides were desalted (sep-pak C18, cartridges, Waters), dried and kept at -80°C until used.
Just before injection, peptides were dissolved in 50μL of 0.1% formic acid. 5μL of each sample were injected in triplicate to an Ultimate 3000 nanoHPLC system with a 1cm Acclaim PepMap100 C18 pre-column and a 50cm Acclaim PepMap100 C18 column. Peptides were eluted with a non-linear gradient from 5% to 90% acetonitrile with 0.1% formic acid in 240min. The HPLC system was coupled to the ESI-Ion Trap mass spectrometer Amazon speed (Bruker Daltonics) operated in positive mode, 400-1400 m/z, the 20 most abundant ions were fragmented every 50 msec, single charged ions were excluded. The ion list was constructed with the Data Analysis software (Bruker Daltonics) and used for subsequent protein identification with the Mascot search engine using the SwissProt human database, two missed trypsin cleavages, carbamidomethylation as fixed modification, methionine oxidation as variable modification, and a peptide mass tolerance of 0.6Da. Only those proteins identified in the three replicates were considered as valid identifications. Venn diagrams were drawn with the website from the University of Southern Mississippi (http://genevenn.sourceforge.net/) and functional categories were analyzed with the statistical overrepresentation test of the PANTHER classification system15 (http://pantherdb.org) in which the analyzed list contained those proteins detected exclusively in the presence of vincristine whereas the reference list contained the proteins detected in both groups (with and without vincristine), the Bonferroni correction for multiple testing was not used. Only the functional categories with a fold enrichment value equal or higher than three were considered for further analysis.
3Results3.1After gradual adaptation, the CCRF-SB cell line grew in the presence of 6nM vincristineSudden vincristine exposition for 48h resulted in IC50 of 10nM (Figure 1A), which is similar to previous reports using other types of sensitive cell lines.9,16,17 However, gradual exposition allowed growth and cell duplication at 6nM. The adaptation procedure depicted in Figure 1B resulted to be highly reproducible and allowed that cell density doubled during the last four days (from about 6x105 to 1.5 x106 cells/ml) keeping cell viability above 70%. These adapted cells were used for proteomic studies.
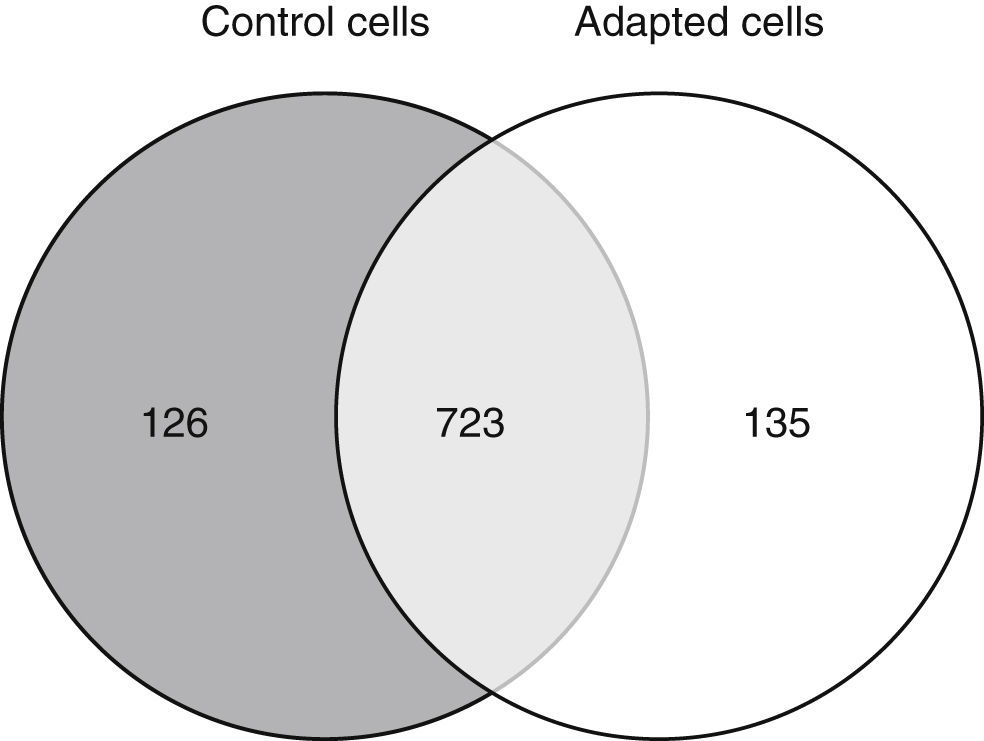
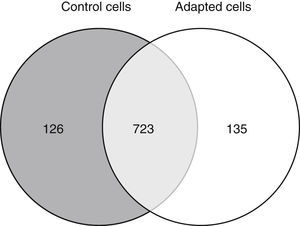
3.2Approximately 15% of the proteome is differentially expressed after adaptation to vincristineThe proteomic approach used resulted in 849 and 858 valid identifications in the control and adapted cells respectively (Figure 2). As expected, most of the proteins (approximately 85%) were present in both conditions. However, 126 proteins were detected exclusively in the absence of vincristine and 135 in the presence of this drug. This result indicated that during the adaptation process, significant changes in the proteome took place.
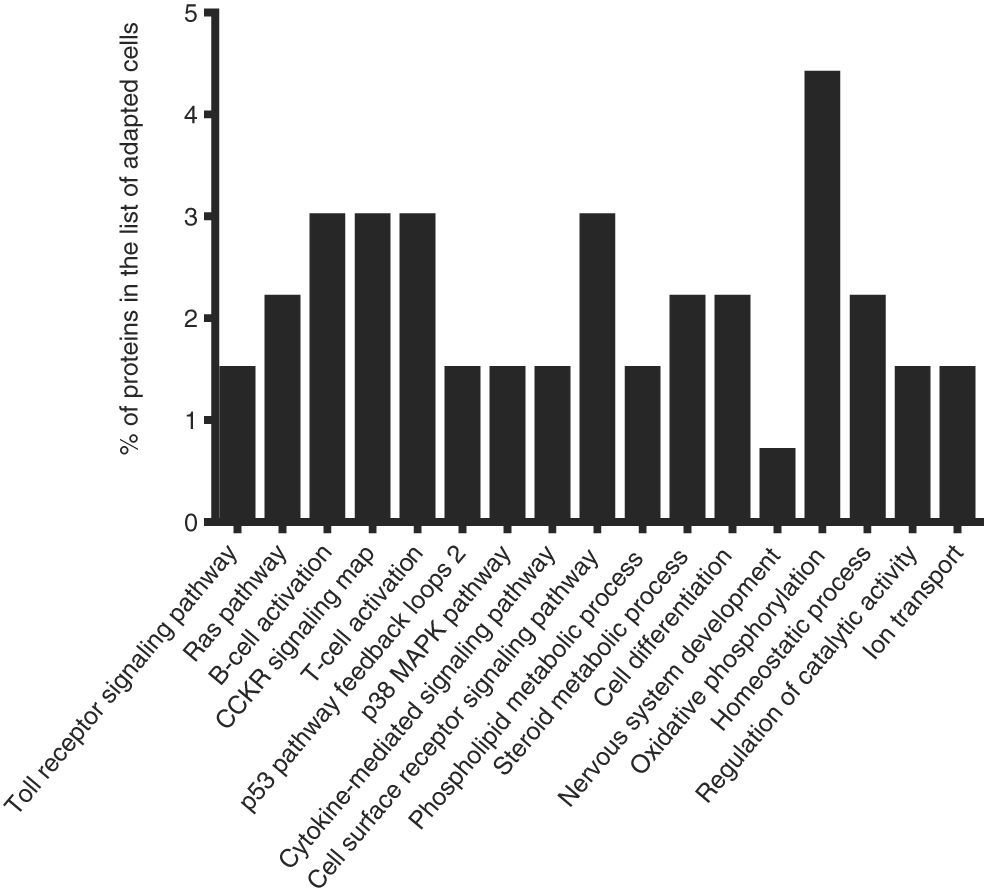
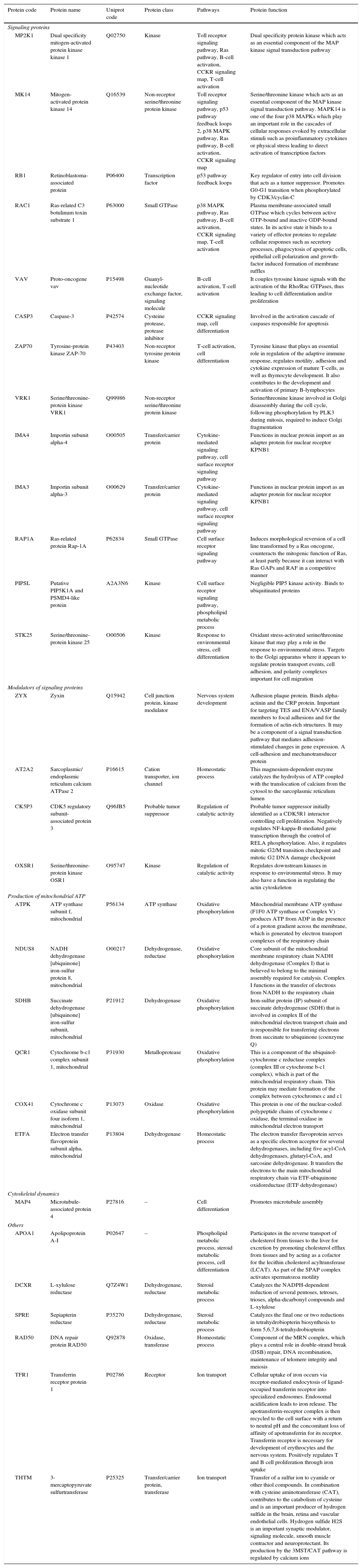
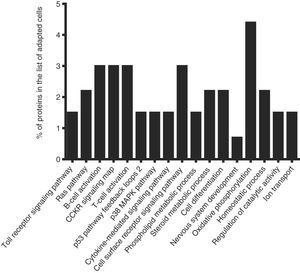
To identify the cellular processes that mainly contributed to drug tolerance, the statistical overrepresentation test was made using the PANTHER classification system15. The 135 proteins identified exclusively in the presence of vincristine were compared with the 723 proteins common to both groups (Figure 2). This resulted in the identification of 17 overrepresented pathways and functions (Figure 3) to which 30 different proteins were assigned (Table 1). Interestingly, 43% of the proteins in Table 1 and 53% of the pathways in Figure 3 were directly involved in signal transduction, whereas 13% of the proteins were involved in regulation of signaling and 20% in oxidative phosphorylation.
Details of the proteins assigned to the overrepresented pathways in adapted cells.
| Protein code | Protein name | Uniprot code | Protein class | Pathways | Protein function |
|---|---|---|---|---|---|
| Signaling proteins | |||||
| MP2K1 | Dual specificity mitogen-activated protein kinase kinase 1 | Q02750 | Kinase | Toll receptor signaling pathway, Ras pathway, B-cell activation, CCKR signaling map, T-cell activation | Dual specificity protein kinase which acts as an essential component of the MAP kinase signal transduction pathway |
| MK14 | Mitogen-activated protein kinase 14 | Q16539 | Non-receptor serine/threonine protein kinase | Toll receptor signaling pathway, p53 pathway feedback loops 2, p38 MAPK pathway, Ras pathway, B-cell activation, CCKR signaling map | Serine/threonine kinase which acts as an essential component of the MAP kinase signal transduction pathway. MAPK14 is one of the four p38 MAPKs which play an important role in the cascades of cellular responses evoked by extracellular stimuli such as proinflammatory cytokines or physical stress leading to direct activation of transcription factors |
| RB1 | Retinoblastoma-associated protein | P06400 | Transcription factor | p53 pathway feedback loops | Key regulator of entry into cell division that acts as a tumor suppressor. Promotes G0-G1 transition when phosphorylated by CDK3/cyclin-C |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 | P63000 | Small GTPase | p38 MAPK pathway, Ras pathway, B-cell activation, CCKR signaling map, T-cell activation | Plasma membrane-associated small GTPase which cycles between active GTP-bound and inactive GDP-bound states. In its active state it binds to a variety of effector proteins to regulate cellular responses such as secretory processes, phagocytosis of apoptotic cells, epithelial cell polarization and growth-factor induced formation of membrane ruffles |
| VAV | Proto-oncogene vav | P15498 | Guanyl-nucleotide exchange factor, signaling molecule | B-cell activation, T-cell activation | It couples tyrosine kinase signals with the activation of the Rho/Rac GTPases, thus leading to cell differentiation and/or proliferation |
| CASP3 | Caspase-3 | P42574 | Cysteine protease, protease inhibitor | CCKR signaling map, cell differentiation | Involved in the activation cascade of caspases responsible for apoptosis |
| ZAP70 | Tyrosine-protein kinase ZAP-70 | P43403 | Non-receptor tyrosine protein kinase | T-cell activation, cell differentiation | Tyrosine kinase that plays an essential role in regulation of the adaptive immune response, regulates motility, adhesion and cytokine expression of mature T-cells, as well as thymocyte development. It also contributes to the development and activation of primary B-lymphocytes |
| VRK1 | Serine/threonine-protein kinase VRK1 | Q99986 | Non-receptor serine/threonine protein kinase | Serine/threonine kinase involved in Golgi disassembly during the cell cycle, following phosphorylation by PLK3 during mitosis, required to induce Golgi fragmentation | |
| IMA4 | Importin subunit alpha-4 | O00505 | Transfer/carrier protein | Cytokine-mediated signaling pathway, cell surface receptor signaling pathway | Functions in nuclear protein import as an adapter protein for nuclear receptor KPNB1 |
| IMA3 | Importin subunit alpha-3 | O00629 | Transfer/carrier protein | Cytokine-mediated signaling pathway, cell surface receptor signaling pathway | Functions in nuclear protein import as an adapter protein for nuclear receptor KPNB1 |
| RAP1A | Ras-related protein Rap-1A | P62834 | Small GTPase | Cell surface receptor signaling pathway | Induces morphological reversion of a cell line transformed by a Ras oncogene, counteracts the mitogenic function of Ras, at least partly because it can interact with Ras GAPs and RAF in a competitive manner |
| PIPSL | Putative PIP5K1A and PSMD4-like protein | A2A3N6 | Kinase | Cell surface receptor signaling pathway, phospholipid metabolic process | Negligible PIP5 kinase activity. Binds to ubiquitinated proteins |
| STK25 | Serine/threonine-protein kinase 25 | O00506 | Kinase | Response to environmental stress, cell differentiation | Oxidant stress-activated serine/threonine kinase that may play a role in the response to environmental stress. Targets to the Golgi apparatus where it appears to regulate protein transport events, cell adhesion, and polarity complexes important for cell migration |
| Modulators of signaling proteins | |||||
| ZYX | Zyxin | Q15942 | Cell junction protein, kinase modulator | Nervous system development | Adhesion plaque protein. Binds alpha-actinin and the CRP protein. Important for targeting TES and ENA/VASP family members to focal adhesions and for the formation of actin-rich structures. It may be a component of a signal transduction pathway that mediates adhesion-stimulated changes in gene expression. A cell-adhesion and mechanotransducer protein |
| AT2A2 | Sarcoplasmic/ endoplasmic reticulum calcium ATPase 2 | P16615 | Cation transporter, ion channel | Homeostatic process | This magnesium-dependent enzyme catalyzes the hydrolysis of ATP coupled with the translocation of calcium from the cytosol to the sarcoplasmic reticulum lumen |
| CK5P3 | CDK5 regulatory subunit-associated protein 3 | Q96JB5 | Probable tumor suppressor | Regulation of catalytic activity | Probable tumor suppressor initially identified as a CDK5R1 interactor controlling cell proliferation. Negatively regulates NF-kappa-B-mediated gene transcription through the control of RELA phosphorylation. Also, it regulates mitotic G2/M transition checkpoint and mitotic G2 DNA damage checkpoint |
| OXSR1 | Serine/threonine-protein kinase OSR1 | O95747 | Kinase | Regulation of catalytic activity | Regulates downstream kinases in response to environmental stress. It may also have a function in regulating the actin cytoskeleton |
| Production of mitochondrial ATP | |||||
| ATPK | ATP synthase subunit f, mitochondrial | P56134 | ATP synthase | Oxidative phosphorylation | Mitochondrial membrane ATP synthase (F1F0 ATP synthase or Complex V) produces ATP from ADP in the presence of a proton gradient across the membrane, which is generated by electron transport complexes of the respiratory chain |
| NDUS8 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 8, mitochondrial | O00217 | Dehydrogenase, reductase | Oxidative phosphorylation | Core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I) that is believed to belong to the minimal assembly required for catalysis. Complex I functions in the transfer of electrons from NADH to the respiratory chain |
| SDHB | Succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial | P21912 | Dehydrogenase | Oxidative phosphorylation | Iron-sulfur protein (IP) subunit of succinate dehydrogenase (SDH) that is involved in complex II of the mitochondrial electron transport chain and is responsible for transferring electrons from succinate to ubiquinone (coenzyme Q) |
| QCR1 | Cytochrome b-c1 complex subunit 1, mitochondrial | P31930 | Metalloprotease | Oxidative phosphorylation | This is a component of the ubiquinol-cytochrome c reductase complex (complex III or cytochrome b-c1 complex), which is part of the mitochondrial respiratory chain. This protein may mediate formation of the complex between cytochromes c and c1 |
| COX41 | Cytochrome c oxidase subunit four isoform 1, mitochondrial | P13073 | Oxidase | Oxidative phosphorylation | This protein is one of the nuclear-coded polypeptide chains of cytochrome c oxidase, the terminal oxidase in mitochondrial electron transport |
| ETFA | Electron transfer flavoprotein subunit alpha, mitochondrial | P13804 | Dehydrogenase | Homeostatic process | The electron transfer flavoprotein serves as a specific electron acceptor for several dehydrogenases, including five acyl-CoA dehydrogenases, glutaryl-CoA, and sarcosine dehydrogenase. It transfers the electrons to the main mitochondrial respiratory chain via ETF-ubiquinone oxidoreductase (ETF dehydrogenase) |
| Cytoskeletal dynamics | |||||
| MAP4 | Microtubule-associated protein 4 | P27816 | – | Cell differentiation | Promotes microtubule assembly |
| Others | |||||
| APOA1 | Apolipoprotein A-I | P02647 | – | Phospholipid metabolic process, steroid metabolic process, cell differentiation | Participates in the reverse transport of cholesterol from tissues to the liver for excretion by promoting cholesterol efflux from tissues and by acting as a cofactor for the lecithin cholesterol acyltransferase (LCAT). As part of the SPAP complex activates spermatozoa motility |
| DCXR | L-xylulose reductase | Q7Z4W1 | Dehydrogenase, reductase | Steroid metabolic process | Catalyzes the NADPH-dependent reduction of several pentoses, tetroses, trioses, alpha-dicarbonyl compounds and L-xylulose |
| SPRE | Sepiapterin reductase | P35270 | Dehydrogenase, reductase | Steroid metabolic process | Catalyzes the final one or two reductions in tetrahydrobiopterin biosynthesis to form 5,6,7,8-tetrahydrobiopterin |
| RAD50 | DNA repair protein RAD50 | Q92878 | Oxidase, transferase | Homeostatic process | Component of the MRN complex, which plays a central role in double-strand break (DSB) repair, DNA recombination, maintenance of telomere integrity and meiosis |
| TFR1 | Transferrin receptor protein 1 | P02786 | Receptor | Ion transport | Cellular uptake of iron occurs via receptor-mediated endocytosis of ligand-occupied transferrin receptor into specialized endosomes. Endosomal acidification leads to iron release. The apotransferrin-receptor complex is then recycled to the cell surface with a return to neutral pH and the concomitant loss of affinity of apotransferrin for its receptor. Transferrin receptor is necessary for development of erythrocytes and the nervous system. Positively regulates T and B cell proliferation through iron uptake |
| THTM | 3-mercaptopyruvate sulfurtransferase | P25325 | Transfer/carrier protein, transferase | Ion transport | Transfer of a sulfur ion to cyanide or other thiol compounds. In combination with cysteine aminotransferase (CAT), contributes to the catabolism of cysteine and is an important producer of hydrogen sulfide in the brain, retina and vascular endothelial cells. Hydrogen sulfide H2S is an important synaptic modulator, signaling molecule, smooth muscle contractor and neuroprotectant. Its production by the 3MST/CAT pathway is regulated by calcium ions |
This study addressed the question of which are the cellular processes that sensitive leukemic cells induced to achieve tolerance to vincristine. To this end, the B-ALL cell line CCRF-SB was gradually exposed until cell proliferation was observed in the presence of 6nM vincristine, and the corresponding proteomic profile was compared to that of cells grown in the absence of the chemotherapeutic drug.
Chemoresistance may be intrinsic or acquired.18 The ability to tolerate high concentrations of chemotherapeutics of an intrinsically resistant cancer cell is not developed as a result of an exposition to the drugs; instead it is the result of genetic abnormalities the cell carries before exposition.18 By contrast, acquired chemoresistance is developed after the cancer cell is exposed to the drug and may be the result of molecular evolution of resistant clones.19 Experimental settings to study acquired chemoresistance include the comparison of matched paired samples at diagnosis and after relapse20 or the comparison of sensitive cell lines and resistant sublines which are obtained after prolonged exposure (several months) to the drug.21 It is likely that resistant clones may have evolved during the gradual adaptation protocol depicted in Figure 1B. However, the low growth rate, the short time of exposure (22 days) and the fact that it was highly reproducible when it was assayed from frozen stocks of sensitive cells suggest that the observed tolerance to vincristine was the result of induction of cellular processes to counteract the cytotoxic effect of the drug rather than the result of events involving mutation, selection, and evolution of resistant clones.
The cytotoxic effects of drugs are dependent on the concentration and the time of exposure. In many studies, cells have been exposed to relatively high concentrations for short times (24 or 48h). In such experimental setting, at concentrations close to the IC50, cells are not growing actively but remain viable and quiescent. Our results showed that even at concentration as low as 1nM for five days, a reduction in cell viability was observed (Figure 1B). The gradual adaptation protocol used allowed cell division even after four days of exposition to 6nM. By contrast, in cells exposed to 6nM vincristine for 48h without previous adaptation, no cellular division was observed (data not shown). In this regard, our results may be complementary to previous studies and may reflect some aspects of leukemic cells in vivo during advanced stages of therapy.
Successful adaptation was also reflected in the absence of overrepresented categories typically associated with cell death (Figure 3).
Interestingly, 43% of the proteins included in the overrepresented categories of adapted cells (namely MP2K1, MK14, RB1, RAC1, VAV, CASP3, ZAP70, VRK1, IMA4, IMA3, RAP1A, PIPSL, and STK25) were involved in signaling pathways like Toll receptor signaling pathway, Ras pathway, B- and T-cell activation, CCKR signaling map, p53 pathway feedback loops, p38 MAPK pathway, cytokine-mediated signaling pathway, cell surface receptor signaling pathway (Table 1); some of them have been reported to be important for growth and proliferation of lymphocytes and chemoresistance.22–24 For example, the MP2K1, MK14 and RAC1 proteins positively regulate the Ras pathway, and the MP2K1 protein is an essential component of the MAP kinase pathway. These signal transduction pathways have been associated with tumorigenesis and chemoresistance.22,25,26 Inhibition of the MP2K1 protein with PD-0325901, BAY86-9766, trametinib, selumetinib, pimasertib or GDC-0973 has been studied in advanced clinical trials.22 The MK14 protein has been reported to be closely associated with drug resistance in leukemia,27 and the RAC1 protein has been proposed as a therapeutic target for gefitinib-resistant non-small-cell lung cancer and pancreatic cancer cells,28–30 and its overexpression has been associated with poor prognosis in non-small-cell lung cancer.30 Deregulation of the VAV protein has been reported in neuroblastoma, melanoma, pancreatic, lung and breast cancers, and B-cell chronic lymphocytic leukemia.31 Furthermore, the proteins ZYX, AT2A2, CK5P3 and OXSR1, which were assigned to categories like Nervous system development, Homeostatic process and Regulation of catalytic activity (Table 1), are modulators of signaling proteins.32,33 By contrast, in the control cells, any pathway resulted to be overrepresented when the statistical overrepresentation test was made comparing the 126 proteins detected exclusively in the absence of vincristine with the proteins detected in both groups (with and without vincristine). This difference may indicate that some signaling proteins and pathways may be crucial for adaptation to vincristine. In this regard, some drugs have been developed to target signaling pathways and to overcome drug tolerance.22,25,34 Further research is necessary to explore this possibility during the gradual adaptation to vincristine.
The cell-adhesion and mechanotransducer protein ZYX was assigned to the Nervous system development category (Table 1). However, it has been reported to participate in signal transduction pathways leading to cell migration, proliferation, and tumourigenesis.32 The ZYX protein is upregulated in human breast cancer and positively correlates with metastasis. It has been proposed as a potential therapeutic target in breast cancer and as an early detection biomarker in non-small cell lung cancer.32,35
Verrills et al. reported the proteomic changes of the T-lineage ALL cell line CCRF-CEM after exposition to 2, 4 and 8nM vincristine for 24h using two-dimensional polyacrylamide electrophoresis and MALDI-TOF mass spectrometry.17 They found that vincristine-induced changes in the expression of 39 proteins and that a resistant subline differentially expressed 42 proteins mainly involved in cytoskeleton metabolism.17 Other differentially expressed proteins resulted to be regulators of apoptosis, chaperones, gene regulators and ribosomal proteins. The involvement of alterations in the metabolism of actin and tubulin during the cellular response to vincristine was confirmed in a subsequent study by the same group using primary cells engrafted into NOD/SCID mice and intraperitoneal vincristine (0.5mg/kg every seven days).36 Results shown in Figure 3 and Table 1 were based in comparisons of those proteins identified exclusively in the adapted cells versus the shared proteins (Figure 2). As cytoskeletal metabolism is very active in every cell, many cytoskeletal regulating proteins were among the 723 shared proteins; this may have prevented the overrepresentation of cytoskeletal metabolism in the adapted cells. To detect changes in the relative abundance of the shared proteins, a quantitative analysis is required. However, the microtubule-associated protein 4 (MAP4) was assigned to the overrepresented pathway Cell differentiation (Table 1). This protein promotes microtubule assembly and may reflect a more active cytoskeletal metabolism in the adapted cells compared to the control cells. Further research is necessary to explore this possibility.
After the combined effect of signaling pathways, Oxidative phosphorylation had the second highest percentage of proteins among the overrepresented pathways (Table 1). This suggested that a higher ATP production is needed to deal with the presence of vincristine. In this regard, modulation of the metabolic peculiarities of cancer cells has been proposed as a promising therapeutic strategy.37,38 The inhibitors metformin, rotenone, α-tocopheryl succinate, benzylisothiocyanate, oligomycin and resveratrol have been used to target mitochondrial energetic metabolism.38 Our results suggest that these compounds may be good candidates to overcome tolerance to vincristine.
Detection of the TFR1 and THTM proteins resulted in the overrepresentation of the Ion transport category in the adapted cells (Table 1 and Figure 3). The protein TFR1 may have contributed to B-cell growth through iron uptake.39 Interestingly, this protein has been implicated in resistance to tamoxifen in a subgroup of ER+/luminal-like breast cancer.40 Calcium is translocated from the cytosol to the sarcoplasmic reticulum lumen by the AT2A2 protein (assigned to the Homeostatic process category in Table 1). Inhibition of this protein has been proposed as a promising strategy to overcome multidrug-resistance in leukemic cells.33 Moreover, calcium signaling modulates the activity of the THTM protein which participates in the production of hydrogen sulfide (Table 1). Interestingly, this compound has been reported to be involved in chemoresistance of lung adenocarcinoma cells, and inhibition of hydrogen sulfide-producing enzymes has been proposed as a strategy to sensitize resistant cells.41
DNA repair protein RAD50 was assigned to the overrepresented category Homeostatic process (Table 1). As part of the MRN complex, this protein has been proposed as a predictor for poor prognosis and chemoresistance in gastric cancer.42
Unexpectedly, p-glycoprotein was not detected in our proteomic analysis of adapted cells. This may be due to the low relative abundance of this protein compared to other cellular proteins. To achieve higher sensitivity, peptide mixture can be fractionated in a previous HPLC run before the nanoHPLC-ESI-MS analysis.
Although monoculture of cell lines is a useful model for in vitro studies, the influence of the bone marrow microenvironment in chemoresistance43 cannot be studied. In this regard, the co-culture of bone marrow stromal cells and leukemic cells44 will certainly give more information of the involved mechanisms.
As a general rule, findings from the omic analyses must be confirmed with traditional techniques like Western blot, flow cytometry, ELISA or RT-PCR. Confirmation would be necessary only for those few genes of interest in a particular study. Although we used stringent criteria for protein identification in the MASCOT search engine, only those proteins identified in the three analytical replicates were considered as valid identifications, and only pathways with a fold enrichment value higher or equal than three were considered as overrepresented. Our results must be validated if they are to be used in further studies aimed to deepen into the molecular mechanisms involved in tolerance to vincristine.
Ethical responsibilitiesProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of dataThe authors declare that they have followed the protocols of their work center on the publication of patient data.
Right to privacy and informed consentThe authors declare that no patient data appear in this article
FundingFederal funding HIM/2015/053 SSA1183, HIM/2016/026 SSA1222 for HQ and HIM/2014/003 SSA1129 for GPL. CONACYT grant 589666 for ALGO.
Conflict of interestThe authors declare no conflict of interest of any nature.